Abstract
The p53 tumor suppressor interacts with its negative regulator Mdm2 via the former’s N-terminal region and core domain. Yet the extreme p53 C-terminal region contains lysine residues ubiquitinated by Mdm2 and can bear post-translational modifications that inhibit Mdm2–p53 association. We show that, the Mdm2–p53 interaction is decreased upon deletion, mutation or acetylation of the p53 C-terminus. Mdm2 decreases the association of full-length but not C-terminally deleted p53 with a DNA target sequence in vitro and in cells. Further, using multiple approaches we demonstrate that a peptide from p53 C-terminus directly binds Mdm2 N-terminus in vitro. We also show that p300-acetylated p53 binds inefficiently to Mdm2 in vitro, and Nutlin-3 treatment induces C-terminal modification(s) of p53 in cells, explaining the low efficiency of Nutlin-3 in dissociating p53-MDM2 in vitro.
INTRODUCTION
The tumor suppressor p53 is the focus of numerous investigations whose goal is reversing or halting tumor progression. The activity of p53 is tightly regulated in cells through posttranslational modifications, localization and degradation1. The murine double minute (Mdm2) protein, a product of a p53 inducible gene, is a RING (Really Interesting New Gene) type E3 ubiquitin ligase responsible for proteasomal degradation of p532,3. As an additional regulatory mechanism, Mdm2 binds directly to the first transactivation domain (TAD-I; amino acids 20–40) of p53, inhibiting its ability to interact with transcriptional co-activators4. X-ray crystallography and NMR studies have characterized the interaction between a hydrophobic pocket within the N-terminal region of Mdm2 and a peptide spanning the TAD-I domain of p535. Biologically active inhibitors of the said interaction (such as Nutlin-3) have been developed with the goal of dissociating p53 from Mdm2 and activating p53 in abnormally proliferating cells6–8.
Upon various forms of DNA damage the TAD-I of p53 is modified by upstream protein kinases on several residues that weaken its interaction with Mdm29,10. Nevertheless, biochemical studies show that phosphorylation of the p53 TAD-I is not sufficient to abolish its interaction with Mdm2, suggesting that in vivo other determinants are required for full p53 activation11–13. Providing additional complexity, the central acidic domain of Mdm2 interacts with the DNA-binding domain of p53 (within conserved regions IV and V, amino acids 234–286) and these contacts are essential for proper p53 ubiquitination14–16. Indeed, p53 lacking its TAD-I region is still able to interact with Mdm2 in pull-down experiments and is efficiently ubiquitinated by Mdm2 in vitro17.
The key lysines of p53 that are ubiquitinated by Mdm2 reside within its extreme C-terminal 30 amino acids (p53-CTD)18. In addition to ubiquitination, Small Ubiquitin-like Modifier (SUMO), Neural precursor cell Expressed Developmentally Down-regulated (NEDD8), acetyl groups and methyl groups can also modify the cluster of lysines in the p53-CTD19. These modifications are deposited and removed from the ε-NH3 groups of the lysines by numerous enzymes including the histone acetylase p300 that interacts with the N-terminal domain of p5319. The ability of Mdm2 to stimulate ubiquitination of the p53-CTD while interacting with distal parts of the protein is still poorly understood, as is the exact mechanism by which RING type E3’s facilitate ubiquitination of their targets.
Previous studies have alluded to the physical separation of Mdm2 and p53 following C-terminal ubiquitination of p5320,21. Li et al21 described mono-ubiquitination as causal of p53 nuclear export, while Carter et al20 expanded on these data to show that Mdm2 dissociates from p53 upon p53 mono-ubiquitination. Primarily this has been shown through changes in localization of a p53-ubiquitin fusion protein from the nucleus to the cytoplasm, although a more recent report demonstrated dissociation between Mdm2–p53 following p53 acetylation22. The authors of the above reports acknowledge that the mechanism connecting modifications of the p53 C-terminal domain with the release of Mdm2 from the complex remains unclear. Two possibilities emerge – one is that p53 modifications lead to the loss of p53-tetramerization revealing the nuclear export signal and facilitating export, and the other is that direct contact between Mdm2 and the p53-CTD exists and is regulated by posttranslational modifications. Based on results showing that either ubiquitinated or acetylated p53 is competent to activate transcription of p53 target genes, an activity which requires p53 tetramer formation, we chose to pursue an investigation of the latter hypothesis22,23.
Here we present evidence of a direct contact between the C-terminus of p53 and the N-terminus of Mdm2. Further, we show that the complex between Mdm2 and p53 can be regulated by modifications of the p53-CTD.
RESULTS
The C-terminus of p53 contributes to the p53–Mdm2 complex
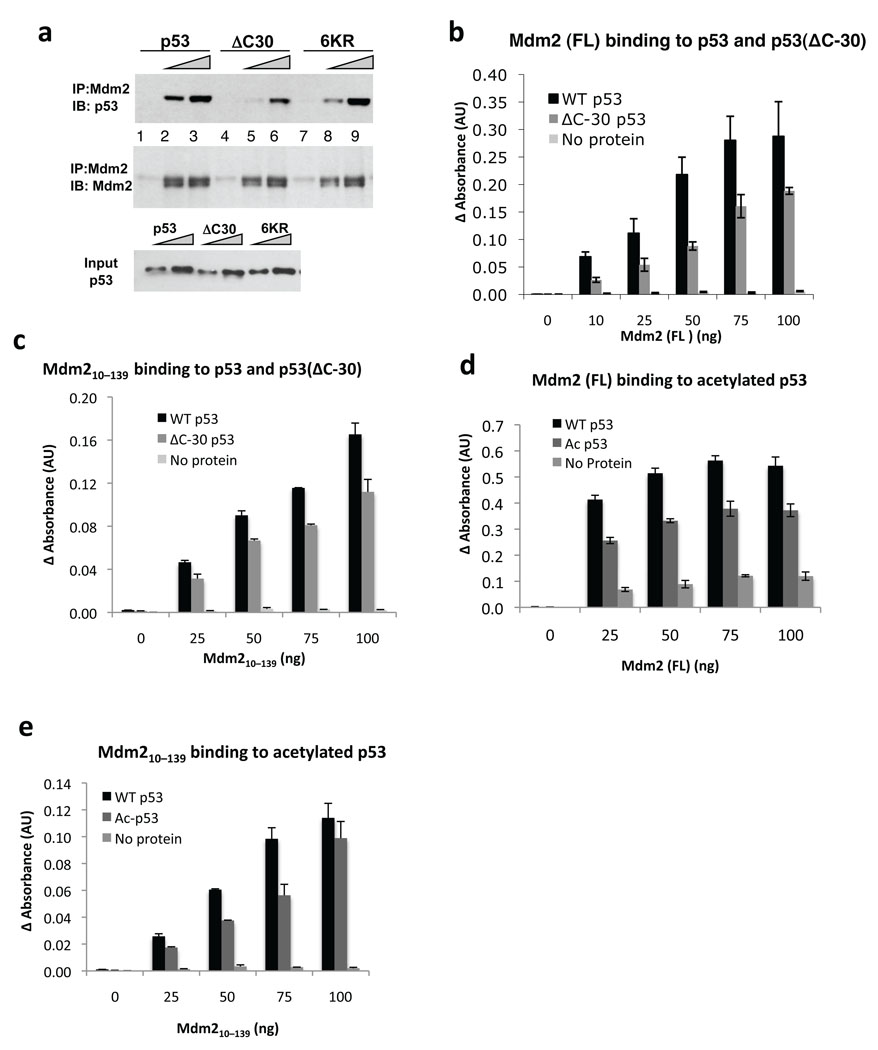
To investigate the role of the p53-CTD in the interaction with Mdm2 we first looked at the ability of wild-type human p53, p53(ΔC30) and p53(6KR) (with the 6 lysines in the CTD substituted for arginine) to bind human Mdm2. These p53 proteins were expressed in H1299 stable cell lines (Fig. 1a and Supplementary Fig. 1a) and partially purified on a heparin column, which allows for the isolation of DNA-binding proteins. Because p53 possesses two nucleic acid binding domains, one at the CTD, the wild-type protein showed higher affinity for heparin than the p53(ΔC30) protein, as expected (Supplementary Fig. 1b). We performed pull-down assays with wild-type and variant p53 and Flag-tagged full-length Mdm2 (Fig. 1a). Wild-type p53 bound Mdm2 better than p53(ΔC30), whereas p53(6KR) was only marginally defective. Because of the similar elution profiles from the heparin column of wild-type p53 and p53(6KR) (Supplementary Fig. 1b), we believe that they have similar overall modifications and thus bind similarly to Mdm2. This is in contrast to previous observations that p53(6KR) has higher affinity for endogenous Mdm2 in cells, which was attributed to the protein’s inability to become modified20.
Figure 1. Contribution of the p53 C-terminus to formation of the p53–Mdm2 complex.
(a) p53(ΔC30) interacts weakly with Mdm2. Partially purified wild-type p53, p53(6KR) or p53(ΔC30) proteins were incubated with Flag-tagged Mdm2 (1 µg) followed by pull-down with anti-Flag beads and detection with anti-p53 and anti-Mdm2 antibodies (see Methods). (b) Wild-type p53 binds Mdm2 more efficiently than p53(ΔC30). His-tagged p53 or p53(ΔC30) proteins were used to coat ELISA plates, followed by addition of Flag-Mdm2. Complexes were detected by anti-MDM2 antibody and enzyme conjugated secondary antibody (see Methods). Each data point is an average of readings from duplicate wells. (c) The N-terminal portion of Mdm2 binds p53(ΔC30) less efficiently; ELISA assay was performed as in 1b, with untagged Mdm2(10–139). (d) Full length Mdm2 binds acetylated p53 less efficiently. ELISA assay was performed as in 1b, with full-length Flag-Mdm2 added to wells pre-coated with unacetylated p53 or p53 co-expressed in SF-9 cells with p300 baculovirus (see Methods). (e) N-terminal portion of Mdm2 binds acetylated p53 less efficiently; ELISA assay was performed as in 1b.
We assessed the binding of Mdm2 to His-tagged p53 and p53(ΔC30) purified from bacteria, in an enzyme-linked immunosorbent assay (ELISA). We coated ELISA plates with p53 or p53(ΔC30) (Supplementary Figs. 1c,d and e) and measured bound Mdm2 (full-length or a truncation corresponding to its N-terminal domain) with an anti-Mdm2 antibody. Mdm2 bound preferentially to the wild-type p53 (Fig. 1b), suggesting that p53-CTD is needed for full interaction with Mdm2. Importantly, we also found that p53(ΔC30) was partially impaired in binding to the N-terminus of Mdm2(10–139) (Fig. 1c). Therefore the N-terminal portion of Mdm2 is sufficient to discriminate between full-length and C-terminally truncated p53. Further, these data indicate the existence of a binding site within the N-terminus of Mdm2 for the p53-CTD.
While the p53-CTD is seldom mutated or deleted in tumors24, it is a site for multiple modifications, including lysines acetylated by p30019. We tested whether modifications in p53-CTD decrease binding to Mdm2, using Flag-p53 purified from insect cells that were co-infected with a baculovirus producing p300. p53 co-expressed with p300 is highly acetylated, as shown by a gain of reactivity with a Pan-acetyl antibody and corresponding loss of reactivity with an antibody PAb 421 that recognizes unmodified p53-CTD residues 372–38225,26 (Supplementary Fig. 2a and b). We found that acetylated p53 was less able to bind full length Mdm2 or Mdm2(10–139), compared to wild-type p53 (Fig. 1d and e). These results are consistent with a direct interaction between Mdm2 N-terminus and the p53-CTD. It should be noted that in addition to multiple C-terminal sites, p300 also acetylates p53 at Lys164 in its core domain22; however, this residue is not near the region that makes contacts with the acidic domain Mdm225, indicating that modification of this site should not contribute to the loss of overall binding.
We further probed the interaction of Mdm2 with acetylated p53 using a Fe2+ localized hydroxyl radical footprinting assay (Supplemental Fig. 3). Addition of Mdm2 to acetylated or nonacetylated p53 generated a different pattern of cleavage products, with an exclusive cleavage site in unmodified p53. These data suggest that acetylated and non-acetylated p53 bind differently to Mdm2.
The C-terminus of p53 interacts with Mdm2
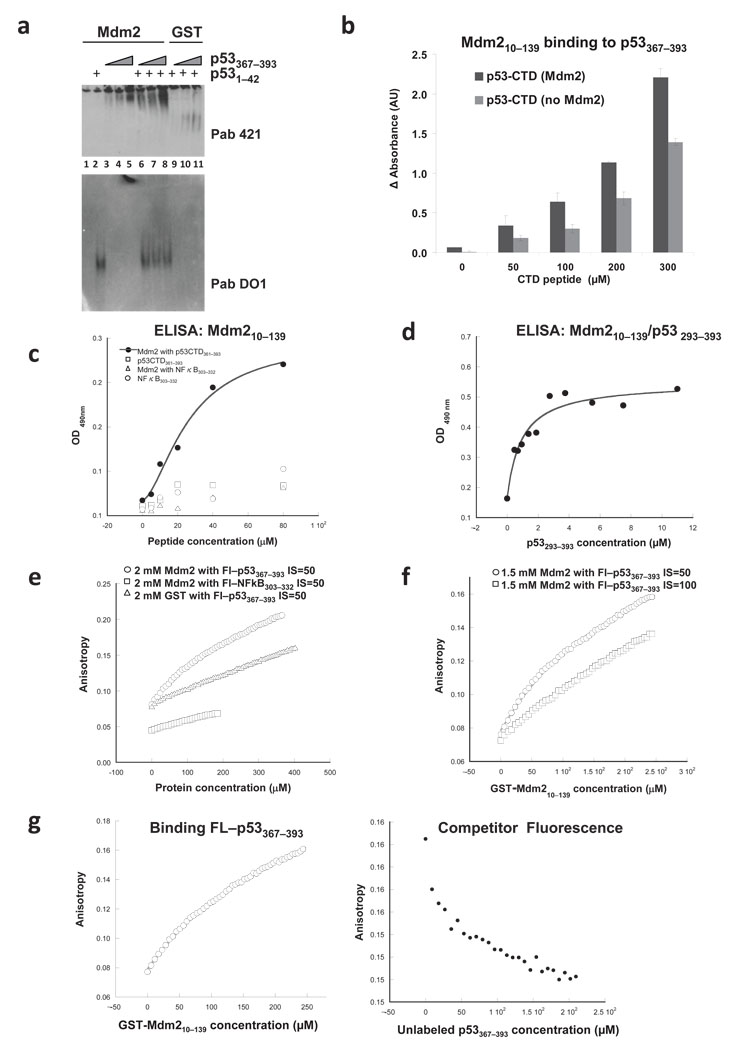
To confirm that the p53-CTD binds directly to the Mdm2 N-terminus, we performed 3 distinct binding assays. First, we assayed the binding of GST-Mdm2(10–139) to a p53-CTD(367–397) peptide in a native gel (Fig. 2a). The basic p53-CTD peptide (pI ≈10.0) does not enter the gel under the buffer conditions we used (pH 8.5) (Fig. 1f, lanes 9–11). Strikingly, in the presence of GST-Mdm2(10–139), we were able to detect markedly enhanced migration of the p53-CTD into the native gel (by immunoblotting with PAb 421), consistent with the formation of a complex. The acidic p53-TAD-I(1–42) peptide transverses the gel too rapidly for retention without Mdm2 but was retained in the gel in the presence of Mdm2 (Fig. 2a, lanes 2, 6–8). Further, the p53-CTD(367–393) bound Mdm2 even in the presence of p53-TAD-I(1–42) peptide, suggesting that the two regions of p53 bind to the N-terminus of Mdm2 non-competitively (Fig. 2a, lanes 6–8). Thus, binding to Mdm2 changes the behavior of the p53 peptides in this assay. Note that in this assay neither the CTD nor the TAD peptides appear at the migration position of Mdm2. The fact that the two peptides appear in different positions on the gel is a reflection of their overall charge and not of the binding to two different pools of Mdm2.
Figure 2. Analysis of the binding between the N-terminus of Mdm2 and p53-CTD.
(a) Mdm2 binds p53-CTD in a native gel. 2.5 µg Mdm2(10–139) (lanes 1–8) or GST (lanes 9–11) in complex with p53-TAD-I(1–42) (20 µg) or p53-CTD(367–393) (15, 20 or 40 µg) were separated in a native gel. p53-CTD and p53-TAD-I were visualized by immunoblotting with PAb 421 (top panel) or PAb DO1 (bottom panel). (b) p53-CTD binds Mdm2 in an ELISA. Mdm2(10–139) was incubated with increasing concentrations of p53-CTD(367–393); the bound p53-CTD was detected by PAb 421 (see Methods). (c) His-tagged p53-CTD(361–393) binds Mdm2 in an ELISA. Immobilized Mdm2(10–139) was incubated with His-p53-CTD(361–393) or His-NFκB(303–322) peptides; the bound peptides were detected with anti-His antibody. (d) Mdm2(10–139) binds longer C-terminal portion of p53 with higher affinity. Immobilized Mdm2(10–139) was incubated with p53(293–393) and the bound fraction of the latter was detected with anti-p53 antibody. (e) GST-Mdm2 binding to a p53-CTD peptide assessed using fluorescence assays. Fluorescein-labeled peptides either FL-p53(367–393) or NFκB(303–332) were mixed with unlabeled GST or GST-Mdm2(10–139) at indicated concentrations in the buffer containing 50 mM NaCl (IS=50) (f) Mdm2 interaction with the p53-CTD is electrostatic in nature. Unlabeled GST-Mdm2(10–139) was added to Fl-p53(367–393) at indicated NaCl concentrations. (g) Unlabeled p53-CTD peptide competes for binding Mdm2. For fluorescence anisotropy competition experiments, unlabeled p53-CTD(367–393) peptide was titrated into a pre-formed complex of Fl-p53-CTD(367–393) and GST-Mdm2(10–139). Left panel shows the binding curves for complex formation, and the right panel describes the competition.
Second, the association of a p53-CTD(367–393) peptide with Mdm2(10–139) was measured in a quantitative ELISA (Fig. 2b). We also detected binding of the CTD peptide to full-length Mdm2 in a parallel ELISA (Supplementary Fig. 4). The interpretation of these data, however, is complicated by the potential contribution of non-specific interaction of the basic CTD peptide with the acidic domain of Mdm2. Additionally, we used a slightly longer His-tagged p53(361–393) peptide in the ELISA experiments along with a His-tagged NFκB(303–322) peptide of similar length, which is also very basic, PI ≈ 9.77 (Fig. 2c). The basic NFκB peptide did not exhibit binding Mdm2 above background. Fitting the curve to a binding equation with a Hill coefficient of around 2, we calculated the Kd from 4 independent experiments to be about 30 µM for the binding between the p53-CTD and the N-terminus of Mdm2. Addition of the tetramerization domain p53(293–393) markedly improved the binding of p53 to Mdm2 N-terminus in similar ELISA experiments (Fig 2d). The calculated Kd was around 1 µM. Please note that data in Figure 2d is shown following background subtraction to clearly demonstrate binding saturation. In all, the binding of Mdm2(10–139) and three different versions of the p53-CTD was detected by three different antibodies, reveal a specific interaction with binding affinities in the low micromolar range.
As a third approach, we measured Mdm2 interaction with the p53-CTD using fluorescence anisotropy. We compared the binding of GST alone or GST-Mdm2(10–139) to the fluorescein-labeled Fl-p53(367–393) peptide, with the binding of GST-Mdm2(10–139) to a fluorescein-labeled Fl-NFκB(303–322) peptide. Only GST-Mdm2 and Fl-p53(367–393) binding produced a rounded cure characteristic of specific binding (Fig. 2f). GST protein, when incubated with increasing amounts of p53-CTD, did not exhibit such a binding curve. Similarly GST-Mdm2 protein did not demonstrate any measurable affinity for the basic Fl-NFκB(303–322) peptide. The inability of GST-Mdm2 to bind to the basic NFκB peptide (PI ≈ 10) is particularly notable because the binding of the p53-CTD with the N-terminus of Mdm2 is electrostatic in nature. The affinity of GST-Mdm2 for the p53(367–393) peptide was reduced when the ionic strength (IS) of the buffer was increased from 50 to 100 mM NaCl (Fig. 2f). This result implies that the p53-CTD is unlikely to interact with the hydrophobic pocket of Mdm2 and thus would not compete for the binding to the N-terminus of Mdm2 with the TAD-I domain of p53. We also used fluorescence anisotropy to confirm that GST-Mdm2(10–139) and Mdm2(10–139) proteins used in the majority of our biochemical assays are properly folded by measuring their binding to the p53-TAD-I peptide. In both cases we obtained Kd values in the nanomolar range, consistent with previously published data (Supplementary Fig. 5).
As an additional specificity control we performed a competition fluorescence anisotropy experiment. Increasing amounts of unlabeled p53(367–393) were titrated into a pre-formed complex of GST-Mdm2 and Fl-p53(367–393) at IS = 50 mM NaCl (Fig. 2g). The decrease in anisotropy back to the initial values suggested efficient competition.
Mapping the Mdm2–p53-CTD interaction
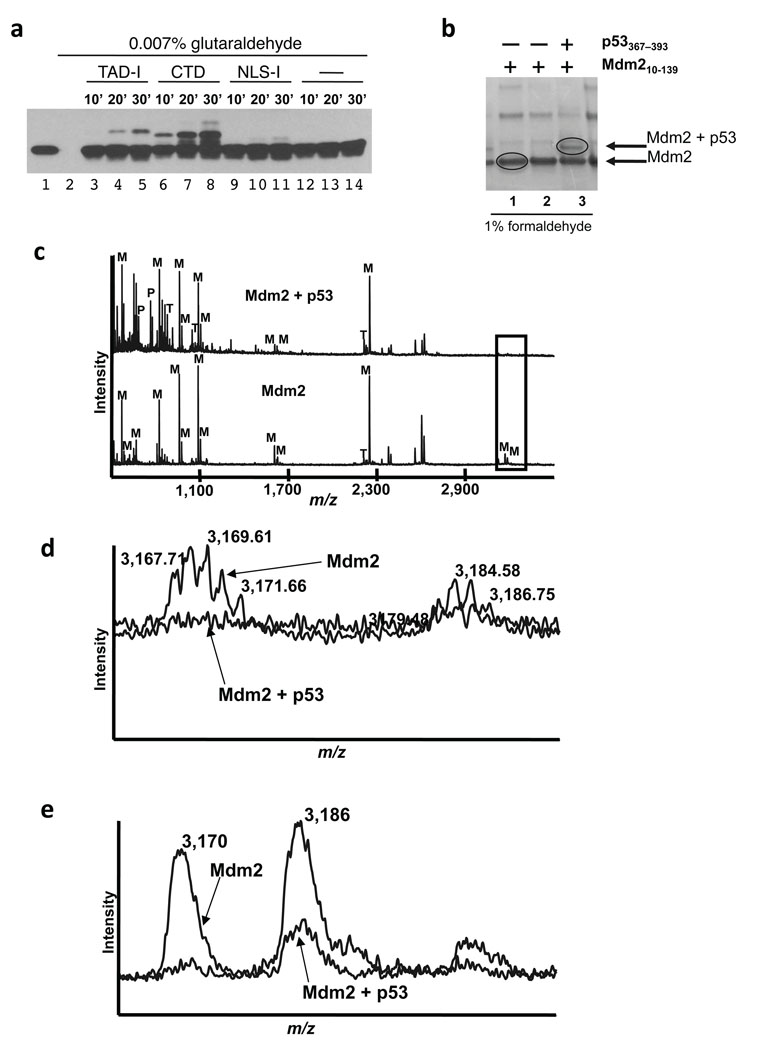
We next performed formaldehyde and glutaraldehyde crosslinking experiments as an additional test of binding. Here we subjected 8 µM Mdm2(10–139) to glutaraldehyde crosslinking alone or in the presence of 35 µM p53-TAD-I, p53-CTD or p53-NLS-I peptides (Fig 3a). Crosslinking resulted in the appearance of novel bands, upshifted by about 5 and 3 KDa in the TAD-I and CTD samples, respectively. The decrease in mobility corresponded to the added molecular weight of the peptide. We also used a p53(305–322) NLS-I peptide that spans a similarly basic region of p53 (PI = 11.3). The intensity of the crosslinked bands corresponds to the probability of crosslink, which is dependent on the number of lysines in each peptide. The p53-TAD-I peptide with only one lysine would crosslink less well then the CTD peptide that has 6 lysines. However, the fact that p53-TAD-I was crosslinked with much higher efficiency than the p53-NLS-I peptide that contains 4 lysines, serves as a specificity control.
Figure 3. Crosslinking and mass spectrometry experiments implicate the N-terminal portion of Mdm2 in binding to the p53-CTD.
(a) p53-CTD can be specifically crosslinked to Mdm2(10–139). Mdm2 (10–139) alone or in the presence of NLS-1, TAD-I or CTD peptides was crosslinked with gluteraldehyde. Samples were analyzed by immunoblotting with anti-Mdm2 antibody (N-20). (b) Reactions containing 1% formaldehyde and either no peptide or p53 CTD or p53 TAD-I peptides visualized by staining the gel with Bio-Safe™ Coomassie stain (BioRad). The indicated bands were then cut out of the gel, and subjected to trypsin digestion prior to mass spectrometry analysis. (c) MALDI-TOF mass spectrum (lower trace) of gel band of Mdm2 (see Fig. 3b, lane 1) compared to the spectrum (upper trace) from putative crosslinked Mdm2 and p53 (see Fig. 3b, lane 3). Peptide masses identified are labeled as follows: M- Mdm2 derived peptide, P- p53 derived peptide and T- trypsin derived peptide. Note loss of peaks at 3167.71 and 3183.70 m/z (box) that are expanded in 3d and 3e below. (d) Overlay of the Mdm2 and Mdm2–p53 spectra suggests that the N-terminal most part of Mdm2 contacts the p53-CTD. Detail of boxed region in Fig. 3c showing loss of monoisotopic masses at 3167.71 and 3183.70 m/z of Mdm2 in the crosslinked samples. (e) Using linear method of analysis the corresponding average masses at 3170 m/z, and 3186 m/z were not completely eliminated suggesting some tailing in the gel from the main Mdm2 band into the cross-linked Mdm2–p53 band.
We then performed preparative crosslinking experiments using 1% formaldehyde26. The bands corresponding to Mdm2 and potentially the Mdm2–p53(367–393) crosslinked heterodimer were excised, subjected to trypsin digestion and MALDI-TOF analysis (Fig. 3b). For the Mdm2 only band (circled in Fig. 3b, lane 1), masses of at least 13 peptides were matched by Mascot at 22 ppm RMS error (68 peptides were searched) with a significant Mascot score of 111. For the band with putative Mdm2 and p53 (Fig. 3b, circled in lane 3), Mdm2 was detected with a Mascot score of 75 (12 peptides matched out of 96 peptides searched). Masses of 670.34 and 763.41 m/z support the presence of peptides WSHLK and KGQSTSR, respectively, from p53 in this gel band (Fig. 3c). Furthermore, formaldehyde cross-linking in this band resulted in the loss of a monoisotopic mass (3167.71 m/z) corresponding to Mdm2 peptide GSMTDGAVTTSQIPASEQETLVRPKPLLLK (aa 10–36 of Mdm2) and a monoisotopic peak at 3183.70 m/z corresponding to the same mass with oxidized methionine (Fig. 3d and e). The loss of this peptide suggests that it spans a potential site of the interaction between the p53-CTD and Mdm2.
Mdm2 inhibition of p53 DNA binding requires the p53 C-terminus
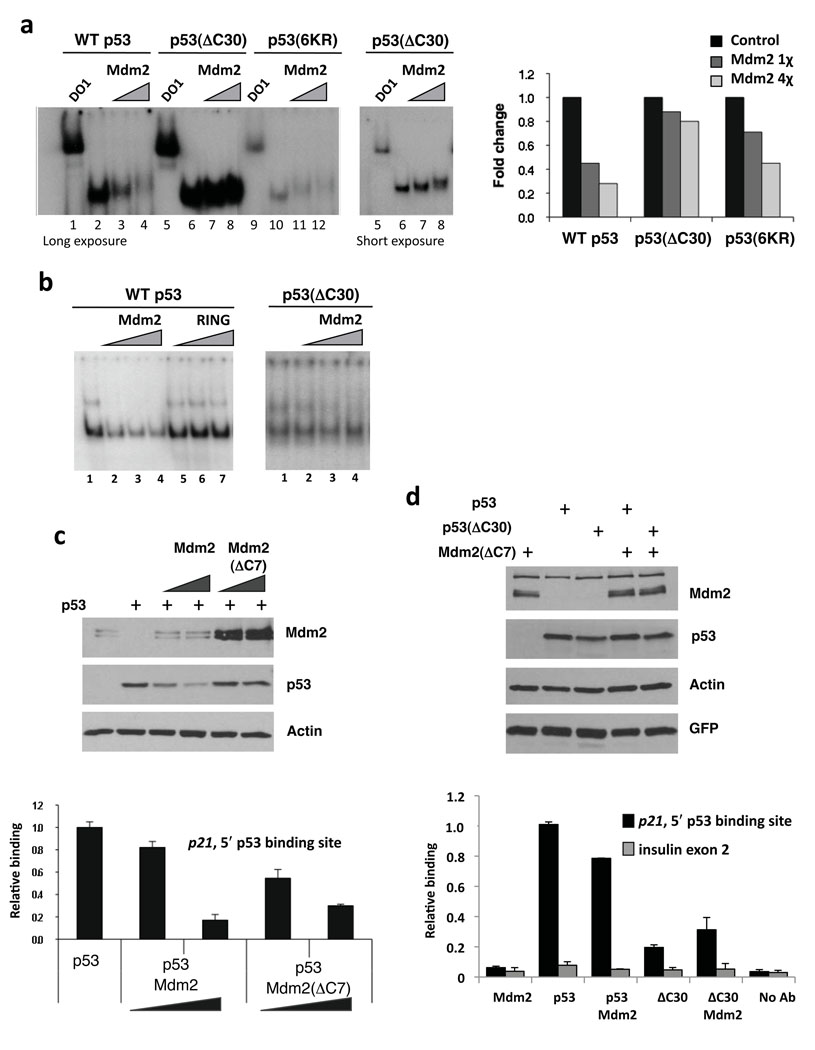
The p53-CTD plays multiple roles in p53 function, serving as a regulatory region and as a non-sequence specific nucleic acid interacting domain27. If Mdm2 is indeed making contacts with this part of p53 it could also alter the ability of p53 to interact with DNA. To test this hypothesis, we first looked at the binding of heparin-purified p53, p53(ΔC30) or p53(6KR) as well as bacterial p53 and p53(ΔC30) to a p53-response element from the p21 promoter in an electrophoretic mobility shift assay (EMSA). Mdm2 was able to inhibit the p53–DNA interaction in a matter that depended on the presence of the intact CTD (Fig. 4a and b). Further, the ability of Mdm2 to inhibit p53–DNA interactions was alleviated by the addition of PAb 421 (Supplementary Fig. 6). As we previously demonstrated that the off-rate kinetics of p53 and p53(ΔC30) are very similar28, we believe that the observed changes in p53–DNA association are caused by interaction with Mdm2. In addition to the loss of p53–DNA complex, we detected a small up-shift of the p53 band in the presence of Mdm2, as has been shown previously22. The RING domain portion of Mdm2, GST-Mdm2(410–491), did not affect the p53–DNA interaction supporting the likelihood that functional contacts are being made in the N-terminal region of Mdm2 (Fig. 4b).
Figure 4. The p53 C-terminus is required for Mdm2 to inhibit p53 DNA binding in vitro and in cells.
(a) Deletion of the C-terminus of p53 largely alleviates the ability of Mdm2 to inhibit the p53–DNA interaction. Left panel: An EMSA performed with heparin-purified p53, p53(ΔC30) and p53(6KR) proteins. p53 bound to a 44-bp fragment spanning the 5′-p53 binding site from the p21-promoter in the presence Mdm2 (.5 or 1 µg). Right panel: graph representing data in left panel. (b) Mdm2 RING domain does not inhibit p53–DNA interaction. EMSAs were performed as in 4a. p53 (left panel) or p53(ΔC30) (right panel) (250 ng) bound DNA in the presence of Flag-tagged Mdm2 (0.5, 1 or 1.5 µg) or GST-Mdm2(410–491) (RING) (0.5, 1 or 1.5 µg). (c) Mdm2(ΔC7) impairs p53 association with p21 promoter in cells without lowering p53 protein levels. 2KO mouse cells (p53−/−;Mdm2−/−) transfected with Mdm2 or Mdm2(ΔC7) (10 and 20 µg) and p53 (4 µg). Protein expression was determined by immunoblotting with anti-Mdm2 (Smp14, 3G5 and 2A10), anti-p53 (PAb 1801 and DO1) and anti-actin antibodies (top 3 panels). Chromatin samples were amplified by Q-PCR for the 5′-p53 binding site within the p21 promoter. (d) Mdm2(ΔC7) reduces the association of p53 with the p21 promoter. 2KO cells were transfected with plasmids expressing Mdm2(ΔC7) (10 µg), p53 (4 µg), p53(ΔC30) (0.4 µg) and GFP (0.1 µg). Extracts were analyzed as in 4c and insulin exon 2 was used as a negative control.
To determine if Mdm2 can affect p53–DNA interactions in cells, we looked at the ability of transfected p53 to interact with the endogenous p21 promoter in the presence of co-expressed Mdm2 by a chromatin immunoprecipitation (ChIP) assay. Co-expression of p53 with Mdm2 in p53/Mdm2 doubly null MEFs (2KO) reduced p53 promoter association (Fig. 4c, graph), although the levels of p53 were also reduced as a result of the E3 ubiquitin ligase function of Mdm2 (Fig. 4c, immunoblot). In order to co-express p53 and Mdm2 without p53 degradation we used an E3-deficient mutant of Mdm2, Mdm2(ΔC7), lacking the last 7 amino acids29. Mdm2(ΔC7) did not degrade p53, but was nevertheless able to reduce p53 association with the p21 promoter (Fig. 4c). To establish the role of the p53-CTD in the observed inhibition we co-expressed p53 or p53(ΔC30) with Mdm2(ΔC7). Due to differences in stability between these two proteins we transfected different amounts of wild-type p53 DNA and p53(ΔC30) DNA to assure equivalent expression. Reproducing our published data, p53(ΔC30) was worse than wild-type p53 at DNA binding in cells30. Furthermore, in accord with our in vitro DNA binding data, Mdm2 did not affect p53(ΔC30) binding to the p21 promoter (Fig. 4d). These data suggest that the interaction of the p53 C-terminus with Mdm2 can have functional consequences that extend beyond facilitating its ubiquitination by Mdm2.
Nutlin-3 and TAD-I region of p53 bind Mdm2 N-terminus differently
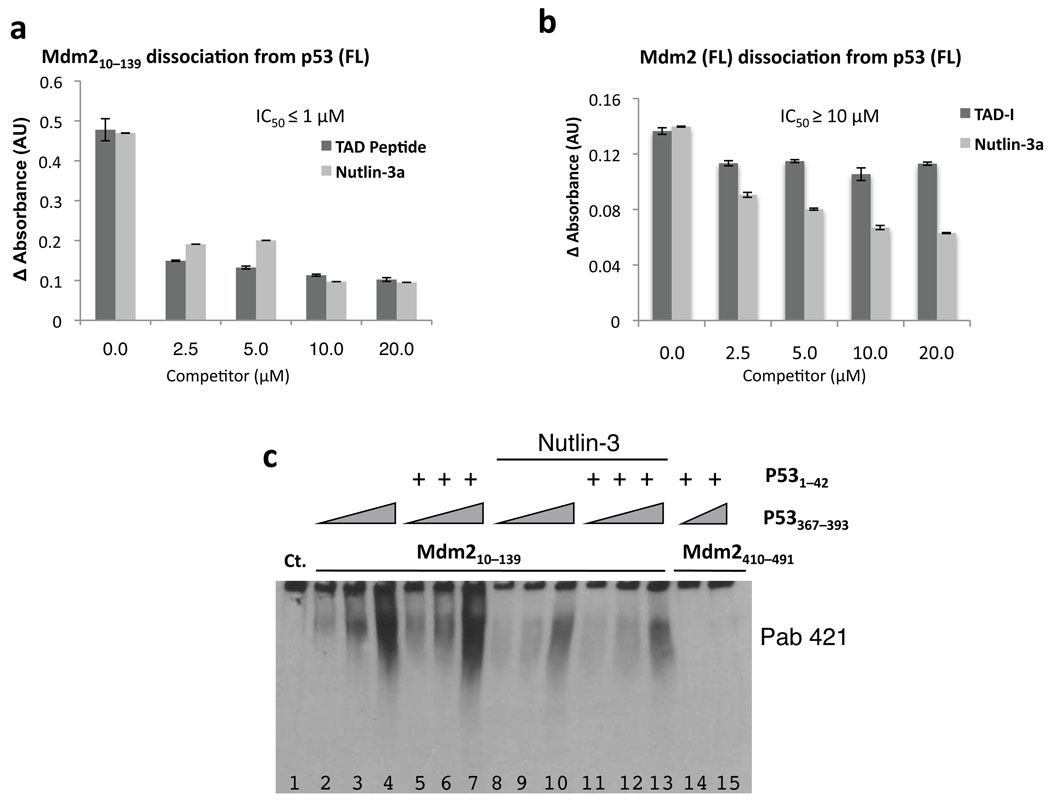
Because multiple contacts contribute to the Mdm2–p53 complex, we wanted to determine the relative contributions of the Mdm2 acidic domain and the p53-CTD to the overall binding. To this end, we tested the ability of Mdm2 and p53 to interact in the presence of Nutlin-3 or p53-TAD-I(1–42). Both of these reagents have been reported to have nanomolar affinities for Mdm2 and to effectively block complex formation between Mdm2(25–128) and p53(1–312)6,31. Accordingly, Nutlin-3 completely blocked the interaction between Mdm2(10–139) and the p53 TAD-I(1–42) peptide in native gel experiments (Supplementary Fig. 7). In ELISA-dissociation experiments, however, both Nutlin-3 and p53-TAD-I(1–42) peptide inhibited binding between p53 (FL) and Mdm2(10–139) with IC50’s of around 1 µM (Fig. 5a). Because earlier reported competition studies were performed with p53 lacking its C-terminus, we believe that the difference in affinity we observed (nM vs. µM IC50’s) is due to the contribution of the contacts made by the p53-CTD to the Mdm2–p53 complex. Indeed, when testing the ability of Nutlin-3 and the p53-TAD-I(1–42) peptide to block the binding of full-length p53 and full-length Mdm2, we observed that the IC50 for Nutlin-3 was about 10 µM while the TAD-I peptide showed very limited inhibition of the interaction (Fig. 5b). Based on these data we postulate that in vitro both the core domain of p53 and the p53-CTD make appreciable energetic contributions to the overall complex. This implies that even when the interaction between the N-termini of these proteins is disrupted, additional modification of the acidic domain of Mdm2 and/or the CTD of p53 may be necessary to completely dissociate Mdm2 and p53 in vivo.
Figure 5. Nutlin-3 inhibits Mdm2–p53 complex formation and leads to modification of the p53 C-terminus.
(a) Nutlin-3 and a p53 N-terminal peptide (TAD-I) each disrupt the interaction of p53 with the N-terminus of Mdm2. An ELISA assay was carried out as described in 1c; 6 |His-p53 (100 ng) and untagged Mdm2(10–139) (70 ng) was added to the p53-coated wells alone or presence of increasing amounts of Nutlin-3 or TAD-I(1–42). Mdm2, bound to p53 in the presence of competitors, was measured by anti-Mdm2 N-20 reactivity. (b) Neither Nutlin-3 nor a p53-TAD-I peptide can completely disrupt the interactions between full length Mdm2 and full-length p53. An ELISA assay was carried out as in 5a. (FL) Flag-Mdm2 (100 ng) was added to each well alone or in the presence of the indicated concentrations of Nutlin-3 or TAD-I(1–42) peptide. The amount of Mdm2 retained in each well was determined by reactivity with the N-20 anti-Mdm2 antibody. (c) Nutlin-3 reduces binding of the p53-CTD to the N-terminus of Mdm2. Reaction mixtures with 450 ng GST-Mdm2(10–139) (lanes 2–13) or GST-Mdm2(410–491) (lanes 14 and 15) contained increasing amounts of p53(367–393) (40, 80, 160 µM) and p53(1–42) (25 µM) where indicated. Additionally, mixtures in lanes 8–13 included 25 µM Nutlin-3, which was dissolved in DMSO. Final volume of DMSO in all reaction mixtures was 10%.
We tested at the ability of the p53 CTD(367–393) peptide to interact with Mdm2(10–139) in a native gel alone or in the presence of Nutlin-3 or the p53 TAD-I(1–42) peptide. The addition of the TAD-I(1–42) peptide did not disrupt the p53 CTD(367–393) interaction with Mdm2. Nevertheless, Nutlin-3 reduced the binding between the CTD and Mdm2 in this assay (Fig. 5c). These data fall in line with an NMR investigation, which compared the structures of Mdm2 N-terminal domain alone, p53-bound or bound to Nutlin-3 and showed that Nutlin-3 binds to Mdm2 via a mechanism distinct from p5332. Thus, it is possible that Nutlin-3-induced changes, which are perhaps refractory to CTD binding, are further contributing to the in vivo efficacy of this compound.
Nutlin-3 and p14ARF induce modification of the p53-CTD
Having accumulated a substantial amount of data demonstrating the interaction between the p53-CTD and the N-terminus of Mdm2, we were faced with a contradictory issue. In vivo inhibitors of the binding between the N-terminus of p53 and the N-terminus of Mdm2 (such as Nutlin-3) are able to effectively stabilize p53 and activate p53 function8,33–37. Nutlin-3 was reported to cause p53 stabilization without p53 being phosphorylated at several known stress inducible sites38. If Nutlin-3 was able to completely inhibit p53-Mdm2 complex formation without induction of any p53 post-translational modifications, then our in vitro binding data would not be relevant under physiological conditions. However, if dissociation of the N-terminal binding domains leads to increased C-terminal modifications (such as acetylation) of p53 in vivo, it is possible that Nutlin-3 treatment would affect not only the N-terminal interaction but would also decrease the C-terminal interaction as well.
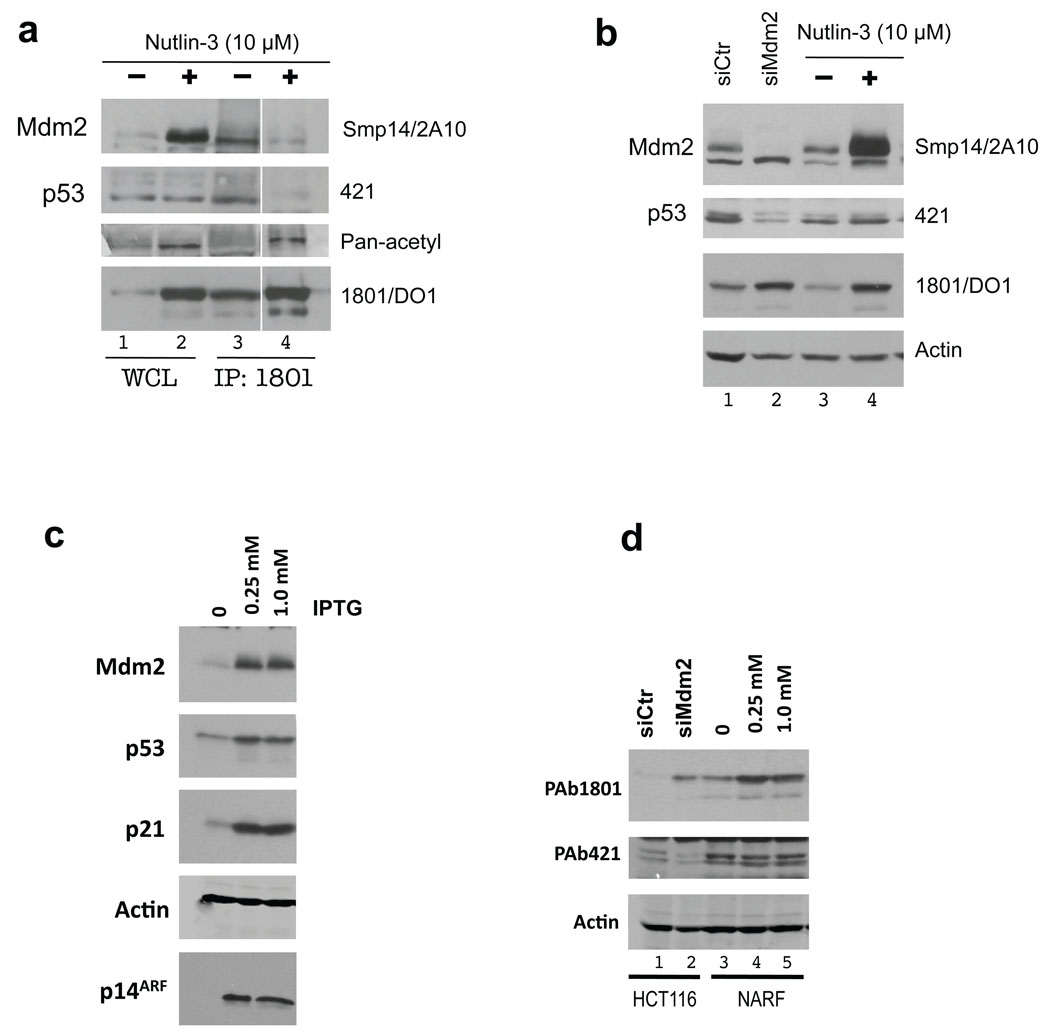
We treated HCT116 cells with 10 µM Nutlin-3 and, as expected, levels of p53 and Mdm2 increased when detected in whole cell extracts with a mixture of p53 N-terminal antibodies (PAb 1801 and DO1) and Mdm2 antibodies (SMP14 and 2A10) respectively (Fig. 6a, left panel lanes 1 and 2; right panel lanes 3 and 4). Consistent with the proposed mechanism of action of Nutlin-3, p53 in cells treated with this compound was not able to interact with Mdm2 as we were only able to detect Mdm2 in p53 immunoprecipitates from untreated cells (Fig. 6a, lanes 3 and 4). Strikingly, when p53 in these cells was detected with modification sensitive PAb 421, there was no increase in reactivity after Nutlin-3 treatment (Fig. 6a, lanes 1 and 2; Fig. 6B, lanes 3 and 4). Ablating Mdm2 by siRNA in these cells also caused an overall p53 accumulation and a decrease in 421-detectable p53, confirming the validity of using PAb 421 reactivity as readout of p53-CTD modifications (Fig. 6b, lanes 1 and 2). This result correlates with our earlier finding that having excess Mdm2 in cells leads to a decrease in p53 C-terminal acetylation39. Using a Pan-acetyl antibody, we saw an increase in signal from p53 after treatment (Fig. 6a). Thus, Nutlin-3 induces modifications of p53 at the C-terminus (including acetylation) and this likely diminishes the interaction of the p53-CTD with Mdm2.
Figure 6. Nutlin-3, p14ARF and Mdm2 siRNA induce C-terminal modifications of p53.
(a) HCT116 cells were treated in parallel with DMSO or Nutlin-3 (10 µM) for 4 hrs. Soluble extracts (75 µg; WCL) were subjected to immunoblotting with the antibodies indicated at right. p53 was immunoprecipitated with PAb 1801 from 1 mg of the same extracts (IP:1801). Lane 3 contained 5| volume of IP sample loaded in lane 4, to attempt to normalize for the amount of p53 on the gel. For p53 detection the blot was probed with PAb 421, stripped and re-probed with PAb 1801-DO1 mixture, then stripped again and re-probed with the Pan-acetyl antibody. (b) HCT116 cells, treated with Nutlin-3 (lanes 3 and 4) as in 6a or transiently transfected for 72 hrs with either control or Mdm2 siRNA (lanes 1 and 2). p53 was first detected by PAb 421, stripped and re-probed with PAb 1801-DO1 mixture. (c) U2OS cells stably expressing p14ARF under the control of IPTG (NARF) were grown exponentially and p14ARF was induced for 24 hrs. Following treatment lysates (75 µg) were resolved by SDS-PAGE and subjected to immunoblotting with the indicated antibodies. (d) 75 µg of soluble extracts from HCT116 cells treated with siMdm2 (lanes 1,2), NARF cells induced by IPTG (lanes 3–5) were resolved by SDS-PAGE and subjected to immunoblotting with the indicated antibodies. For p53 detection the blot was first probed with PAb 421, stripped and re-probed with PAb 1801-DO1 mixture.
It has been previously reported that p14ARF expression is able to stabilize p53 in the absence of N-terminal modifications40. To test the effects of p14ARF on the p53-CTD modifications we used NARF cells41. p14ARF induction led to stabilization of p53 and accumulation of p21 and Mdm2 (Fig. 6c). We then compared PAb 421 and PAb 1801-DO1 reactivity of p53 in NARF cells. Similarly to Nutlin-3 treatment, the amount of p53 detected by a mixture of PAb 1801-DO1 antibodies increased following p14ARF induction, while PAb 421 reactivity remained unchanged (Fig. 6d, lanes 3–5). Thus, three distinct DNA-damage-independent Mdm2 inhibitors were able to stimulate modifications of p53-CTD, most likely by shifting binding equilibrium away from Mdm2 and toward other modifying enzymes.
DISCUSSION
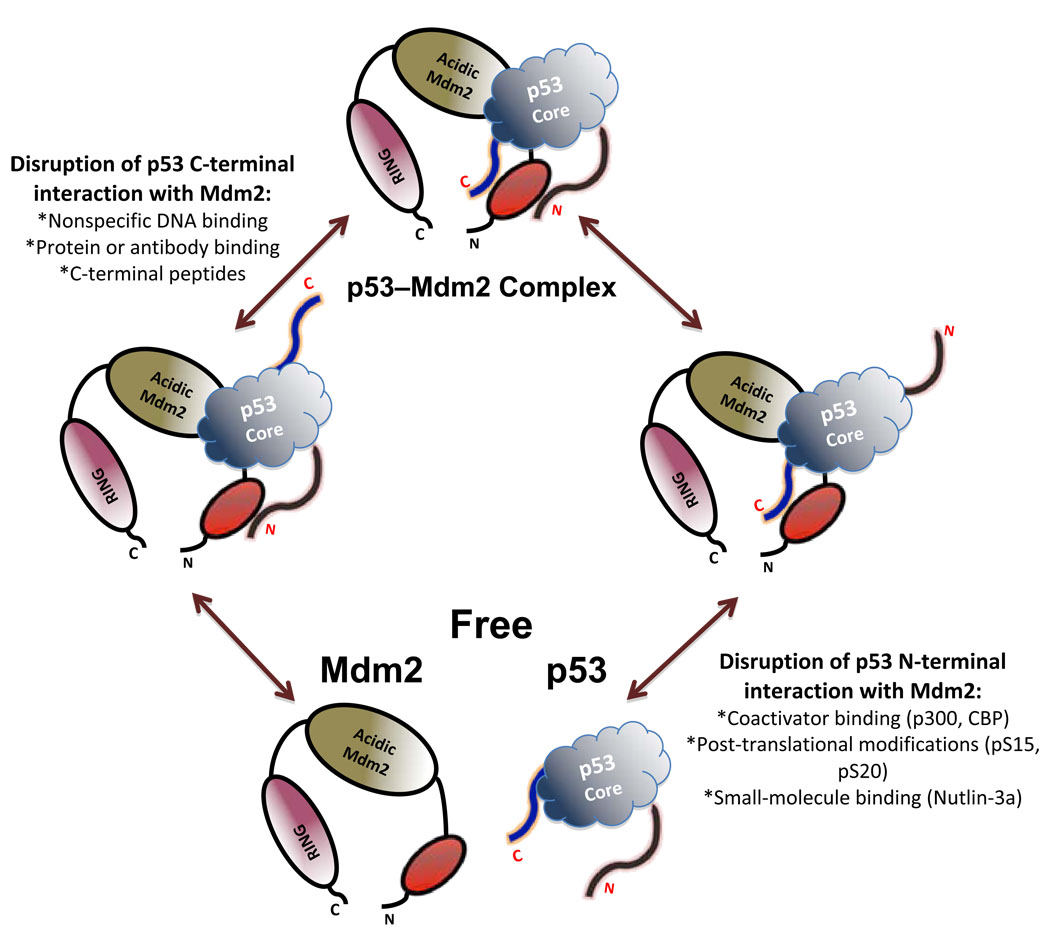
The interaction of Mdm2 and p53 is a highly regulated fine-tuned process. Here we describe an additional level of modulation of this association. Using different approaches, we show that the p53-CTD interacts directly with the Mdm2 N-terminus. C-terminal modifications of p53 can contribute to the full dissociation of Mdm2–p53 complex in vivo, and Nutlin-3 treatment or p14ARF overexpression can induce C-terminal modifications of p53 in cells. We propose a model where multiple interactions between p53 and MDM2 act in concert for optimal regulation of p53 (Fig. 7).
Figure 7. Proposed mechanism for the function of the p53 C-terminus and its modifications in Mdm2 complex formation.
Model postulates interactions between three p53 and three Mdm2 regions. Within the N-terminal ~ 140 amino acids of Mdm2 are predicted to be two discrete surfaces that interact with the N- and C-termini of p53 respectively. Within the central regions of p53 (core domain) and Mdm2 (the acidic region) are third interacting surfaces. Modifications at the N-terminus of p53 or disruption of its contacts with Mdm2 in vivo facilitate further modifications at the C-terminus leading to full disruption of the complex. Alternatively, modification(s) within the C-terminus of p53 could destabilize the Mdm2–p53 complex, favoring its dissociation.
We calculated the binding constant for the Mdm2–p53-CTD complex to be in the low micromolar range, which is similar to the affinity of peptides from the Mdm2 acidic region for p53 core domain (in the range of 10–100 µM14–16) but much weaker than the interaction of Mdm2 with the p53 N-terminus (nanomolar range). While we were unable to obtain evidence for complex formation by NMR (Supplemental Fig. 8), multiple other assays confirmed direct binding, and our data indicate that CTD–Mdm2 interaction is important for overall Mdm2–p53 binding. Indeed, the rather low affinity of the CTD for Mdm2 may actually be essential to allow this interaction to be modulated by modifications of the lysines in this region of the protein.
The interaction between the p53-CTD and Mdm2 described here provides a potential mechanism for published observations linking C-terminal modifications of p53 and the dissociation of the Mdm2–p53 complex20,22. While we have only addressed the effects of C-terminal acetylation of p53 on the Mdm2–p53 complex formation, the global regulatory scheme is likely to be far more complex. Mdm2 is extensively phosphorylated within its acidic domain19,42,43. The interaction of the Mdm2 acidic domain with the Box IV and V regions of p53 is essential for proper p53 ubiquitination by Mdm214. Our results that the Mdm2–p53 complex involves multiple contacts are consistent with the observations that Nutlin-3, TAD-I peptides or the removal of the 116 N-terminal amino acids of p53 do not block the ability of Mdm2 to ubiquitinate p53 in vitro15,17. It would be exciting to investigate effects of posttranslational modifications of Mdm2 and p53 from the perspective of their ability to stimulate or inhibit the formation of the multiple Mdm2/p53 interfaces.
The data (see Fig. 2a and 5c) showing that the binding of p53-CTD and TAD-I domains to Mdm2 are not mutually exclusive presuppose that different surfaces within the Mdm2 N-terminus contact the TAD-I and p53-CTD. Indeed, our preliminary MALDI-TOF analysis, suggests that the p53-CTD interacts with the flexible lid region at the N-terminus of Mdm2. This is of considerable interest, as Apo-Mdm2 predominantly exists in the closed conformation, which disfavors p53 interaction32,44. Potentially, binding of the basic p53-CTD to this region would disrupt the intra-molecular contacts made by the lid region and facilitate the conversion of Mdm2 to an open conformation thereby stimulating the binding of the TAD-I domain. Our findings also predict that the N- and C-termini of p53 are in spatial proximity, at least while both are bound to Mdm2. The putative closeness of the N- and C-termini of p53 in the context of the intact p53 tetramer could provide additional mechanisms of p53 regulation and facilitate Mdm2-mediated ubiquitination of this region. Perhaps modifications of the p53-CTD disrupt contacts not only with Mdm2 but alter the spatial relationship between the TAD-I and the p53-CTD, causing long-range conformational changes in the overall protein structure. Such changes could contribute to the loss of Mdm2 binding and have other functional consequences for p53-mediated transcription.
Nutlin-3 activates p53 efficiently in vivo. Based on our data we propose that multiple cooperating factors are needed for a robust p53 response. According to our model, Nutlin-3 disrupts the N-terminus of p53 from binding with Mdm2, thus freeing p53 N-terminus to bind to other C-terminal modifiers such as p30045,46. Consequent modifications of the p53 C-terminus further destabilize Mdm2–p53 complex. Thus, Nutlin-3 treatment shifts the competition between positive and negative regulators of p53 toward the positive regulators (Fig. 7)19,47. Both Nutlin-3 treatment and p14ARF expression lead to accumulation of p53 protein that lacks DNA damage-inducible phosphorylation marks at multiple N-terminal sites38,40. In this light, the induction of C-terminal modifications by both of these pathways seems highly noteworthy as such modifications could dictate differential transcriptional programs and lead to varying outcomes depending on cellular context. It has been reported that phosphorylation of p53 at key N-terminal residues induces association with p300, leading to C-terminal modification48,49. However, un-phosphorylated p53 is still able to bind to p300, and we believe that this interaction would be favored in cells where Mdm2 is either sequestered in complex with p14ARF or inhibited by Nutlin-3. Additionally, it is possible that Nutlin-3 and other p53 activating compounds are more effective in cancer cells6,8, because these cells may already have a low level of stress induced modifications on p5350,51, so the dissociation of Mdm2 and p53 is more easily accomplished in such cells.
Finally, our finding of a third interface between p53 and Mdm2 allows for a more detailed view of the complex and could provide additional avenues for therapeutic activation of wild-type p53 in cells.
METHODS
Plasmids and antibodies
Plasmids encoding human Mdm2, Mdm2(ΔC7), GST-Mdm2(410–491) and GST-Mdm2(10–139) were previously described29. The HA-p53 and p53(ΔC30) were cloned for mammalian expression from the CMV promoter in PcDNA-3 vector. Mdm2 proteins were detected with hybridoma supernatants (SMP14, 2A10, 3G5) or polyclonal antibody N20 (SantaCruz). p53 proteins were detected with hybridoma supernatants (PAb 1801, DO1 and 421) as indicated. Pan-acetyl antibody was purchased from Cell Signaling. p53 in ELISA experiments was detected with either purified PAb 421, rabbit polyclonal p53 FL-393 (SantaCruz) or His-probe conjugated HRP (SantaCruz) antibodies as indicated.
EMSA
Electrophoretic Mobility Shift Assay (EMSA) experiments were performed as previously described52.
ELISA
Enzyme-Linked Immunosorbent Assay (ELISA) experiments were performed as previously described53. Briefly, proteins in 200 µl of PBS were used to coat wells in a 96-well Pro-bind plate (Falcon) at 4°C overnight. The wells were then washed three times in PBS containing 0.05% Tween and then incubated in blocking buffer (PBS containing 0.05% Tween and 1% (w/v) bovine serum albumin) for 1 hr at 4°C. Increasing amounts of binding protein (as indicated) were added to the wells in PBS for 2 hr at 4°C. The wells were washed three times with PBS containing 0.05% (v/v) Tween, at which time binding protein specific antibody in blocking buffer was added and incubated for 1 hr at room temperature. After three washes, a 1:2000 dilution of monoclonal anti-rabbit or anti-mouse IgG antibody conjugated to alkaline phosphatase (Sigma) in blocking buffer was added and incubated at room temperature for 30 min. After five washes, 10 mM p-nitrophenol phosphate (Sigma) in 100 mM 2-amino-2-methyl-1,3-propanediol (Sigma) was added to the wells. Absorbance at 490 nm was measured at 1-min intervals using a Victor3 Plate Reader (PerkinElmer).
Fluorescence Spectrometry
Titration of Mdm2 into the fluorescein-labeled peptides was performed in 20 mM Hepes at pH 7.3 with ionic strength (IS) of 50 mM or 100 mM. Fluorescence was measured with excitation at 480 nm and emission at 530 nm. The bandwidths were changed depending on the amount of the labeled molecule used. The labeled peptide was placed in the cuvette in a volume of 1 ml, at a concentration of 100 nM, and 100–200 µl of GST-Mdm2 or GST (1.4–2 mM) were placed in the dispenser. Additions of 4 µl were titrated at 1 min intervals, the solution was stirred for 10 s, and the fluorescence and anisotropy were measured. The data were fit to 1:1 binding model54. In the competition binding assay, the conditions were as follows: 100 µl of unlabeled p53(367–393) (2724 mM) were added to a 1200 µl mixture of 100 nm fluorescein-labeled p53(367–393) and 1.4 mM GST-Mdm2 at ionic strength of 50 mm. Measurements were performed as previously described54.
Native Gel Experiments
4% Acrylamide-Tris/Borate gels without SDS were run at pH 8.5 in 1χ TB running buffer (90 mM Tris, 90 mM Boric acid). Following 2 hr resolution at 150 V at 4°C the gels were transferred to nitrocellulose membrane and subjected to immunoblotting.
Protein Crosslinking and Mass Spectrometry
Crosslinking was performed in buffer containing 25 mM Hepes pH 7.3, 250 mM NaCl and 2 mM DTT with or without 0.007% guleraldehyde or 1% formaldehyde (w/v). Reactions were terminated by the addition of 3χ SDS-PAGE buffer, denatured and resolved on 10% SDS-PAGE or 4–12% gradient MES gel respectively. Following resolution the gels were either subjected to immunoblotting with N-20 anti-Mdm2 antibody or were stained with Bio-Safe™ Coomassie stain (BioRad). Indicated bands were digested with trypsin and spotted on stainless steel targets with α-cyano-4-hydroxycinnamic acid as described previously55. All digests were analyzed by MALDI-TOF MS with a Voyager DE-Pro mass spectrometer (Applied Biosystems) equipped with a 337 nm nitrogen laser operated in the positive ion, reflectron mode with delayed extraction over the m/z range of 500–4000. Spectra were collected with 400 laser shots averaged to yield a peptide mass fingerprint. Spectra were processed using Mascot Wizard and a Mascot server (Matrix Science Ltd.) for protein identifications of Mdm2. Searches were made against the NCBI non-redundant database of 08/25/09 with human taxonomic filter (209358 sequences). Other spectra were recorded in positive linear mode in an effort to detect weak peptide signals.
ChIP Assays and Q-PCR
ChIP and Q-PCR analysis was carried out using p53−/−;mdm2−/− (2KO) cell extracts transfected with the indicated constructs as previously described56. Graphs are representative of at least three biological replicate experiments. Standard curves containing 0.1–27 ng of 2KO cell genomic DNA were run alongside the samples for each primer pair. Results were analyzed by the absolute quantification method. Error bars indicate the standard deviation of three PCR amplifications of one experiment. Primer sequences are as follows:
Insulin exon2
F: TGGCTTCTTCTACACACCCAAG; insulin exon2; R:ACAATGCCACGCTTCTGC;
p21 5′ distal p53 binding site:
F:TGGCCTTCAGGAACATGTCTT; R:CACCACCCTGCACTGAAGC)
Peptide synthesis
All peptides were synthesized on Rink amide MBHA resin on a Liberty MAPS (Microwave-Assisted Peptide Synthesizer, CEM) using standard Fmoc (9-fluorenylmethoxycarbonyl) chemistry. Trp was added at the peptides’ N-termini for UV spectroscopy. The peptides were purified on a Gilson HPLC (high pressure liquid chromatography, Middelton, WI) using a reverse-phase C8 semi-preparative column (ACE) with a gradient from 5% to 60% acetonitrile in water (both containing 0.001% (v/v) trifluoroacetic acid).
Cell lines, transfections, immunoblotting and protein purification are described in Supplementary Methods.
Supplementary Material
Acknowledgements
We are exceedingly grateful to Ella Freulich for her expert technical assistance and members of the Prives’ laboratory for their helpful suggestions. This work was supported by grant CA58316 from the NIH to CP.
Footnotes
Author Contributions:
M.V.P., A.F. and C.P. designed research; M.V.P., C.K., O.L., M.L., J.A., I.L.B., J.C.N., R.G., M.M., A.Z. and L.M.B. performed research and analyzed data; M.V.P. and C.P. wrote the manuscript.
REFERENCES
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 4.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 5.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 6.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–888. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 7.Klein C, Vassilev LT. Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer. 2004;91:1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issaeva N, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 9.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 10.Appella E, Anderson CW. Signaling to p53: breaking the posttranslational modification code. Pathol Biol (Paris) 2000;48:227–245. [PubMed] [Google Scholar]
- 11.Kane SA, et al. Development of a binding assay for p53/HDM2 by using homogeneous time-resolved fluorescence. Anal Biochem. 2000;278:29–38. doi: 10.1006/abio.1999.4413. [DOI] [PubMed] [Google Scholar]
- 12.Sakaguchi K, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 13.Lai Z, Auger KR, Manubay CM, Copeland RA. Thermodynamics of p53 binding to hdm2(1–126): effects of phosphorylation and p53 peptide length. Arch Biochem Biophys. 2000;381:278–284. doi: 10.1006/abbi.2000.1998. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu H, et al. The conformationally flexible S9–S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446–28458. doi: 10.1074/jbc.M202296200. [DOI] [PubMed] [Google Scholar]
- 15.Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251–263. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 16.Yu GW, et al. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci U S A. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J, et al. A second p53 binding site in the central domain of Mdm2 is essential for p53 ubiquitination. Biochemistry. 2006;45:9238–9245. doi: 10.1021/bi060661u. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter S, Vousden KH. Modifications of p53: competing for the lysines. Curr Opin Genet Dev. 2009;19:18–24. doi: 10.1016/j.gde.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 21.Li M, et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 24.Hainaut P, et al. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 27.Ahn J, Prives C. The C-terminus of p53: the more you learn the less you know. Nat Struct Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- 28.Cain C, Miller S, Ahn J, Prives C. The N terminus of p53 regulates its dissociation from DNA. J Biol Chem. 2000;275:39944–39953. doi: 10.1074/jbc.M002509200. [DOI] [PubMed] [Google Scholar]
- 29.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. Embo J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 31.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 32.Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Bruschweiler R. Quantitative lid dynamics of MDM2 reveals differential ligand binding modes of the p53-binding cleft. J Am Chem Soc. 2008;130:6472–6478. doi: 10.1021/ja800201j. [DOI] [PubMed] [Google Scholar]
- 33.Ding K, et al. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc. 2005;127:10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Echeverria C, Chene P, Blommers MJ, Furet P. Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J Med Chem. 2000;43:3205–3208. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 35.Duncan SJ, Cooper MA, Williams DH. Binding of an inhibitor of the p53/MDM2 interaction to MDM2. Chem Commun (Camb) 2003:316–317. doi: 10.1039/b211889k. [DOI] [PubMed] [Google Scholar]
- 36.Bottger V, et al. Identification of novel mdm2 binding peptides by phage display. Oncogene. 1996;13:2141–2147. [PubMed] [Google Scholar]
- 37.Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3:419–421. [PubMed] [Google Scholar]
- 38.Thompson T, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 39.Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem. 2006;281:16943–16950. doi: 10.1074/jbc.M601388200. [DOI] [PubMed] [Google Scholar]
- 40.de Stanchina E, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stott FJ, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hay TJ, Meek DW. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 2000;478:183–186. doi: 10.1016/s0014-5793(00)01850-0. [DOI] [PubMed] [Google Scholar]
- 43.Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy MA, Gesell JJ, Senior MM, Wyss DF. Flexible lid to the p53-binding domain of human Mdm2: implications for p53 regulation. Proc Natl Acad Sci U S A. 2003;100:1645–1648. doi: 10.1073/pnas.0334477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito A, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. Embo J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci U S A. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahl GM. Mouse bites dogma: how mouse models are changing our views of how P53 is regulated in vivo. Cell Death Differ. 2006;13:973–983. doi: 10.1038/sj.cdd.4401911. [DOI] [PubMed] [Google Scholar]
- 48.Feng H, et al. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure. 2009;17:202–210. doi: 10.1016/j.str.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkins LM, et al. Two distinct motifs within the p53 transactivation domain bind to the Taz2 domain of p300 and are differentially affected by phosphorylation. Biochemistry. 2009;48:1244–1255. doi: 10.1021/bi801716h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 51.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 52.Lokshin M, Li Y, Gaiddon C, Prives C. p53 and p73 display common and distinct requirements for sequence specific binding to DNA. Nucleic Acids Res. 2007;35:340–352. doi: 10.1093/nar/gkl1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T, Prives C. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J Biol Chem. 2001;276:29702–29710. doi: 10.1074/jbc.M011326200. [DOI] [PubMed] [Google Scholar]
- 54.Rotem S, et al. The structure and interactions of the proline-rich domain of ASPP2. J Biol Chem. 2008;283:18990–18999. doi: 10.1074/jbc.M708717200. [DOI] [PubMed] [Google Scholar]
- 55.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckerman R, et al. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baptiste N, Friedlander P, Chen X, Prives C. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene. 2002;21:9–21. doi: 10.1038/sj.onc.1205015. [DOI] [PubMed] [Google Scholar]
- 58.Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci U S A. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.