Abstract
Malaria parasites utilize a short N-terminal amino acid motif termed the Plasmodium export element (PEXEL) to export an array of proteins to the host erythrocyte during blood stage infection. Using immunoaffinity chromatography and mass spectrometry, insight into this signal-mediated trafficking mechanism was gained by discovering that the PEXEL motif is cleaved and N-acetylated. PfHRPII and PfEMP2 are two soluble proteins exported by Plasmodium falciparum that were demonstrated to undergo PEXEL cleavage and N-acetylation, thus indicating that this N-terminal processing may be general to many exported soluble proteins. It was established that PEXEL processing occurs upstream of the brefeldin A-sensitive trafficking step in the P. falciparum secretory pathway, therefore cleavage and N-acetylation of the PEXEL motif occurs in the endoplasmic reticulum (ER) of the parasite. Furthermore, it was shown that the recognition of the processed N-terminus of exported proteins within the parasitophorous vacuole may be crucial for protein transport to the host erythrocyte. It appears that the PEXEL may be defined as a novel ER peptidase cleavage site and a classical N-acetyltransferase substrate sequence.
Keywords: Malaria, Plasmodium falciparum, Protein export, N-acetylation, Endoplasmic reticulum, Brefeldin A
1. Introduction
The asexual blood stage of the apicomplexan parasite Plasmodium falciparum is responsible for clinical manifestations associated with the most virulent form of malaria in humans [1]. As the parasite matures within a parasitophorous vacuole (PV) inside the red blood cell (RBC), it extensively modifies the RBC to make it amenable for nutrient uptake and to prevent clearance by the spleen [2,3]. The parasite accomplishes this remodeling of the terminally differentiated RBC by exporting proteins across the parasite plasma membrane and the parasitophorous vacuolar membrane (PVM) to the red cell cytosol (RCC) or red cell membrane (RCM). A major example of a parasite-induced modification to the host RBC is the formation of knobs on the outer surface of the RCM [3]. These large protrusions consist of the parasite-encoded knob-associated histidine-rich protein (KAHRP) anchoring the immunovariant adhesin P. falciparum erythrocyte membrane protein 1 (PfEMP1) to the RBC cytoskeleton [4–6]. These protein complexes are implicated in the cytoadherance of infected RBCs to the host endothelium leading to their sequestration in the peripheral vasculature [7]. Therefore, the exported proteins involved in the formation of these knobs and the actual machinery involved in their transport are not only major virulence factors in severe disease pathology, they are also crucial to the survival of P. falciparum.
A highly conserved 5-amino acid motif (RxLxE/Q/D) termed the Plasmodium export element (PEXEL) or host-targeting signal was recently identified through bioinformatic approaches and experimentally verified to mediate the translocation of parasite-derived proteins across the PVM and into the host RBC [8,9]. Site-directed mutagenesis of the conserved residues R, L, or E/Q/D with alanine abolished the export of green fluorescent protein (GFP) chimeras to the RCC. The discovery of the PEXEL has allowed for the in silico annotation of the P. falciparum “exportome” or “secretome” that has brought to light novel protein families exported by the parasite [10]. Conservation of the PEXEL motif and its corresponding pathway across the genus Plasmodium indicates the existence of common trafficking components in malaria parasites. The importance and uniqueness of this trafficking signal has generated considerable interest in the identification of the machinery that interacts with the PEXEL because of the potential for drug design.
With the discovery of this signal-mediated mechanism in P. falciparum, an updated model for the export of proteins to the host RBC involves recruitment into the parasite endoplasmic reticulum (ER) [11,12], default secretion into the PV lumen [12–15], and PEXEL-mediated translocation across the PVM [8,9,14,15]. In other eukaryotic cells, proteins destined for export enter the secretory pathway by co-translational translocation across the ER membrane mediated by the recognition of a N-terminal signal peptide and concomitant cleavage of this signal peptide by the signal peptidase complex (SPC) [16]. It has been assumed that P. falciparum proteins exported to the host RBC have their ER-type signal peptides cleaved by the parasite SPC but this type of N-terminal processing has never been definitively characterized. Here, we show that exported parasite proteins do undergo N-terminal processing that involves cleavage and N-acetylation of the PEXEL. The two examples of PEXEL processing described here indicate that this N-terminal cleavage and acetylation are likely to occur in many soluble proteins exported by P. falciparum. Probing these N-terminal processing events with brefeldin A (BFA) reveal that PEXEL cleavage and N-acetylation occurs in the parasite ER. The PEXEL processing of a PV-trapped GFP chimera suggests a recognition event in the PV may be crucial for export beyond the PVM. The dissection of the PEXEL as a novel ER peptidase cleavage site and a classical N-acetyltransferase substrate sequence is also discussed.
2. Materials and methods
2.1. In vitro culturing of P. falciparum
P. falciparum strain 3D7 and transfected parasites expressing the transgene KAHRP(-His)-GFP were obtained from the Malaria Research and Reference Reagent Resource Center (MR4) [12]. For clarification purposes, this construct has been renamed as KAHRP(1-60)-GFP. The transfected 3D7 line expressing the PfHRPIImyc transgene was kindly provided by Dr. Kasturi Haldar (Northwestern University, Chicago, IL) [17]. Parasites were cultured in human RBCs maintained at 2% hematocrit in RPMI 1640 medium supplemented with 25 mM HEPES pH 7.4, 0.2% sodium bicarbonate, 30 mg/L hypoxanthine, 0.5% AlbuMAX II (Invitrogen), and 0.1 mg/mL gentamycin in an atmosphere of 90% N2, 5% CO2, and 5% O2 at 37 °C [18]. Synchronization was maintained by serial treatment of ring stage parasites with 5% d-sorbitol (Sigma) [19]. Parasitemia was determined from thin smears prepared on microscope slides and stained with Hema Wright-Giemsa stain (Fisher). The transfected parasite lines expressing the transgenes PfHRPIImyc and KAHRP(-His)-GFP were episomally maintained with 25 ng/mL pyrimethamine (Sigma) and 5 nM WR99210, respectively.
2.2. Plasmid construction, transfection, and microscopy of PfEMP2-GFP
The GFPmut2 sequence was PCR-amplified with primers 5′-GACTAGGATCCCATCAC CATCACCATCATATGAGTAAAGGAGAAGAACTTTTCACTGG-3′ and 5′-GATA CTCGAGTTATTTGTATAGTTCATCCATGCCATG-3′ from pfGNr (MR4) to introduce a His6 linker N-terminal to GFPmut2 and the PCR product was inserted into BamHI/XhoI-restricted pET-MALcH to obtain His6-GFPmut2 pET-MALcH [20]. The nucleotide sequence encoding for the first 87 amino acids of PfEMP2 was PCR-amplified from the PfEMP2 open reading in a 3D7 26-h cDNA library (MR4) with primers 5′-CGCTAGAG CTCCATGGAGGTAATTTGTAGAAA-3′ and 5′-GACTAGAATTCGCCTCTAAACTC ATCGGTGGTT-3′ and inserted into SacI/EcoRI-restricted His6-GFPmut2 pET-MALcH to place the PEXEL processing site 19 amino acids upstream of a thrombin cleavage site (TCS) and generate PfEMP2(1-87)-TCS-His6-GFPmut2 pET-MALcH. The subcloned PfEMP2-GFP construct was PCR-amplified with primers 5′-GGTGGATACTCGAGAT GGAGGTAATTTGTAGAA-3′ and 5′-GATACTCGAGTTATTTGTATAGTTCATCCA TGCCATG-3′ and inserted into the XhoI site in pHC1 (MR4) to obtain PfEMP2-GFP pHC1 [21].
Parasites (3D7) were transfected with PfEMP2-GFP pHC1 by the DNA-loaded RBC method as described with some modifications [22]. Briefly, 100 μg of Qiagen-purified supercoiled plasmid DNA was mixed with 0.250 mL of fresh RBCs in incomplete cytomix and loaded into a 0.2-cm electroporation cuvette and subjected to Bio-Rad Gene Pulser II settings of 0.31 kV and 950 μF. DNA-loaded RBCs were then washed with culture media and innoculated with 3D7 schizont-infected RBCs achieve a final parasitemia of ∼1%. A selection pressure of 25 ng/mL pyrimethamine was applied to the culture after parasitemia had reached ∼5% rings and maintained to select for stable transfectants. Transfected parasites reached ∼1% parasitemia after 21 days of selection.
The expression and export of PfEMP2-GFP was confirmed by fluorescence microscopy. The nuclei of P. falciparum-infected RBCs were stained with 5 μM DRAQ5 (Alexis) and live imaging was carried out by a custom-built, wide-field fluorescence microscope with a 100× oil immersion objective (1.4 NA; Olympus). Z-sections of 0.2 μm were taken and three-dimensional stacks were subjected to constrained iterative deconvolution according to the previous method [23].
2.3. Purification of P. falciparum exported proteins by immunoaffinity chromotography
Polyclonal rabbit antibodies raised to a C-terminal repeat (HHAADA) of PfHRPII (Invitrogen) were coupled to Sepharose CL-4B (Amersham) by cyanogen bromide activation to couple 1.8 mg antibody/mL Sepharose. RBCs infected with late trophozoite stage (∼33–36 h) 3D7 parasites were harvested and lysed with 10 volumes of 0.1% saponin (Sigma) at room temperature for 5 min to release the soluble contents of the PV lumen and RCC. The lysis mixture was then diluted with PBS and spun down to obtain the saponin supernatant and parasite pellet. The resulting saponin supernatant was applied to Ni-NTA Sepharose (Qiagen) to enrich for PfHRPII and eluted with 1 M imidazole (Sigma). The Ni-NTA eluate was then flowed over 2 mL of the prepared anti-PfHRPII Sepharose and PfHRPII was eluted with 50 mM triethanolamine pH 12 and neutralized with 1 M NaOAc pH 4.8.
To enrich for PfHRPIImyc from the RCC, saponin supernatant was flowed over 5 mL EZview Red Anti-c-Myc Affinity Gel (Sigma), washed with a modified RIPA buffer (50 mM Tris–HCl, 500 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10% glycerol, pH 8), and eluted with the same high pH buffer used previously. ER-trapped PfHRPIImyc was enriched from ring stage parasites treated with 5 μg/mL BFA (Sigma) for 24 h. The parasite pellet remaining after saponin lysis was solubilized with modified RIPA buffer to extract the PfHRPIImyc trapped in the ER and purified the same way as the RCC purification.
PfEMP2-GFP exported to the RCC was enriched from saponin supernatant using 400 μL agarose conjugated anti-GFP (MBL). The PfEMP2-GFP accumulating in the parasite ER from BFA treatment for 24 h was once again extracted from the parasite pellet with modified RIPA buffer and enriched using the anti-GFP resin. KAHRP(1-60)-GFP accumulating in the PV lumen was enriched from trophozoite-infected RBC saponin supernatant by also utilizing anti-GFP agarose.
2.4. SDS-PAGE and Western blot analysis
Protein samples eluted from the immunoaffinity resin were solubilized in 6× SDS-PAGE load buffer and separated on Novex 10–20% Tris–glycine gels (Invitrogen). Gels were stained with Coomassie R-250 stain (Sigma) and Western blotting to 0.45 μm nitrocellulose (Schleicher and Schuell) was performed according to standard protocols. Antibodies used were polyclonal anti-PfHRPII (1:10,000), polyclonal anti-GFP antibodies (1:5000; Invitrogen), stabilized goat anti-rabbit, and anti-mouse HRP-conjugated secondary antibodies (1:1000; Pierce). SuperSignal West Femto maximum sensitivity substrate (Pierce) in conjunction with a Fluor-S MultiImager (Bio-Rad) was used for the visualization of all Western blots.
2.5. Proteolytic digests and mass spectrometry (MS) analysis of peptide fragments
Endogenous PfHRPII purified from 3D7 saponin supernatant (∼2 μg) was buffered exchanged into 50 mM ammonium bicarbonate pH 8 and the in-solution digest was initiated with 20 ng Asp-N (1:100 wt/wt; Roche). After 2 h at 37 °C, the digest was stopped by acidification with the addition of trifluoroacetic acid (TFA) up to 0.2% final concentration. The resulting peptide fragments generated from the proteolytic digest were concentrated and desalted using a C18 ZipTip (Millipore) and eluted with 4 μL of a 50% MeCN:49.9% H2O:0.1% TFA solution.
PfHRPIImyc elutions from the anti-c-Myc affinity resin was resolved by SDS-PAGE and the gel band was excised and prepared for in-gel digestion [24]. Gel bands were incubated with Asp-N (1:25) overnight (∼22 h) at 37 °C and peptides were then extracted with the 50% MeCN:49.9% H2O:0.1% TFA solution. The volume of the resulting solution was reduced by SpeedVac and the extracted peptides for each sample were treated with a C18 ZipTip.
PfEMP2-GFP eluted from agarose-conjugated anti-GFP was buffered exchanged into the previous ammonium bicarbonate buffer. About 3 μg of the enriched GFP chimera was treated with thrombin (1:1000) and incubated at 37 °C overnight. The resulting sample was desalted with a C18 ZipTip for mass spectrometry analysis.
KAHRP(1-60)-GFP enriched with anti-GFP agarose was subjected to SDS-PAGE and the gel band was excised for an Asp-N (1:10) in-gel digest overnight at 37 °C and the peptides were extracted and treated with a C18 ZipTip.
After all samples were desalted with a C18 ZipTip, 0.8 μL of each eluate was mixed with an equal volume of saturated HCCA solution (Agilent) containing 13.2 mM ammonium citrate, spotted on the MALDI plate, and allowed to dry. A full-scan mass spectrum for all samples was recorded on an AB 4700 MALDI tandem TOF instrument in reflector mode. The peptide ions of interest were then isolated in MS1 and individual CID spectra were acquired. Air was used as the collision gas at 1.0 keV (lab frame) collision energy. The spectra were internally calibrated with known molecular masses of peptides from the digests.
3. Results and discussion
3.1. PEXEL of PfHRPII is cleaved and N-acetylated
To address whether exported Plasmodium proteins undergo N-terminal processing during trafficking to the host RBC, P. falciparium histidine-rich protein II (PfHRPII) was purified and its N-terminus was characterized. PfHRPII is a soluble parasite protein known to be exported to the RCC [13,17,25,26]. RBCs infected with 3D7 parasites were harvested at the late trophozoite stage (∼33–36 h after invasion) because of the maximal expression and export of PfHRPII during this period [17]. The infected RBCs were lysed with 0.1% saponin to release the soluble components of the PV lumen and RCC. Nickel chelate chromatography was used to enrich for PfHRPII (histidine content 32%) from this resulting saponin supernatant and then was purified to ≥95% purity by anti-PfHRPII immunoaffinity chromatography (Fig. 1A). The N-terminus of the purified protein was analyzed by Edman degradation but the lack of signal indicated that the N-terminus of PfHRPII was blocked (data not shown). Therefore, PfHRPII purified from saponin supernatant was subjected to Asp-N proteolysis and analyzed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS (Fig. 1A). The protein was digested and confirmed to be PfHRPII (70% sequence coverage) (Table S1). After accounting for all predicted Asp-N proteolysis products, the peptide fragment containing the mature N-terminus of PfHRPII was then sought after. Tandem MS analysis with collision-induced dissociation (CID) of the peptide at m/z 1953.87 identified it as the most N-terminal fragment corresponding to the sequence Ac-48HETQAHVDDAHHAHHVA64 based on its fragmentation pattern (Fig. 1B). The de novo sequencing of this peptide shows that the PEXEL in PfHRPII is cleaved (45RLL↓) and N-acetylated (Ac-HE49). There is no evidence of an uncleaved and/or unacetylated PEXEL, suggesting that PfHRPII is quantitatively processed during export to the host RBC. An uncleaved PEXEL would generate a peptide at m/z 2224.28 for amino acids 1–21 and a cleaved but unacetylated PEXEL would generate a peptide at m/z 1911.86 for amino acids 48–64 without the monoisotopic mass change of 42.01 for N-acetylation. Characterization of the N-terminus of a C-terminally c-Myc-tagged PfHRPII protein (PfHRPIImyc) overexpressed under the parasite cam promoter and exported to the RCC also showed identical PEXEL processing as the endogenous protein to indicate that overexpression does not negatively impact N-terminal processing of the PfHRPII PEXEL (Fig. S1, Table S2). This is consistent with previous data showing that the overexpression of PfHRPIImyc does not have an effect on bulk protein export to the host RBC and ultrastructural organization of the parasite [17].
Fig. 1.
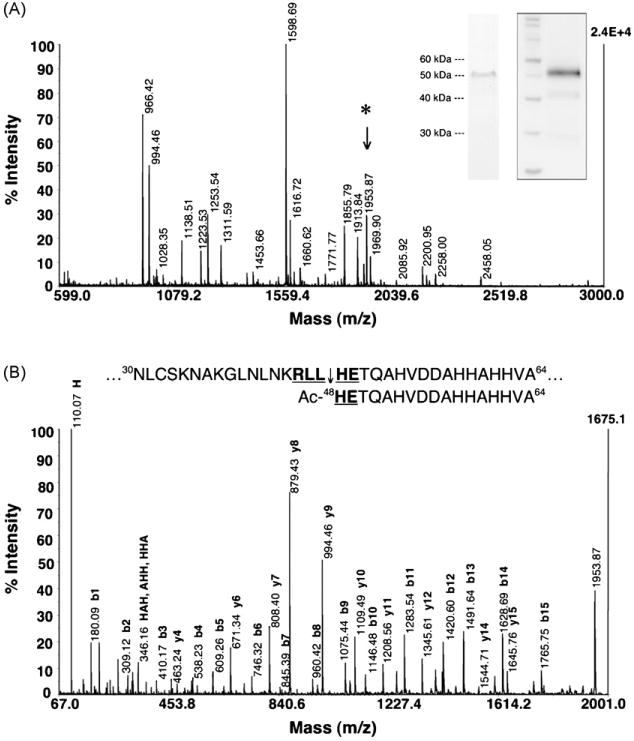
PEXEL of PfHRPII is cleaved and N-acetylated. (A) Native PfHRPII purified from the saponin supernatant of trophozoite-infected RBCs was subjected to an Asp-N solution digestion and analyzed by MALDI-TOF MS. The inset Coomassie-stained gel (left) and anti-PfHRPII Western blot (right) confirms the purified protein. The peptide fragment at m/z 1953.87 (*) was selected for tandem MS analysis. (B) De novo sequencing of PfHRPII m/z 1953.87. CID analysis reveals that m/z 1953.87 is the most N-terminal peptide fragment and demonstrates the PEXEL in PfHRPII (bold and underlined) is cleaved after residue 47 (45RLL↓) and the mature N-terminus is N-acetylated (Ac-HE49). The absolute intensity of the highest signal for each spectrum is labeled in bold on the right vertical axis (2.4E+4 and 1675.1, respectively).
3.2. PEXEL of PfEMP2 is cleaved and N-acetylated
We also examined whether this N-terminal processing occurs in the P. falciparum erythrocyte membrane protein 2 (PfEMP2) which is a soluble protein also exported by the parasite to the host RBC and deposited on the cytoplasmic face of the RCM under knobs [25–27]. Amino acids 1–87 of PfEMP2 were appended to the N-terminus of GFP to create a PfEMP2-GFP chimera (Fig. 2C). A thrombin cleavage site was placed 19 amino acids from the predicted PEXEL processing site (75RIL↓SE79) to simplify the subsequent biochemical analysis. If the PEXEL of PfEMP2-GFP is fully processed, thrombin cleavage of the processed protein would generate a single peptide at m/z 2095.04. This construct was cloned into the pHC1 Plasmodium expression vector and overexpressed under the parasite cam promoter that is maximally active in the trophozoite stage [21].
Fig. 2.
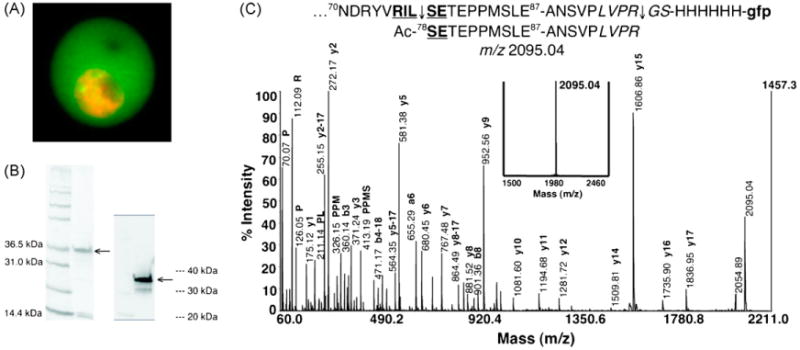
PEXEL of PfEMP2 is cleaved and N-acetylated. (A) PfEMP2-GFP is exported to the host RBC. PfEMP2-GFP (green) was imaged live and the nuclei of the parasites were stained with DRAQ5 (orange). (B) PfEMP2-GFP was enriched from trophozoite-infected RBC saponin supernatant. The Coomassie-stained gel (left) and anti-GFP Western blot (right) confirms the enriched protein (arrows). The contaminating proteins at ∼15 kDa are likely the α and β subunits of hemoglobin. (C) PfEMP2-GFP was treated with thrombin to generate a single peptide at m/z 2095.04. The PfEMP2-GFP chimera has the first 87 amino acids of PfEMP2 containing the PEXEL 75RILSE79 (bold and underlined), a thrombin cleavage motif (italicized), and a His6 linker fused to the N-terminus of the GFP reporter (lower case). A peptide with a predicted m/z 2095.04 is expected to be produced after PEXEL processing and thrombin cleavage. Upon treatment of the enriched PfEMP2-GFP with thrombin, a single peptide at m/z 2095.04 was indeed generated (bold inset spectrum). Tandem MS analysis by CID produced a fragmentation pattern that confirms the sequence of the generated peptide to reflect the cleavage (75RIL↓) and N-acetylation (Ac-SE79) of the PEXEL in PfEMP2.
Live imaging of the resulting RBC infected with the transfected parasite line verifies that the PfEMP2-GFP is correctly trafficked to the RCC (Fig. 2A). The extensive nuclear staining is indicative of the onset of nuclear division in the late trophozoite/early schizont stage. Exported PfEMP2-GFP protein was enriched from the saponin supernatant of trophozoite-infected RBCs by anti-GFP immunoaffinity chromatography (Fig. 2B) and treated with thrombin. The thrombin cleavage produced a single peptide at m/z 2095.04 and the CID spectrum confirmed the sequence to be Ac-SETEPPMSLEANSVPLVPR (Fig. 2C), thus demonstrating the cleavage (75RIL↓) and N-acetylation (Ac-SE79) of the PEXEL in PfEMP2. Given the likelihood that soluble parasite proteins containing a PEXEL utilize the same protein trafficking pathway and interact with the same secretory machinery as they are exported to the host RBC, our two examples of PEXEL processing suggest that this N-terminal cleavage (RxL↓) and N-acetylation (Ac-xE/Q/D) may occur in many soluble proteins exported by P. falciparum to the host RBC.
3.3. PEXEL processing occurs in the parasite ER
To determine the site of PEXEL-mediated N-terminal processing in the P. falciparum export pathway, the cleavage and N-acetylation events were probed with BFA, a fungal metabolite that prevents anterograde transport from the ER to the Golgi in P. falciparum causing exported proteins to accumulate in the parasite ER [17,28]. BFA treatment of ring stage parasites can be used to determine if PEXEL cleavage and/or N-acetylation occur upstream or downstream of the BFA-sensitive trafficking step by characterizing the N-termini of ER-trapped PfHRPIImyc and PfEMP2-GFP.
Trafficking of endogenous PfHRPII and transgenic PfHRPIImyc to the RCC are both BFA-sensitive [17]. Thus, ring-infected RBCs (0–4 h post-invasion) expressing PfHRPIImyc under the parasite cam promoter were treated with 5 μg/mL BFA before the onset of promoter activity in the early ring stage, then incubated with BFA for 24 h while the transgene was expressed, and finally the parasite pellet was harvested after saponin lysis. ER-trapped PfHRPIImyc was extracted and enriched from the parasite pellet (Fig. 3A) and subjected to an Asp-N in-gel digestion. MALDI-TOF MS analysis confirmed the PfHRPIImyc with 88% sequence coverage (Table S3). After accounting for all predicted Asp-N proteolysis products, two peptide fragments at m/z 863.40 and 1953.87 (Fig. 3A) were confirmed by CID to have the sequences Ac-48HETQAHV54 (Fig. 3B) and Ac-48HETQAHVDDAHHAHHVA64 (data not shown), respectively. This data shows that the PfHRPIImyc PEXEL is cleaved and N-acetylated upstream of the BFA-sensitive trafficking step in the P. falciparum export pathway.
Fig. 3.
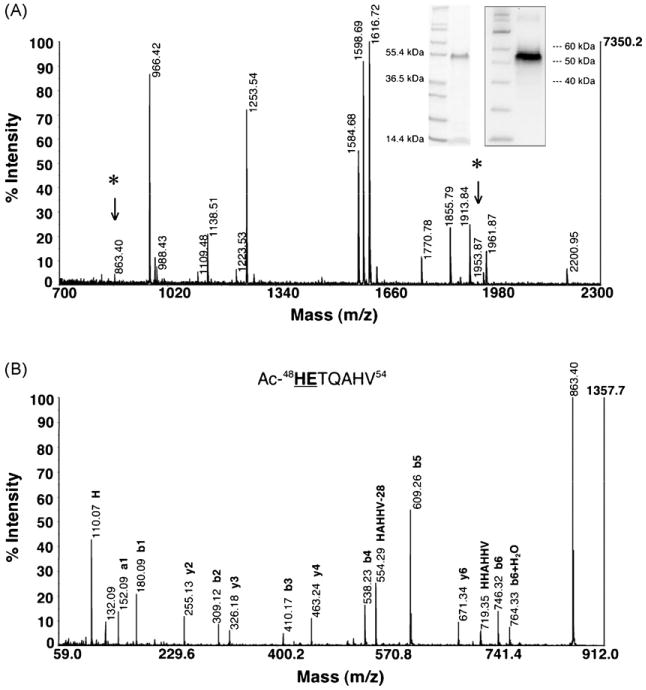
PfHRPIImyc PEXEL is processed upstream of the BFA-sensitive trafficking step. (A) PfHRPIImyc trapped in the ER from BFA treatment was extracted and enriched from the resulting parasite pellet after saponin lysis. The inset Coomassie-stained gel (left) and anti-c-Myc Western blot (right) confirms the enriched protein. An Asp-N in-gel digestion was performed because of contaminating hemoglobin (∼15 kDa). After accounting for all predicted Asp-N proteolysis products, the peptide fragments at m/z 863.40 (*) and 1953.87 (*) were selected for tandem MS analysis. (B) De novo sequencing of PfHRPIImyc m/z 863.40. CID analysis reveals that m/z 863.40 is one of the N-terminal peptide fragments along with m/z 1953.87 (not shown) to demonstrate that the PEXEL processing of PfHRPIImyc occurs upstream of the BFA-sensitive trafficking step. Residues in the sequenced peptide that are bold and underlined are a part of the PfHRPIImyc PEXEL.
Since the trafficking of PfEMP2-GFP has never been established to be BFA-sensitive, the BFA-sensitivity of PfEMP2-GFP trafficking was determined by fluorescence microscopy. Upon treatment with 5 μg/mL BFA for 24 h, the GFP signal was exclusively localized to a perinuclear compartment indicative of the parasite ER (Fig. 4A), thus showing that the trafficking of PfEMP2-GFP is BFA-sensitive. The accumulated protein was extracted and enriched from BFA-treated ring parasites (Fig. 4B) and treated with thrombin. The MS data for PfEMP2-GFP shows that a single peptide at m/z 2095.04 with the sequence Ac-SETEPPMSLEANSVPLVPR was generated by the thrombin cleavage (Fig. 4C). The fact that the PEXEL motifs of both exported proteins are cleaved and N-acetylated prior to ER-to-Golgi transport establishes that both PEXEL processing events occur in the ER. Since there is precedence that N-terminal cleavage and acetylation of eukaryotic proteins are co-translational events [29,30], PEXEL processing could be co-translational as exported proteins are translocated across the parasite ER membrane.
Fig. 4.
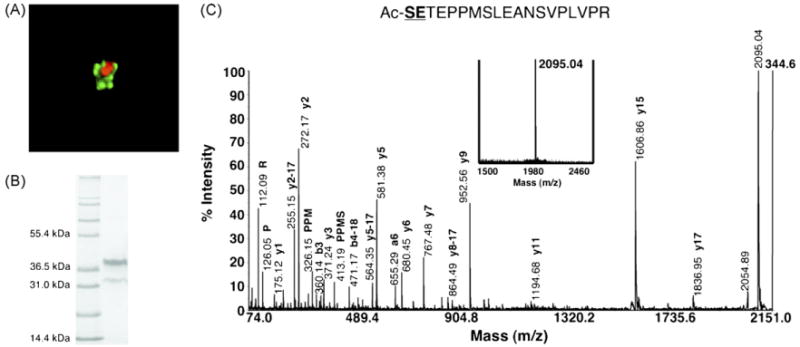
PEXEL processing occurs in the parasite ER. (A) Transfected parasites expressing PfEMP2-GFP were treated with 5 μg/mL BFA for 24 h. Live imaging of the BFA-treated parasites indicates that the trafficking of PfEMP2-GFP (green) is BFA-sensitive. Since the parasite nucleus is stained with DRAQ5 (orange), chimeric protein is exclusively localized to a perinuclear compartment indicative of the parasite ER. (B) PfEMP2-GFP trapped in the ER from BFA treatment was enriched from the parasite pellet after saponin lysis. The predominant upper band at ∼35 kDa is the full-length chimera while the lower band at ∼30 kDa is a GFP degradation product seen previously in the anti-GFP Western blot in Fig. 2B. (C) Thrombin cleavage of the ER-trapped PfEMP2-GFP produced a single peptide at m/z 2095.04 (bold inset spectrum). This peptide was confirmed by CID to correspond to the fragment generated after PEXEL processing and thrombin cleavage. The cleavage and N-acetylation of the PEXEL prior to anterograde transport between the ER and Golgi reveals that PEXEL processing occurs in the parasite ER.
3.4. Transport across the PVM mediated by the recognition of the processed N-terminus
In a previous study, it was observed that some GFP chimeras with a full PEXEL sequence are trapped in the PV and are not trafficked beyond the PVM [31]. Further analyses indicate that there is a spatial requirement of at least 10 amino acids between the PEXEL motif and GFP N-terminus that allows for export beyond the PVM. This observation is further supported by two more examples subsequent to this study as well as the two examples of this phenomenon published prior to the discovery of the PEXEL [12–15]. Therefore, we characterized the PEXEL processing of a PV-trapped GFP chimera to determine if blockage of transport across the PVM is due to a defect in either PEXEL cleavage and/or N-acetylation or a defect in a recognition event in the PV.
A transgenic P. falciparum parasite line expressing a GFP chimera, KAHRP(1-60)-GFP, a construct that contains the first 60 amino acids of KAHRP fused to the N-terminus of GFP (Fig. 5A) was shown to be trapped in the PV lumen despite having a full PEXEL sequence of 54RTLAQ58 [12]. In contrast, a second GFP construct containing the first 123 amino acids of KAHRP at the N-terminus was properly exported to the host RBC illustrating the spatial requirement between the KAHRP PEXEL and the GFP N-terminus. KAHRP(1-60)-GFP accumulating in the PV was enriched from trophozoite-infected RBC saponin supernatant. The ∼30 kDa band was confirmed as the target protein by an anti-GFP Western blot (Fig. 5B) and the gel band was excised for an Asp-N in-gel digestion. MALDI-TOF MS analysis of the digest mixture confirmed the identity of the KAHRP(-His)-GFP chimera with 76% sequence coverage (Fig. 5B, Table S4). De novo sequencing of the peptide fragment at m/z 2580.43 by CID revealed the N-terminal peptide sequence of Ac-AQKQPRSKGEELFTGVVPILVEL (Fig. 5C), therefore, demonstrating that the PEXEL in KAHRP(1-60)-GFP is cleaved (54RTL↓) and N-acetylated (Ac-AQ58).
Fig. 5.
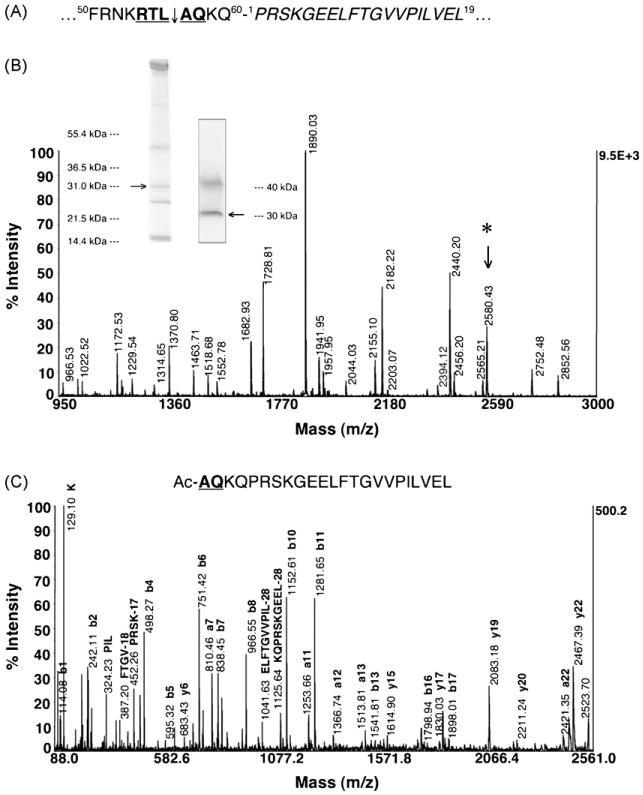
Transport across the PVM may be mediated by the recognition of the processed PEXEL. (A) KAHRP(1-60)-GFP traffics to the PV lumen despite having a full PEXEL motif (bold and underlined) because of its close proximity to the N-terminus of GFP (italicized). (B) KAHRP(1-60)-GFP trapped in the PV of trophozoite parasites was enriched from saponin supernatant and subjected to an Asp-N in-gel digestion. The inset Coomassie-stained gel (left) and anti-GFP Western blot (right) confirms the enriched protein (arrows) at ∼31 kDa while the upper band (∼40 kDa) is a cross-reactive protein. The peptide fragment at m/z 2580.43 (*) was selected for CID analysis. (C) De novo sequencing of m/z 2580.43 by CID establishes that the PEXEL of KAHRP(1-60)-GFP is cleaved (54RTL↓) and N-acetylated (Ac-AQ58), thus indicating that PEXEL processing in the ER is not affected by the close proximity of the GFP reporter to the PEXEL, but there may be a failure in the recognition of the processed PEXEL (Ac-xE/Q/D) by a putative translocase at the PVM.
The fully processed KAHRP PEXEL represents a third example of this N-terminal processing, providing further evidence for the conservation of PEXEL cleavage and N-acetylation in soluble proteins exported by P. falciparum to the host RBC. The cleaved and N-acetylated PEXEL in KAHRP(1-60)-GFP demonstrates that recruitment into the ER and the downstream N-terminal processing are not affected by the PEXEL motif distance to the N-terminus of GFP. This finding indicates that the recognition of the processed N-terminus, lacking the first three residues of the PEXEL (RxL), may be occurring at the PVM by a putative translocase and the inability of the parasite to traffic KAHRP(1-60)-GFP beyond the PVM could be due to a defect in this proposed recognition event. The crystal structure of GFP shows that the first nine amino acids are involved in the formation of an α-helix that plays an integral part in capping one end of the protein and may aid in folding events or in protecting the fluorophore [32,33]. It is possible that the close proximity of the processed PEXEL to this N-terminal α-helical cap of GFP obstructs the recognition of the processed N-terminus of the KAHRP chimera by a putative PVM translocase. This structural requirement is illustrated by the insertion of an unstructured polyalanine spacer between the PEXEL and the GFP N-terminus of PV-trapped chimeras resulting in the restoration of export to the RCC [14,31].
3.5. Dissection of the PEXEL motif
The discovery of the PEXEL has allowed for the prediction of proteins that might be exported by the parasite to the host RBC. However, the mechanism of how the PEXEL functions within the parasite secretory pathway has not been described [8,9]. The characterization of the conserved cleavage and N-acetylation of the PEXEL in the parasite ER reported here leads to a model for the signals that may be encoded in this unique Plasmodium motif and leads further towards speculation on the machinery responsible for PEXEL processing (Fig. 6). Starting at the gateway into the parasite's export pathway at the ER membrane, the PEXEL motif can be ruled out as an ER translocation signal because all GFP chimeras containing a mutation in the first, third, or fifth conserved amino acid position in the PEXEL (R, L, or E/Q/D, respectively) are translocated into the parasite ER [8,9,14]. Since the first three PEXEL residues (RxL) are proteolytically removed in the parasite ER, this highly conserved motif possibly represents a novel ER peptidase cleavage site. Mutation of the conserved R or L residue prevents the export of GFP chimeras [8,9,14], thus indicating that PEXEL cleavage by this unknown ER peptidase may be required for trafficking to the host RBC. Since the only known ER peptidase in the classical secretory pathway involved in N-terminal processing is the SPC, the parasite SPC could possibly have an additional role in signal peptide cleavage and also be responsible for PEXEL cleavage. If the parasite SPC is indeed responsible for PEXEL cleavage, then the substrate specificity of this novel ER peptidase would have to be able to accommodate both PEXEL cleavage (RxL↓) along with classical signal peptide cleavage (AxA↓) [34,35]. Analysis of the P. falciparum genome reveals homologues of the SPC21 catalytic subunit (PlasmoDB ID: MAL13P1.167, GenBank accession no. NP_705217) and the SPC22/23 subunit (PlasmoDB ID: PFI0215c, GenBank accession no. CAD51729) of the classical SPC. Biochemical studies of these predicted SPC subunits to test this hypothesis are currently underway.
Fig. 6.
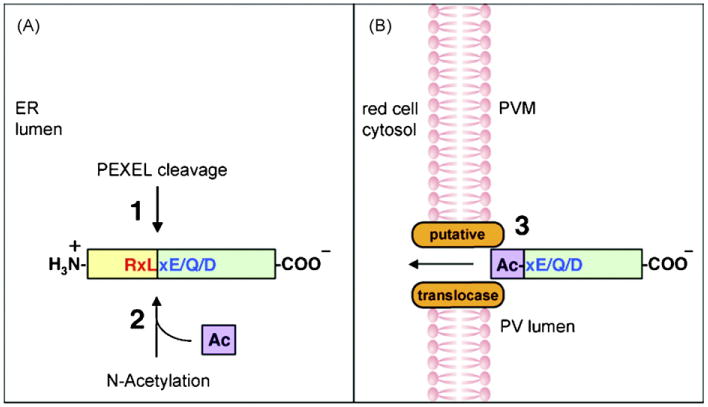
Model illustrating PEXEL processing and the definition of the PEXEL. (Panel A) Soluble proteins entering the P. falciparum secretory pathway undergo PEXEL cleavage (1) and N-acetylation (2) in the parasite ER lumen. (Panel B) The cleaved, N-acetylated protein is recognized by the putative translocase at the PVM (3) for export to the host RBC. The PEXEL may be defined as a novel ER peptidase cleavage site (RxL) (red) and a classical N-acetyltransferase substrate sequence (xE/Q/D) (blue).
Cleavage of the PEXEL after the third residue in the motif (RxL↓) leaves the last two amino acids of xE/Q/D at the N-terminus, where most often x = S, A, or T in the previously published sequence logos [8,9]. These last two residues may be a classical N-acetyltransferase (Nat) substrate sequence because they resemble a motif from a subclass of NatA substrates in higher eukaryotes, which is designated as NatD substrates in yeast [36]. After methionine cleavage, the most common N-terminal residue for N-acetylation by NatA is S, A, or T. The addition of the amino acid E or D penultimate to that N-terminal residue then classifies them as NatD substrates. The P. falciparum genome does encode for the homologues of NatD proteins Ard1p (PlasmoDB ID: PF10_0036, GenBank accession no. NP_011877) and Nat3p (PlasmoDB ID: PFA0465c, GenBank accession no. NP_015456), indicating that this pathway may be active in the parasite. N-acetylation of the PEXEL by these putative NatD components could be important for trafficking beyond the PVM as mutating the last amino acid of the PEXEL prevents the export of GFP chimeras [8,9,14]. With the requirement of the penultimate acidic residue in NatD substrates, mutation of the E/Q/D in the PEXEL may cause a defect in N-acetylation and thus have a downstream effect on the recognition of the processed N-terminus by a putative PVM translocase. It is possible that N-acetylation of the positively charged N-terminus of P. falciparum exported proteins may allow for the putative translocase to discriminate between proteins destined for export beyond the PVM and those that reside in the PV.
The PEXEL processing in soluble proteins exported by P. falciparum reported here leads to the proposal that the PEXEL contains a novel ER peptidase cleavage site and classical N-acetyltransferase substrate sequence. We further hypothesize that the new N-terminus resulting from PEXEL processing may facilitate protein export from the PV to the host RBC. The PEXEL processing of membrane proteins exported by P. falciparum has yet to be determined and, therefore, needs to be addressed because of the possibility that the membrane protein trafficking pathway in P. falciparum may be distinct from the trafficking pathway utilized by soluble proteins. From our data, it is also not possible to say whether the atypical PEXEL sequence in PfEMP1 is N-terminally processed and, thus, forms the basis of further studies [8,9]. Another step in further elucidating this novel pathway is to characterize how the parasite uses PEXEL cleavage and/or N-acetylation to properly sort exported proteins along the P. falciparum secretory pathway. This can be accomplished by determining how PEXEL processing is affected by mutation of the conserved residues in the PEXEL. These PEXEL mutants may allow for the definition of the requirements of these post-translational modifications for proper compartmentalization. The transfection of PEXEL mutants of PfEMP2-GFP was attempted but was unsuccessful possibly because the strong parasite cam promoter may not be amenable to the expression of these constructs due to toxicity (data not shown). Transfection of these constructs expressed under a weaker promoter and the characterization of other PEXEL mutant parasite lines are currently being pursued.
Supplementary Material
Acknowledgments
We thank Dr. Sharleen Zhou and Dr. David S. King for Edman sequencing and mass spectrometry work. We also thank Dr. Thomas Akompong and Dr. Kasturi Haldar for providing the transfected parasite line expressing PfHRPIImyc. H.H.C was the recipient of a National Science Foundation Graduate Fellowship.
Abbreviations
- BFA
brefeldin A
- CID
collision-induced dissociation
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- KAHRP
knob-associated histidine-rich protein
- MALDI-TOF
matrix-assisted laser/desorption/ionization time-of-flight
- MS
mass spectrometry
- Nat
N-acetyltransferase
- PEXEL
Plasmodium export element
- PVM
parasitophorous vacuolar membrane
- PV
parasitophorous vacuole
- PfEMP1
P. falciparum erythrocyte membrane protein 1
- PfEMP2
P. falciparum erythrocyte membrane protein 2
- PfHRPII
P. falciparum histidine-rich protein II
- RBC
red blood cell
- RCC
red cell cytosol
- RCM
red cell membrane
- SPC
signal peptidase complex
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molbiopara.2008.04.011.
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–9. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Kirk K. Membrane transport in the malaria-infected erythrocyte. Physiol Rev. 2001;81:495–537. doi: 10.1152/physrev.2001.81.2.495. [DOI] [PubMed] [Google Scholar]
- 3.Kilejian A. Characterization of a protein correlated with the production of knob-like protrusions on membranes of erythrocytes infected with Plasmodium falciparum. Proc Natl Acad Sci USA. 1979;76:4650–3. doi: 10.1073/pnas.76.9.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pologe LG, Ravetch JV. A chromosomal rearrangement in a P. falciparum histidine-rich protein gene is associated with the knobless phenotype. Nature. 1986;322:474–7. doi: 10.1038/322474a0. [DOI] [PubMed] [Google Scholar]
- 5.Baruch DI, Pasloske BL, Singh HB, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 6.Waller KL, Cooke BM, Nunomura W, Mohandas N, Coppel RL. Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) J Biol Chem. 1999;274:23808–13. doi: 10.1074/jbc.274.34.23808. [DOI] [PubMed] [Google Scholar]
- 7.Craig A, Scherf A. Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol Biochem Parasitol. 2001;115:129–43. doi: 10.1016/s0166-6851(01)00275-4. [DOI] [PubMed] [Google Scholar]
- 8.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–3. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 9.Hiller NL, Bhattacharjee S, van Ooij C, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–7. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 10.Sargeant TJ, Marti M, Caler E, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickham ME, Rug M, Ralph SA, et al. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–49. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Estrano C, Bhattacharjee S, Harrison T, Haldar K. Cooperative domains define a unique host cell-targeting signal in Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci USA. 2003;100:12402–7. doi: 10.1073/pnas.2133080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przyborski JM, Miller SK, Pfahler JM, et al. Trafficking of STEVOR to the Maurer's clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 2005;24:2306–17. doi: 10.1038/sj.emboj.7600720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knuepfer E, Rug M, Klonis N, Tilley L, Cowman AF. Trafficking determinants for PfEMP3 export and assembly under the Plasmodium falciparum-infected red blood cell membrane. Mol Microbiol. 2005;58:1039–53. doi: 10.1111/j.1365-2958.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- 16.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–51. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akompong T, Kadekoppala M, Harrison T, et al. Trans expression of a Plasmodium falciparum histidine-rich protein II (HRPII) reveals sorting of soluble proteins in the periphery of the host erythrocyte and disrupts transport to the malarial food vacuole. J Biol Chem. 2002;277:28923–33. doi: 10.1074/jbc.M201968200. [DOI] [PubMed] [Google Scholar]
- 18.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 19.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–20. [PubMed] [Google Scholar]
- 20.Pryor KD, Leiting B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expres Purif. 1997;10:309–19. doi: 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 21.Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:131–44. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- 22.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–3. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–77. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Gormley JA, Howard RJ, Taraschi TF. Trafficking of malarial proteins to the host cell cytoplasm and erythrocyte surface membrane involves multiple pathways. J Cell Biol. 1992;119:1481–95. doi: 10.1083/jcb.119.6.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haeggstrom M, Kironde F, Berzins K, Chen Q, Wahlgren M, Fernandez V. Common trafficking pathway for variant antigens destined for the surface of the Plasmodium falciparum-infected erythrocyte. Mol Biochem Parasitol. 2004;133:1–14. doi: 10.1016/j.molbiopara.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Howard RJ, Lyon JA, Uni S, et al. Transport of an Mr approximately 300,000 Plasmodium falciparum protein (PfEMP2) from the intraerythrocytic asexual parasite to the cytoplasmic face of the host cell membrane. J Cell Biol. 1987;104:1269–80. doi: 10.1083/jcb.104.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumgartner F, Wiek S, Paprotka K, Zauner S, Lingelbach K. A point mutation in an unusual Sec7 domain is linked to brefeldin A resistance in a Plasmodium falciparum line generated by drug selection. Mol Microbiol. 2001;41:1151–8. doi: 10.1046/j.1365-2958.2001.02572.x. [DOI] [PubMed] [Google Scholar]
- 29.Wollenberg MS, Simon SM. Signal sequence cleavage of peptidyl-tRNA prior to release from the ribosome and translocon. J Biol Chem. 2004;279:24919–22. doi: 10.1074/jbc.C400018200. [DOI] [PubMed] [Google Scholar]
- 30.Driessen HP, de Jong WW, Tesser GI, Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- 31.Knuepfer E, Rug M, Cowman AF. Function of the Plasmodium export element can be blocked by green fluorescent protein. Mol Biochem Parasitol. 2005;142:258–62. doi: 10.1016/j.molbiopara.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–51. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 33.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 34.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 35.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–38. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polevoda B, Sherman F. Nalpha-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–82. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


