Abstract
Upregulation of structurally homologous oncoproteins Hdm2 and Hdmx has been linked to the depletion or inactivation of their common regulation target the tumor suppressor p53 protein leading to the progression of cancer. The restoration of the p53 function, rendered suppressed or dormant by these negative regulators, establishes, therefore, a unique opportunity for a targeted induction of apoptosis in cancers that retain wild-type p53. While several small molecules have been reported to rescue the tumor suppressor by antagonizing the Hdm2–p53 interaction, these agents displayed limited application scope by being ineffective in tumors enriched with active Hdmx. Here, we describe the use of a genetic selection system and encoded library of conformationally preorganized peptides to perform functional profiling of each regulator revealing specific recognition features that guide the antagonism of Hdm2–p53 and Hdmx–p53 interactions. Structure-activity relationship analysis of the most effective leads identified functional and structural elements mediating selective recognition of the two structurally related regulators, while providing convenient starting points for further activity optimization.
Keywords: Hdm2, Hdmx, Protein-protein interactions, Genetic selection, Cyclic peptides
1. Introduction
The tumor suppressor p53 protein plays a vital role in maintaining genome stability by functioning as a transcriptional activator for genes mediating several corrective or antiproliferative biological outcomes, such as cell cycle arrest, senescence, DNA repair, and apoptosis, among others.1 Loss of p53 function through genetic or regulatory causes has, therefore, been linked to malignant transformations of cells.2 While deletions and mutations of the p53 gene account for approximately half of all human cancers,1 upregulations of the Hdm2 (also known as MDM2) and Hdmx (also known as MDMX or MDM4) proteins have emerged as significant additional mechanisms for the impairment of the tumor suppressor, leading to a variety of malignancies.3,4
Hdm2, one of the transcriptional targets of the tumor suppressor, and constitutively expressed Hdmx5 combine to form a regulatory circuit that exerts a sensitive control over the cellular function of p53. Both regulators can interfere with the intranuclear activities of p53 by occluding its transcriptional activation domain (TAD) essential for mounting stress-induced responses. Structural analysis and directed mutagenesis have indicated that the helical TAD of p53 relies on three co-directionally projected hydrophobic residues—Phe19, Trp23, and Leu26—for the recognition of structurally complimentary subsites within the binding pockets of both Hdm2 and Hdmx.6,7 Despite this similarity in the mode of binding to p53, the current mechanistic hypothesis assigns non-redundant roles to the Hdm proteins in the regulation of p53,8 since, at low Hdm2 levels, Hdmx merely sequesters the tumor suppressor into a transcriptionally inactive complex that serves as an immediately available pool of p53 for initiating a rapid stress-response cascade.4 In contrast, Hdm2 negatively regulates the stability of the tumor suppressor by acting as a p53-specific E3 ubiquitin-ligase that stimulates nuclear export and proteosome-dependent degradation of p53. When concentration of Hdm2 increases, Hdmx can reverse its p53-preserving function by augmenting the Hdm2-mediated degradation of p53 through the formation of a heterodimeric Hdmx–Hdm2 complex capable of enhanced trans-ubiquitination.9,10
Deregulation of the p53 pathway in malignancies expressing wild-type (wt) p53 has been linked to several Hdm2- and Hdmx-dependent events resulting from overexpression, amplification, reduced clearance, or changes in activation status of either regulator.11–13 Tumor-specific induction of apoptosis through the rescue of wt p53 from complexes with the Hdm proteins has, therefore, been proposed as an attractive strategy for tumor-specific chemotherapy,4 and is currently being actively explored through rational design and high-throughput discovery of Hdm2 and Hdmx antagonists.14–17 Here we describe our studies to characterize functional differences between Hdm2 and Hdmx and to ascertain the possibility of developing both highly discriminating recognition motifs as well as bispecific ones. We employed the combination of a vast pool of structurally constrained peptides and a high-throughput genetic selection system to retrieve compact sequences capable of disrupting the interactions between p53 and Hdm proteins. The sequences that displayed optimal activities were further analyzed through site-directed mutagenesis and focused diversification to yield key insights into elements needed for effective antagonism of these structurally related oncoproteins.
2. Results and discussion
2.1. Discovery of Hdm2 and Hdmx antagonists through genetic selection
2.1.1. Genetic system for monitoring Hdm2–p53 and Hdmx–p53 interactions
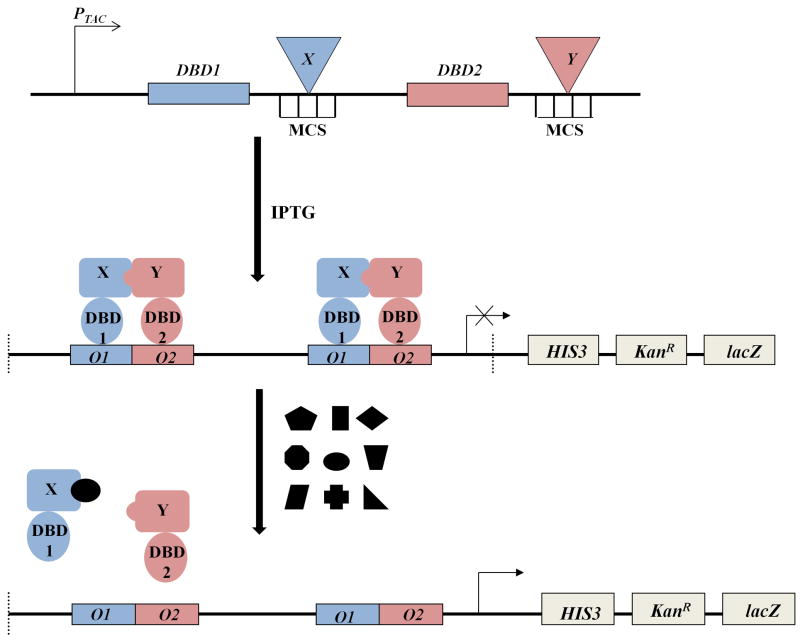
Since rational structure-based efforts have not yet provided Hdmx-selective agents, we chose to perform a functional analysis of p53-binding pockets in both Hdm2 and Hdmx using a previously disclosed Escherichia coli (E. coli)-based genetic selection system designed for rapid identification of agents perturbing protein-protein interactions.18 In this system (Figure 1), a repressor engineered from a pair of interacting proteins blocks the transcription of reporter genes in the tricistronic HIS3-KanR-lacZ operon that expresses i) imidazole glycerol phosphate dehydratase (IGPD) from Saccharomyces cerevisiae, compensating for a deleted homolog of the host, ii) aminoglycoside 3’-phosphotransferase, responsible for the resistance to kanamycin, and iii) β-galactosidase, respectively. The first two reporter genes can be used in tandem to provide a dramatically improved dynamic range of a host survival response toward conditions of histidine-starvation and antibiotic stress, resulting in a greatly reduced number of false positives.18 In addition, the HIS3 marker enables sensitive chemical tuning of selection stringency with 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of the yeast IGPD enzyme.19 The transcriptional repressor system is expressed from a separate isopropyl-β-D-thiogalactoside (IPTG)-regulated cassette as a pair of fusions with DNA-binding domains (DBD1 and DBD2 in Figure 1) possessing distinct recognition properties for cognate operator half-sites (O1 and O2).20 Upon expression, the interacting pair forms a heterodimeric complex that cooperatively recognizes the corresponding chimeric operator sites embedded within the promoter, and, thereby, blocks the expression of the reporter cassette. This combination of the orthogonal DBDs and the operator half-sites allows specific interrogation of heterodimeric interfaces in higher order complexes, and any interference with these interactions from a co-expressed modulator or a cell-permeable agent should lead to repressor disassembly and ultimately to the rescue of host’s viability from the starvation and antibiotic stress applied during selection.
Figure 1.
Schematic representation of the genetic system for selection of agents targeting protein-protein interactions. A pair of interacting proteins, designated as X and Y, are inserted downstream of genes coding for DNA-binding domains (DBD1 and DBD2) possessing orthogonal operator affinities (top). Upon induction of PTAC promoter with IPTG, the expressed fusions form a functional heterodimeric repressor that recognizes the corresponding chimeric O1–O2 operator embedded within the promoter region of the tricistronic HIS3-KanR-lacZ reporter operon (middle). Perturbation of the repressor complex with an agent selected from a library results in a relief of the promoter blockade and expression of the reporter genes (bottom).
The extended TAD region of p53 (residues 1–83) was subcloned into one of two multiple cloning sites of the previously reported pTHCP14 expression plasmid18 for in-frame fusion with the preinstalled DBD gene. Insertion of the N-terminal domain of Hdm2 (residues 1–188) and the corresponding region of Hdmx (residues 1–189) into the remaining cloning site of modified pTHCP14, provided two bicistronic expression plasmids for expressing Hdm2-DBD1 and Hdmx-DBD1 fusions as repressor counterparts of p53-DBD2.
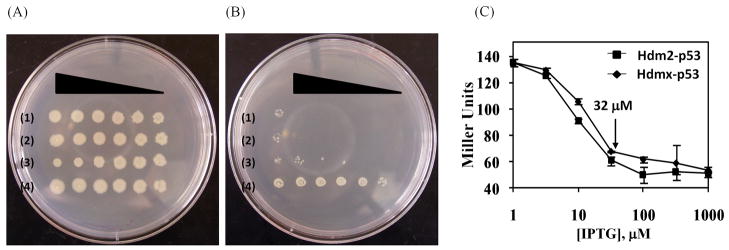
The resulting expression constructs were subsequently integrated21 into the chromosome of HIS3-KanR-lacZ reporter strain SNS12618 to stabilize expression of the repressor halves under conditions of selection stress. The integrated strains, referred henceforth as ‘Hdm2–p53’ and ‘Hdmx–p53’, were evaluated for growth trends via a droplet inoculation technique, whereby serially diluted cell suspensions are arrayed on non-selective and selective media for side-by-side comparison of resulting phenotypes at different colony densities. The Hdmx–p53 and Hdm2–p53 strains (rows 1 & 2, respectively, in Figures 2A & B) displayed the following phenotypes: i) fast growth on non-selective media indistinguishable from constitutively repressed (row 3)22 and unrepressed (row 4) control strains, and ii) significantly reduced survival on selective media compared to the unrepressed control. In addition, the gene-reporter assay, involving hydrolysis of o-nitrophenyl-β-D-galactoside (ONPG) by the third reporter β-galactosidase,23 was used to provide a quantitative measure of inducer-dependent repression exhibited by the two strains (Figure 2C). This assay was used to identify the minimal level of induction required for nearly complete (≈ 90%) repression to be applied in the subsequent genetic selections (32 μM IPTG for both targets).
Figure 2.
Intracellular analysis of repressor pairs simulating Hdm2–p53 and Hdmx–p53 interactions in the E. coli reporter strain. (A & B) Growth analysis through droplet inoculation of serially diluted strains (10–106 colony forming units (cfu)) expressing the Hdmx–p53 pair of DBD fusions (row 1), the Hmd2–p53 pair of DBD fusions (row 2), a constitutively active repressor control (row 3)[ref. 22]) and an unrepressed (DBDs only) control (row 4) on a (A) non-selective inducing medium (LB with 100 μM IPTG) and (B) selective inducing medium (LB with 100 μM IPTG and 25 μg/mL kanamycin). (C) Reporter-gene (β-galactosidase) activity analysis of Hdm2–p53 and Hdmx–p53 strains. The arrow indicates the expression level (32 μM) for achieving approximately 90% reduction in the reporter expression to be used in the selection procedure.
2.1.2. Implementation of genetic selection for discovery of Hdm2 and Hdmx antagonists
The requisite molecular diversity for high-throughput selection was provided by the SICLOPPS (Split Intein-mediated Circular Ligation of Peptides and ProteinS) genetic system producing, upon induction with L-(+)-arabinose, a pool of polypeptides programmed to form backbone cyclic structures inside the expression host.24 Specifically, a hexamer ‘C+5’ library encoding an invariable nucleophilic cysteine followed by five random amino acids (Figure S1)18 was chosen to identify functional motifs competitive with p53 for the Hdm proteins. Backbone cyclic hexapeptides, which tend to exist preferentially as turn-containing conformers,25 were expected to yield conformationally constrained and functionally compact leads facilitating structure-activity relationship (SAR) analyses for further optimization.26–28 Notably, display of the p53-binding epitope from a cyclic core has been shown to be as effective as linear helical presentations derived from native p53.16,29
The ‘C+5’ SICLOPPS library was transformed into the repressor-modified reporter strains providing approximately 108 individual library members in each system for subsequent selections. The transformants were then inoculated at the density of approximately 3 × 106 cfu onto minimal media plates supplemented with inducers, L-(+)-arabinose (13 μM) and IPTG (32 μM), and selecting agents, kanamycin (75 μg/mL) and 3-AT (10 mM). The seeded plates were incubated for 72 hours at 37 °C until surviving colonies (ca. 300 from 20 plates) could be reliably harvested from the background cell lawn for further analysis. The plasmids isolated from the selected strains were then transformed into the original selection hosts and subjected to phenotypical screens at different levels of arabinose—0, 13, and 23 μM—corresponding to the conditions of zero, selection level, and high induction of SICLOPPS, respectively. Strains that did not reproduce selective advantage, survived on L-(+)-arabinose-free media, or displayed significant toxicity upon overexpression were eliminated from further consideration.
2.2. Characterization of selected sequences
The plasmids isolated from the best performing strains were sequenced to reveal the amino acid composition of their variable regions (Table S1). All variable inserts in the SICLOPPS peptides that promoted survival of the Hdm2–p53 strain converged onto two aromatic-rich motifs—ΦWWYΦ and ΦFYYΦ—where Φ is an aliphatic amino acid (Table 1). This functional bias, as well as a double emergence of sequence CLWWYM (shown with the flanking invariable cysteine), indicated a highly discriminating nature of ligand recognition by Hdm2 and, at the same time, validated the power of genetic selection to yield repeatedly rare individuals with unique properties. The high level of aromatic content is also indicative of convergence onto existing amino acid-based Hdm2 recognition motifs. Thus, the tyrosine-tryptophan dyad has repeatedly emerged in phage display-optimized α-helical peptides30,31 and miniature proteins,32 whereas the tryptophan-free recognition has been previously seen with the Hdm2-binding HIF1α fragment homologous to the p53 TAD.33 The sequences identified against Hdmx, on the other hand, appeared to feature more functional and sequence variability than the corresponding anti-Hdm2 leads. The notable preference for aliphatic leucine by Hdmx is consistent with Leu26 in p53 playing a significantly greater role in the recognition of Hdmx than Hdm2.34 In addition, charged amino acids, particularly basic residues (Arg, Lys, and His), were present in almost every isolated candidate (Table 1).
Table 1.
Translated sequences of variable inserts, occurrence frequency, and β-galactosidase activities of SICLOPPS leads selected as antagonists of Hdm2–p53 and Hdmx–p53 interactions.
| Target interaction | SICLOPPS Isolate (# of occurrences) | Sequence in linear form | % derepression (Hdm2)a | % derepression (Hdmx)a |
|---|---|---|---|---|
| Hdm2–p53 | I-1 (2) | CLWWYM | 18 ± 2 | 41 ± 4 |
| II-1 (1) | CIWWYM | 12 ± 2 | 49 ± 3 | |
| II-7 (1) | CIFYYV | 51 ± 5 | 42 ± 3 | |
| IV-7 (1) | CMFYYI | 18 ± 3 | 37 ± 3 | |
| Hdmx–p53 | X1-25 (1) | CLFFNY | 18 ± 2 | 47 ± 2 |
| X2-1 (1) | CKAVLF | 7 ± 3 | 51 ± 3 | |
| X4-40 (1) | CHLRWL | 10 ± 3 | 58 ± 4 | |
| X4-46 (1) | CDLRWF | 6 ± 2 | 76 ± 4 | |
| X6-19 (1) | CRLLDF | 7 ± 2 | 49 ± 3 |
% derepression is determined as the percentage of β–galactosidase activity enhancement in the selected SICLOPPS strain versus that in the unrepressed control (see Table S2 for details).
The activity ranking was established through the ONPG assay, which quantitatively reported on β-galactosidase production induced by the co-expressed SICLOPPS leads (Table 1). In addition to the original or ‘native’ strain/plasmid combinations, we have also evaluated the sequenced constructs for target selectivity by testing the inhibitory potential of the anti-Hdm2 leads in the Hdmx–p53 strain and vice versa. Transcriptional activities of the selected peptides in the ‘non-native’ combinations generally did not surpass the original performances (Table 1). Although the anti-Hdm2 leads exhibited relatively high reporter derepression in the strain regulated by the Hdmx-p53 hybrids, the only anti-Hdmx peptide with a moderate level of activity in the Hdm2–p53 strain was the sole uncharged CLFFNY lead. These observations appear to highlight the presence of charged residues in the remaining anti-Hdmx peptides as the important element for target differentiation. On the basis of both activity and apparent selectivity in the gene-reporter assay, two sequences—tryptophan-free CIFYYV and doubly charged CDLRWF—emerged as the top candidates against Hdm2 and Hdmx, respectively.
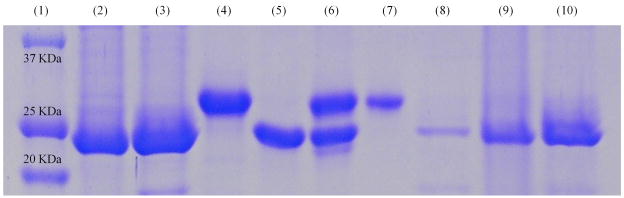
Because the indirect and artifact-prone nature of activity assessments by the gene-reporter assay, we next performed a cell-free analysis of target selectivities through an affinity capture–elution assay, whereby the corresponding SICLOPPS polypeptides fused to a chitin-binding domain (CBD) were overexpressed at a reduced temperature in an unspliced form and immobilized as intact fusions on chitin beads. The resulting affinity probes were exposed to an equimolar mixture of purified Hdm2 and Hdmx (1 mM each). Protein material retained by the immobilized SICLOPPS probes was then incubated with p53-derived decapeptide ETFSDLWKLL,34,35 and the eluates, as well as other components of the assay, were analyzed by SDS-PAGE (Figure 3). Highly specific interactions displayed by both lead sequences were evidenced by selective retention of the ‘native’ target from the mixture (lanes 7/9 and 8/10 in Figure 3). Since target selectivity was not the criterion for the assay and is, in fact, a mere artifact of the high-throughput selection, the observed level of target discrimination suggested the presence of substantial differences in the ligand-recognition mode of the two receptors. At the same time, the specific elution of the ‘native’ Hdm protein with the p53-derived peptide in both instances points to the competitive mode of inhibition exhibited by the discovered leads.
Figure 3.
SDS-PAGE analysis of the affinity capture-elution assay performed with the selected SICLOPPS hits. The unspliced constructs containing sequences CIFYYV and CDLRWF were immobilized via chitin-binding domain (CBD) fusion fragments on chitin beads and incubated with an equimolar mixture of Hdm2 or Hdmx (1 mM). Retained materials were subsequently treated with a solution (1 mM) of a p53-derived peptide (ETFSDLWKLL), and the eluate as well as other components of the assay were analyzed by SDS-PAGE. The lane assignments are as follows: Lane 1 is a protein ladder; Lanes 2 and 3 correspond to the CIFYYV and CDLRWF SICLOPPS constructs, respectively, isolated by chitin beads from crude overexpression lysates; Lanes 4 and 5 contain purified Hdm2 and Hdmx, respectively; Lane 6 corresponds to an equimolar mixture of Hdm2 and Hdmx; Lanes 7 and 8 contain protein material eluted with the p53-derived peptide from the affinity supports containing CIFYYV and CDLRWF leads, respectively, which were pretreated with the equimolar mixture of Hdm2 and Hdm; Lanes 9 & 10 were loaded with the post-elution material retained by the chitin beads pre-functionalized with the CIFYYV and CDLRWF constructs, respectively.
2.3. Systematic Mutagenesis Analysis
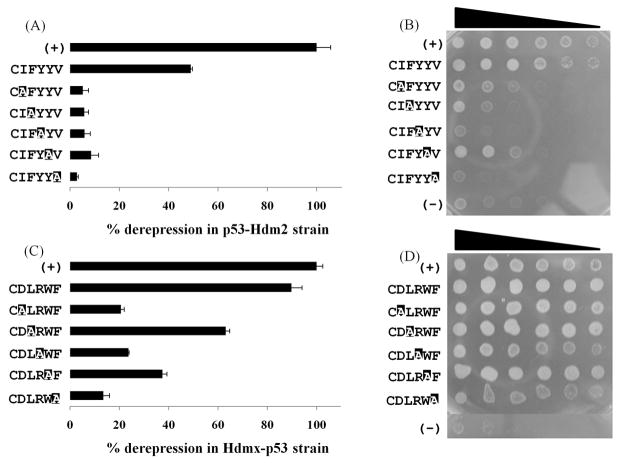
To extract the epitope maps responsible for the observed activities, we performed systematic alanine-scanning mutagenesis of the superior SICLOPPS leads, anti-Hdm2 candidate CIFYYV and anti-Hdmx candidate CDLRWF. The mutants were tested in the corresponding repressor strains by both the ONPG and droplet-inoculation assays. In all cases, the gene-reporter effects generally matched the mutant survival trends (Figure 4), which are likely to be affected also by reporter-independent factors, such as toxicity of the expressed peptides. For the putative antagonist of Hdm2, the replacement of any residue by alanine resulted in essentially inert mutants, indicating that the five side chains combine to produce an integrated epitope in which every functional element contributes to the observed activity (Figures 4A & B). The anti-Hdmx candidate exhibited a more tolerating activity profile, with the mutations of phenylalanine and the charged residues affecting the reporter activity the most (Figures 4C & D).
Figure 4.
Performance of the selected SICLOPPS constructs and the corresponding single-alanine mutants in the reporter-gene and growth rate assays. (A & B) ONPG assay data and droplet inoculation analysis, respectively, of the anti-Hdm2 CIFYYV construct and its mutants in the Hdm2–p53 strain. (C & D) ONPG assay data and droplet inoculation analysis, respectively, of CDLRWF and its mutants in the Hdmx–p53 strain.
2.4 Rationalization of target selectivity
As expected from the similarity in the protein-protein interaction modes, the p53-binding domains of both Hdm proteins display high extent of sequence and structure homology,36 with ten out of thirteen contact residues being identical.8 In fact, various p53-derived peptides have been reported to bind to both Hdm proteins with comparable affinities,31,34 suggesting that development of bispecific inhibitors may be feasible.6 However, the non-redundant functional roles of Hdm2 and Hdmx,8 their common mutual exclusivity in cancers,13 and a danger of stressing normal cells suggest that developing separate, highly selective agents may provide certain advantages over bispecific chemotherapeutics. Moreover, the use of highly discriminating agents could become particularly attractive in anti-cancer strategies individualized for a specific Hdm2/Hdmx composition of a tumor.4 Although an agent that moderately favors Hdmx over Hdm2 as the target for binding has recently been disclosed,17 the functional elements responsible for the selectivity and specific recipes for further optimization remain to be elucidated.
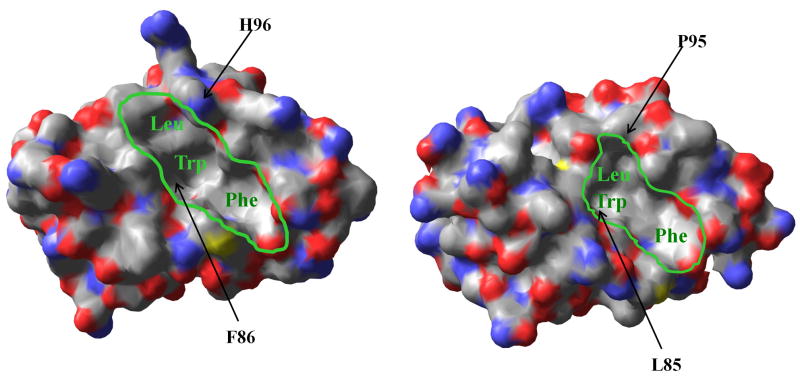
Recent crystallographic studies revealed that subtle sequence variations in the p53-binding regions of Hdm2 and Hdmx combine to create several notable topological distinctions (Figure 5). A larger and deeper binding site in Hdm2,37 has been effectively exploited by protruding haloaryl functionalities present in many Hdm2-selective agents.15,16 The more shallow, crescent-like topology of the binding cleft in Hdmx, which resists the antagonism by the existing small-molecule inhibitors of Hdm2,38–41 suggests that a somewhat distinct functional motif may be necessary for its modulation.
Figure 5.
Surface representations of Hdm2 (PDB: 1YCR; left) and Hdmx (PDB: 3DAB; right) in bound states with ligands (p53 residues 15–29) not shown for clarity. The proteins are colored according to the elemental make-up (C = grey, O = red, N = blue, S = yellow). The p53-binding pockets with labeled Leu, Trp and Phe subsites (green) are outlined to highlight the topological differences in the respective binding pockets. The residues proposed to be responsible for differences in ligand recognition patterns (F86 and H96 in Hdm2; L85 and P95 in Hdmx) are indicated by arrows.
Alignment-assisted structural analysis of the Hdm proteins indicates that the most prominent differences in the p53-binding pockets involve two sites, namely, aromatic Phe86 at the base of the Trp subsite and solvent-exposed His96 in Hdm2, which are replaced by aliphatic Leu85 and Pro95 respectively in Hdmx (Figure 5). While the former difference is responsible for a shallower Trp subsite of Hdmx,6 the latter alteration may account for the electrostatic effect, whereby the His96 residue (likely to be present in a charged form) creates the observed forbidding environment for the recognition of positively charged SICLOPPS ligands by Hdm2. The more relaxed target specificity displayed by the only neutral anti-Hdmx selectant CLFFNY supports this electrostatic hypothesis. Both changes appear to contribute to the formation of a deeper and less permissive cleft in Hdm2,6,37 which appears to require more precise orientations of functional groups projected from a ligand for effective recognition. This is consistent with both the identification of more conserved anti-Hdm2 binding motifs and non-permissive mutagenesis of the optimal sequence. On the contrary, the shallow, less segregated cleft in Hdmx that is known to allow the cross-talk between the Leu and Trp subsites,6 could, for example, be responsible for i) the relaxed functional maps observed in the selected candidates, ii) the more tolerating mutagenesis, and iii) the preference for flexible leucine over more protruding and rigid aromatic residues. Collectively, these observations indicate that the structurally related oncogenic receptors possess sufficiently distinctive binding sites and, thus, development of bispecific agents with high affinities toward both regulators of p53 may not be a trivial task. In fact, our attempt to identify a more effective bispecific peptide by performing sequential selections against both oncogenic targets failed to yield a sufficiently cross-reactive agent.42 The failure to generate such a ligand suggests that structural flexibility featured, for example, by linear p53-derived peptides could be the pre-requisite for cross-reactivity by allowing topological adaptation to the binding sites in Hdm2 and Hdmx. Thus, our findings support the development of highly selective antagonists of Hdm2 and Hdmx for chemotherapeutic strategies designed to target specific compositional differences between normal and transformed cells.
2.5. Biochemical Evaluation of Synthetic Inhibitors of Hdm2 and Hdmx
To validate the selected sequences as viable antagonists of the p53 regulators, we proceeded to synthesize the lead cyclic candidates and their linear derivatives. A modified thioester strategy43,44 was employed to provide resin-immobilized tert-butyloxycarbonyl (Boc)-terminated pentapeptides, corresponding to the variable SICLOPPS inserts (see Supporting Information for synthesis details). In addition, truncates of the two sequences were collected after each coupling iteration. Silver(I)-assisted cleavage of the resulting oligomers with a solution-phase amine (ammonia or piperidine) generated linear variants of the selected leads. To produce cyclic architectures the cysteine residues required for the intein-splicing chemistry were replaced by chemically inert alanine residues, and the resulting hexamers were exposed to silver(I) trifluoracetate in anhydrous N,N’-dimethylformamide (DMF). The release of target peptides from the solid support was monitored by reverse-phase (RP) HPLC.
Following RP-HPLC purification, the synthesized peptides were subjected to a protein-protein inhibition assay employing a competitive enzyme-linked immunosorbant assay (ELISA) format. Table 2 shows inhibition data for representative derivatives of the anti-Hdm2 lead CIFYYV, and the p53-derived control decapeptide (ETFSDLWKLL). The truncated variant, tetrapeptide 2a, displayed no activity toward Hdm2–p53 complex at concentrations of up to 560 μM. The reconstitution of the original pentapeptide sequence (2b) was indeed required to reproduce the antagonism of p53–Hdm2 interaction, albeit at a relatively high concentration (560 μM). This finding matched our systematic mutagenesis results indicating that all five residues in the anti-Hdm2 candidate combine to create an integrated functional epitope. The inhibitory activity was further enhanced by constraining this epitope through backbone cyclization, yielding peptide 3 that displayed the antagonism of the Hdm2–p53 complex in a dose-dependent manner (Figure S8A), with the IC50 value of 209 ± 14 μM in our assay. The enhanced level of activity displayed by the cyclic variant validates the use of the SICLOPPS library in the search for antagonists of protein–protein interactions, and provides a convenient conformationally organized template for further optimization using tools of peptidomimicry. Although the level of activity is approximately 13-fold lower than that of the decapeptide control, one of the most potent p53 truncates,35 the cyclic inhibitor did not antagonize the Hdmx–p53 complex when tested at a wide range of concentrations (up to 1 mM), unlike the control peptide.34
Table 2.
ELISA-generated Hdm2 inhibition data for the derivatives of the CIFYYV lead and the control p53 peptide.
| Compound | Name | % Inhibition at 560 μM | IC50 (μM)a |
|---|---|---|---|
| 1 | p53 (17–26) b | 100 ± 4 | 16 ± 1 |
| 2a | Boc–FYYV–N(CH2)5 | No Inhibition | ND c |
| 2b | Boc–IFYYV–N(CH2)5 | 16 ± 1 | ND c |
| 3 | c-(AIFYYV) | 78 ± 1 | 209 ± 14 |
Calculated from weighted non-linear fits of dose-dependent responses (see Supporting Information for details).
Not determined due to low activity and poor solubility at high concentrations.
A collection of compounds derived from anti-Hdmx lead CDLRWF was also prepared for biochemical evaluation to confirm the results observed in the whole cell assay. In the process of synthesizing linear variants of the anti-Hdmx lead, we have discovered that installation of piperidinamide at the C-termini resulted in a significant activity boost for all tested peptides (compare series a and b, Table 3). As predicted by systematic mutagenesis (Figures 4C & 4D), the C-terminal truncates of the DLRWF sequence exhibited residual activities against the Hdmx–p53 complex, with even tryptophan-phenylalanine dimer 4b displaying measurable inhibition. Iterative elongation reconstituting the original sequence led to activity improvements of the piperidinamide-terminated peptides. Thus, addition of arginine, the second most important functional contributor according to the mutagenesis data, provided trimer 5b, which was found to exhibit dose-dependent activity with the IC50 value of 463 ± 45 μM (Figure S8B). Further elongation with leucine, the least important residue of the sequence according to the alanine-scanning analysis, yielded nevertheless improved inhibitor 6b with an IC50 of 192 ± 27 μM. The notable exception in this trend was pentamer 7b, in which installation of the charged aspartic acid residue resulted in a dramatic reduction of activity. A similar outcome was observed with cyclic hexapeptide 8, thus advancing the smaller trimer and tetramer variants as the most effective leads for further elaboration as antagonists of Hdmx. Gratifyingly, neither tripeptide 5b nor tetrapeptide 6b displayed any measurable inhibition of the Hdm2–p53 complex at concentrations of up to 1 mM, establishing that the truncated leads maintained the selectivity of the original SICLOPPS construct. While the observed drop in the activity of the cyclic hexapeptide is unexpected, the discovery of truncates maintaining the selective antagonism of Hdmx–p53 suggests that the selection results are valid, albeit the active species in the whole cell environment may not have been the cyclic product of post-translational ligation but perhaps the initial SICLOPPS aptamer or a splicing byproduct.45
Table 3.
ELISA-generated Hdmx inhibition data for the derivatives of the CDLRWF lead and the control p53 peptide.
| Compound | Name | % inhibition at 320 μM | IC50 (μM)a |
|---|---|---|---|
| 1 | p53 (17–26) b | 98 ± 2 | 8 ± 1 |
| 4a | Boc–WF–NH2 | No Inhibition | ND c |
| 4b | Boc–WF–N(CH2)5 | 13 ± 1 | ND c |
| 5a | Boc–RWF–NH2 | No Inhibition | ND c |
| 5b | Boc–RWF–N(CH2)5 | 42 ± 3 | 463 ± 45 |
| 6a | Boc–LRWF–NH2 | 14 ± 2 | ND c |
| 6b | Boc–LRWF–N(CH2)5 | 82 ± 4 | 192 ± 27 |
| 7a | Boc–DLRWF–NH2 | No Inhibition | ND c |
| 7b | Boc–DLRWF–N(CH2)5 | 13 ± 1 | ND c |
| 8 | c-(ADLRWF) | 12 ± 1 | ND c |
Calculated from weighted non-linear fits of dose-dependent responses (see Supporting Information for details).
KD of 1 has been reported to be 390 ± 20 nM (ref. 34).
Not determined due to low activity and poor solubility at high concentrations.
2.6. Optimization of the Hdmx-selective lead
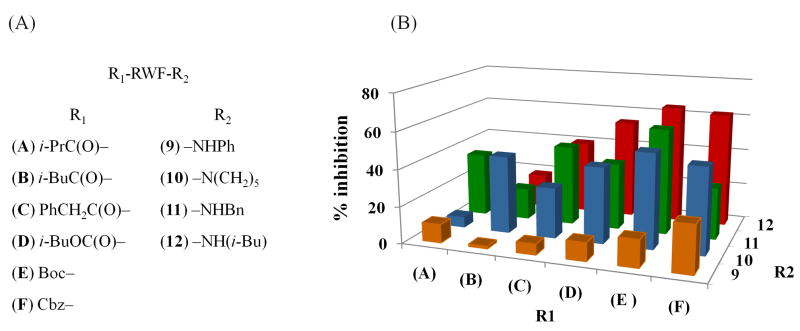
We proceeded to perform an SAR analysis of the anti-Hdmx tetrapeptide lead by unraveling the roles that the flanking aliphatic functionalities—N-terminal Boc-leucine and C-terminal piperidinamide—play in the recognition of Hdmx. To accomplish this, we designed a focused library based on the RWF tripeptide with chemical diversification directed toward both termini (Figure 6A). The tripeptide was generated as a side-chain-unprotected thioester, and a 6-member panel was produced by either leaving the Boc-functionalized N-terminus intact or capping it with activated carboxylic acids (isobutyric, valeric, phenylacetic) or chloroformates (benzyl, isobutyl). Each of the resulting derivatives was then cleaved from the support with four different amines (aniline, piperidine, benzylamine, isobutylamine) to furnish linear peptides with diverse aliphatic and aromatic functionalities at the termini. The resulting library members were then tested in parallel using the competitive ELISA (see Table S5). The accumulated inhibition profile of the library is presented in Figure 6B, with superior activities appearing to cluster within the 9-member 10–12/D–E panel featuring non-anilide carboxamides at the C-terminus and carbamates at the N-terminus.
Figure 6.
Optimization of the anti-Hdmx inhibitor using a focused library approach. (A) Chemical structures of RWF tripeptide variants with modified termini. (B) Accumulated inhibition profile of the library members at 320 μM (See Table S3 for % inhibition ± SD values). Abbreviations used: i-Pr = isopropyl, i-Bu = isobutyl, Ph = phenyl, Cbz = carbobenzyloxy, Bn = benzyl.
Four out of twenty-four compounds tested (11E, 12E, 12D, and 12F) were identified to possess activities that matched or surpassed the performance of parental tripeptide 5b (Table 4). Among the top leads, derivative 12D containing isobutyl carboxamide and isobutyl carbamate, proved to be the most effective, yielding the IC50 value of 300 ± 25 μM (Figure S8C). Apparent preference for isobutyl carboxamide at the C-terminus in 12D–12F, coupled with Boc at the N-terminus in three out of four tripeptides tested (10E–12E), is consistent with the observed common occurrence of similarly branched aliphatic leucine residues in the SICLOPPS sequences selected against Hdmx. As with the parental sequences, no activity against Hdm2–p53 complex was observed for these four peptides. The improvement of the activity level, obtained through a relatively small-scale diversification of two flanking functionalities, suggests that further optimization of the tripeptide lead may yield more effective agents against Hdmx.
Table 4.
IC50 values for tripeptides 11E, 12D–12F.a
| Compound | IC50 (μM)a |
|---|---|
| 11E | 307 ± 37 |
| 12D | 300 ± 25 |
| 12E | 339 ± 27 |
| 12F | 348 ± 36 |
Calculated from weighted non-linear fits of dose-dependent responses (see Supporting Information for details).
3. Conclusions
In conclusion, this research describes the implementation of the high-throughput conditional transcription system to profile the functional elements important for the antagonism of Hdm2–p53 and Hdmx–p53 interactions, implicated as causative events in a large proportion of human malignancies. The search for effective inhibitors was carried out by rapidly sifting through a vast library of conformationally constrained peptides to identify sequences that interfered with these interactions. The two separate selection attempts resulted in the identification of leads that yielded important recognition preferences of each oncogenic regulator. The sequence analysis, systematic mutagenesis, and affinity capture–elution assay established that Hdm2 favored highly integrated, neutral, and aromatic-rich epitopes (ΦWWYΦ and ΦFYYΦ, where Φ is an aliphatic amino acid), whereas more functionally relaxed and charged motifs were found to be selective against Hdmx. The biochemical evaluation of synthetic leads and their derivatives validated the intracellular results and allowed further elaboration of the binding epitopes through truncation analysis. Thus, gratifyingly, the combination of the genetic selection and focused synthetic diversification provided highly discriminating leads against the two regulators of p53—a conformationally constrained epitope targeting Hdm2, c-(AIFYYV), and a small tripeptide motif antagonizing Hdmx, i-BuOC(O)–RWF–NH(i-Bu)—both representing convenient starting points for further optimization. In the absence of established functional maps for targeting Hdmx with a high level of discrimination, our findings of functional preferences for cationic and branched aliphatic functional groups should be important for the development of oncoprotein-specific antagonists. Discovery of such agents will allow the implementation of anti-cancer strategies that can target specific compositional differences between normal and transformed cells without the complications associated with conventional genotoxic or antimitotic approaches. Our effort to convert the discovered leads into more effective cell-permeable probes will be disclosed in due course.
4. Experimental
4.1 Materials
The chemical and biochemical reagents were purchased from VWR International, Sigma Aldrich Chemical Company, or Acros Organics unless specified otherwise. Restriction enzymes were purchased from Promega Corporation, New England Biolabs Inc., and Fermentas Inc. DNA polymerases Taq and Pfu were from GenScript Corporation and Stratagene, respectively. Deoxynucleoside triphosphates were purchased from GenScript Corporation. Oligonucleotides were purchased from Sigma-Genosys. BL21(DE3) and TunerTM(DE3) expression strains were purchased from Novagen. Linear p53 peptide ETFSDLWKLL was synthesized by ChemImpex, Inc., in 95% HPLC purity.
4.2. Methods
4.2.1 Culture, media, and growth conditions
Antibiotics were provided at the following concentrations: 50 μg/mL ampicillin, 30 μg/mL chloramphenicol, 50 μg/mL kanamycin, and 12 μg/mL tetracycline. For chromosomal markers, concentrations of antibiotics were reduced 2-fold. Minimal media A (MMA) was used with 0.5% glycerol, 10 μg/mL thiamine, and 1 mM MgSO4. Media used for selections was generated by supplementing MMA with 75 μg/mL kanamycin and 10 mM 3-AT.
4.2.2 Recombinant DNA techniques
PCR conditions used for amplification of genes were: 30 cycles of denaturation at 94 ºC for 30 s, annealing at 59 ºC for 30 s, and extension at 72 ºC. DNA manipulations were carried out in E. coli GM2163 (New England Biolabs Inc.), DH5α-E (Invitrogen) or DH5[α]pir46 strains. Plasmid isolation, PCR purification, and gel extraction kits were purchased from Qiagen. Plasmids were transformed by heat-shock or electroporation. All DNA sequencing was performed at Purdue Genomics Core Facility.
4.2.3. Construction of selection strains
The gene coding for p53(1–83) was amplified from the pGEX4T1hp53wtN(1–83) plasmid,47 a generous gift from Dr. Thanos Halazonetis (Wistar Institute) with primers 5’-GTTGTTCATATGGGATCCGAGGAGCCGCAGTCAGATC-3’ and 5’-GTTGTTGAGCTCTCACGCCGGTGTAGGAGCTGC-3’, introducing BamHI and SacI restriction sites, respectively. The PCR product, digested with BamHI and SacI, was subcloned into pTHCP14,18 linearized with the same restriction enzymes, providing pSD1A. Hdm2(1–188) gene was amplified from the pGEM-HDM2(1–491) plasmid, also a gift from Dr. Thanos Halazonetis, using primers 5’-GTTGTTCTCGAGATGTGCAATACCAACATGTCTGTA-3’ and 5’-GTTGTTGGTACCACTATCAGATTTGTGGCGTTTTC-3’, introducing XhoI and KpnI restriction sites, respectively. Hdmx(1–189) gene was amplified from human placenta cDNA library (BioChain Institute Inc.) with primers 5’-GTTGTTCCATGGCTCGAGACATCATTTTCCACCTCTGCTCAG-3’ and 5’-GTTGTTGGATCCGGTACCTTAACCAAGGTCCAGCCGAGATGTTTCATCTTG-3’. The amplified Hdm2 and Hdmx genes were processed with XhoI and KpnI and subcloned into the pSD1A plasmid, providing pSD6 and pSD10, respectively. BsaBI/SacI fragments from the resulting plasmids were subcloned into the pAH68 CRIM plasmid,21 modified with a tetracycline-resistance marker (pAH68-tet). Integration of the resulting constructs into reporter strain SNS12618 was performed according to the standard protocol.21 The unrepressed control was produced by integration of the insert-free integration plasmid. A derivative of pAH68-tet containing a single-chain repressor22 was integrated to produce a constitutively repressed control.48
4.2.4. ONPG assay
β-Galactosidase activities of the integrated strains were measured using the standard protocol23 with the cells grown in Luria Broth (LB) at different concentrations of IPTG (1, 3.2, 10, 32, 100, 320, and 1000 μM). SICLOPPS-transformed cells were characterized using cells grown on solid LB media containing 32 μM IPTG and 9.7 μM L-(+)-arabinose.
4.2.5. Droplet inoculation analysis
Cells were grown overnight in LB media, and the resulting cell concentrations were obtained using the optical densities at 600 nm (OD600) and the following relationship: OD600 of 1 corresponds to 2 × 108 cells/mL. The concentrations were adjusted with sterile water to obtain stocks of 2 × 108 cells/mL. The resulting suspensions were serially diluted, and 2 μL droplets of each dilution were inoculated onto solid media supplemented with a particular combination of selecting (kanamycin and 3-AT), and inducing agents (IPTG and L-(+)-arabinose) in an array format providing 10–106 cfu range per droplet for each strain. The inoculated plates were incubated at 37 ºC until phenotypical differences could be photographically documented.
4.2.6. Genetic selection and post-selection screens
A SICLOPPS library of pARCBD24-derived ‘C+5’ plasmids18 (64 ng) was transformed through electroporation into Hdmx–p53 and Hdm2–p53 reporter strains. The transformants were rescued with SOC media at 37 ºC for 1 h, washed with liquid MMA, and plated on selecting minimal media supplemented with 13 μM L-(+)-arabinose and 32 μM IPTG. Upon incubation at 37 ºC for 72–96 h, the surviving colonies were harvested, the isolated plasmids were retransformed into the original selection strain, and the resulting transformants were streaked on plates containing the same media with or without L-(+)-arabinose. Colonies dependent on L-(+)-arabinose induction for survival were restreaked on over-inducing selecting media (23 μM L-(+)-arabinose). Variable insert regions within the selected SICLOPPS plasmids were sequenced using products of PCR amplification with primers 5’-TGCCTGACGGTTTTTGCCGCG-3’ and 5’-GTTGTTAAGCTTTCATTGAAGCTGCCACAAGG-3’.
4.2.7. Protein expression and purification
The full-length p53 TAD (1–83) was subcloned into the pET41a(+) vector (Novagen, Inc.) using SpeI and SacI sites, and the resulting plasmid was transformed into BL21(DE3). A single transformed colony was inoculated into LB supplemented with kanamycin, and the suspension was agitated at 37 ºC until OD600 reached 0.6. The cells were induced with 1 mM IPTG, cultivated for 4 h at 37 ºC, and then harvested by centrifugation. The pelleted cells were resuspended in 1X phosphate buffered saline (PBS),pH 7.2 containing 1 mM dithiothreitol (DTT) and 1 mM phenylmethylsulfonyl fluoride (PMSF), and lysed using the BugBusterTM reagent (Novagen, Inc.). After centrifugation for 30 min at 12,000 × g, the lysate was incubated with high-affinity glutathione resin (Genscript Corp.) for 1 h at 4 ºC. Purified p53-GST (glutathione S-transferase) fusion was eluted with 10 mM glutathione in 1X PBS (pH 7.2) containing 1 mM DTT. Genes coding for Hdm2(1–188) and Hdmx(1–189) were subcloned into pET28a(+) expression vector (Novagen, Inc.) using NheI and XhoI sites installing hexahistidine tags on both termini of each protein. The plasmids containing Hdm2 and Hdmx gene fragments were transformed into TunerTM(DE3) and BL21(DE3), respectively. Tagged Hdm2 was overexpressed with 100 μM IPTG at 16 ºC over 16 h, whereas Hdmx was overexpressed with 1 mM IPTG at 30 ºC over 5 h. Upon centrifugation, the harvested cells were lysed in an extraction buffer (1X PBS, 1 mM PMSF, 10 mM imidazole, pH 7.9), supplemented with 1X BugBusterTM, and the lysate was cleared by centrifugation at 12,000 × g for 30 min at 4 ºC. The lysates were incubated with TALON metal affinity resin (BD Biosciences) for 1 h at 4 ºC, washed with the extraction buffer, and treated with 300 mM imidazole in 1X PBS (pH 7.9). Purity of the eluants was assessed by SDS-PAGE, their concentrations were measured by Bradford’s assay (Pierce Biotechnology), and the aliquots were stored in 50% glycerol at −20 ºC.
4.2.8. Affinity capture-elution assay
SICLOPPS fusions were expressed in the corresponding selection strains in LB supplemented with 13 μM L-(+)-arabinose and chloramphenicol at 16 ºC over 16 h. The induced cells were harvested and lysed with an extraction buffer (1X PBS, 1mM PMSF, pH 7.2) containing 1X BugBusterTM. The lysate was incubated with chitin beads (New England BioLabs, Inc.) at 4 ºC for 1 h. The beads were washed with 1X PBS, blocked with 0.1% bovine serum albumin (BSA) in 1X PBS, and incubated for 1.5 h with an equimolar mixture of Hdm2 and Hdmx (1 mM each in 1X PBS, 1 mM DTT, pH 7.2), followed by three PBST (1X PBS, 0.1% Tween-20, pH 7.2) washes. Peptide ETFSDLWKLL (1 mM in 1X PBS, 1 mM DTT, pH 7.2) was incubated with the washed beads for 16 h at room temperature, and the eluants along with the post-elution beads were analyzed by SDS-PAGE.
4.2.9. Alanine-scanning mutagenesis
Alanine mutants of the SICLOPPS candidates were generated using a PCR with the NcoI-containing primer 5’-GTTGTTCCATGGTTAAAGTTATCGGTCG-3’ and the following mutagenic primers: 5’-GTTGTTCTTAAGCAGACGTAGTAGAAGGCACAATTGTGGG-3’, 5’-GTTGTTCTTAAGCAGACGTAGTAGGCGATACAATTGTGG-3’, 5’-GTTGTTCTTAAGCAGACGTAGGCGAAGATACAATTGTG-3’, 5’-GTTGTTCTTAAGCAGACGGCGTAGAAGATACAATTG-3’, 5’-GTTGTTCTTAAGCAGGCGTAGTAGAAGATAC-3’ for the CIFYYV lead, and 5’-GTT GTTCTTAAGCAGAACCACCTCAGGGCACAATTGTGGG-3’, 5’-GTTGTTCTTAAGCAGAACCACCTCGCGTCACAATTGTGG-3’, 5’-GTTGTTCTTAAGCAGAACCACGCCAGGTCACAATTGTG-3’, 5’-GTTGTTCTTAAGCAGAACGCCCTCAGGTCACAATTG-3’, 5’-GTTGTTCTTAAGCAGGCCCACCTCAGGTCAC-3’ for the CDLRWF lead (where the non-complementary nucleotides are underlined and the universal AflII site is italicized). The amplified fragments were digested and ligated back into the NcoI/AflII-linearized SICLOPPS plasmid.
4.2.10. Synthesis and characterization of peptides
See the Supporting Information.
4.2.11. Competitive ELISA
Nunc MaxiSorpTM plates were incubated with purified p53-GST protein (2.5 μg/mL) in a coating buffer (100 mM NaHCO3/Na2CO3, pH 9.6) for 16 h at 4 ºC. The plates were then washed with PBST (400 μL/well, 3×) and blocked with 0.1% BSA and 10% non-fat dry milk in 1X PBS (pH 7.2) at room temperature for 30 min. Purified Hdm proteins (1.5 μM Hdm2 or 200 nM Hdmx) were pre-incubated with different concentrations of peptides in 1X PBS with 1% milk and 10 mM DTT for 30 min, and the resulting mixtures were added to the wells. Upon incubation (30 min at room temperature) and washing with PBST (3×), detection of hexahistidine-tagged Hdm proteins was performed with nickel·nitrilotriacetic acid-horseradish peroxidase conjugate (Pierce Biotechnology) exposed to 3,3',5,5'-tetramethylbenzidine substrate (Bethyl Labs) by monitoring the absorbance change at 450 nm after quenching with 1 M HCl. To obtain IC50 values, dose–response data points were fitted using the standard single-site competitive inhibition equation (percent inhibition = 100[I]/([I] + IC50), where [I] is the concentration of the peptide inhibitor) in KaleidaGraph V.4 software (Synergy Software).
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (1R21AI077482-01) and Walther Cancer Institute FDN Inc. MEB was funded by Purdue Research Foundation grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Toledo F, Wahl GM. Nat Rev. 2006;6:909. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 2.Junttila MR, Evan GI. Nat Rev Cancer. 2009;9:821. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 3.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 4.Wade M, Wahl G. Mol Cancer Res. 2009;7:1. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. FEBS Lett. 1999;447:5. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 6.Kallen J, Goepfert A, Blechschmidt A, Izaac A, Geiser M, Tavares G, Ramage P, Furet P, Masuya K, Lisztwan J. J Biol Chem. 2009;284:8812. doi: 10.1074/jbc.M809096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 8.Toledo F, Wahl GM. Int J Biochem Cell Biol. 2007;39:1476. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badciong JC, Haas AL. J Biol Chem. 2002;277:49668. doi: 10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 10.Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. Cell Cycle. 2005;4:1166. doi: 10.4161/cc.4.9.1981. [DOI] [PubMed] [Google Scholar]
- 11.Onel K, Cordon-Cardo C. Mol Cancer Res. 2004;2:1. [PubMed] [Google Scholar]
- 12.Ramos YFM, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Cancer Res. 2001;61:1839. [PubMed] [Google Scholar]
- 13.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Marine JC. Mol Cell Biol. 2004;24:5835. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chene P. Nat Rev Cancer. 2003;3:102. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 15.Fischer PM. Int J Pept Res Ther. 2006;12:3. doi: 10.1007/s10989-006-9016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray JK, Gellman SH. Pept Sci. 2007;88:657. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 17.Reed D, Shen Y, Shelat A, Arnold A, Ferreira A, Zhu F, Mills N, Smithson D, Regni C, Bashford D, Cicero S, Schulman B, Jochemsen AG, Guy K, Dyer MA. J Biol Chem. 2010;285:10786. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horswill AR, Savinov SN, Benkovic SJ. Proc Natl Acad Sci U S A. 2004;101:15591. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joung JK, Ramm EI, Pabo CO. Proc Natl Acad Sci U S A. 2000;97:7382. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Lallo G, Castagnoli L, Ghelardini P, Paolozzi L. Microbiology. 2001;147:1651. doi: 10.1099/00221287-147-6-1651. [DOI] [PubMed] [Google Scholar]
- 21.Haldimann A, Wanner BL. J Bacteriol. 2001;183:6384. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinqiu Chen J, Pongor S, Simoncsits A. Nucleic Acids Res. 1997;25:2047. doi: 10.1093/nar/25.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; New York: 1972. pp. 352–355. [Google Scholar]
- 24.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Proc Natl Acad Sci U S A. 1999;96:13638. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler H, Gratias R, Hessler G, Gurrath M, Muller G. Pure Appl Chem. 1996;68:1201. [Google Scholar]
- 26.Okumu FW, Pauletti GM, Vander Velde DG, Siahaan TJ, Borchardt RT. Pharm Res. 1997;14:169. doi: 10.1023/a:1012092409216. [DOI] [PubMed] [Google Scholar]
- 27.Ladner RC. Trends Biotechnol. 1995;13:426. doi: 10.1016/S0167-7799(00)88997-0. [DOI] [PubMed] [Google Scholar]
- 28.Adessi C, Soto C. Curr Med Chem. 2002;9:963. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 29.Fasan R, Dias RLA, Moehle K, Zerbe O, Vrifbloed JW, Obrecht D, Robinson JA. Angew Chem Int, Ed. 2004;43:2109. doi: 10.1002/anie.200353242. [DOI] [PubMed] [Google Scholar]
- 30.Bottger V, Bottger A, Howard SF, Picksley SM, Chene P, Garcia-Echeverria C, Hochkeppel HK, Lane DP. Oncogene. 1996;13:2141. [PubMed] [Google Scholar]
- 31.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov S, Lu W. Proc Natl Acad Sci U S A. 2009;106:4665. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kritzer JA, Zutshi R, Cheah M, Ran FA, Webman R, Wongjirad TM, Schepartz A. ChemBioChem. 2006;7:29. doi: 10.1002/cbic.200500324. [DOI] [PubMed] [Google Scholar]
- 33.LaRusch GA, Jackson MW, Dunbar JD, Warren RS, Donner DB, Mayo LD. Cancer Res. 2007;67:450. doi: 10.1158/0008-5472.CAN-06-2710. [DOI] [PubMed] [Google Scholar]
- 34.Chong Li, Pazgier M, Li C, Yuan W, Liu M, Wei G, Lu W, Lu W. J Mol Biol. 2010;398:200. doi: 10.1016/j.jmb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. J Mol Biol. 2002;323:491. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 36.Böttger V, Böttger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, Lane DP. Oncogene. 1999;18:9199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 37.Popowicz G, Czarna A, Rothweiler U, Szwagierczak A, Krajewski M, Weber L, Holak T. Cell Cycle. 2007;6:2386. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 38.Wade M, Wong ET, Tang M, Vassilev LT, Wahl GM. J Biol Chem. 2006;281:33036. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 39.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. J Biol Chem. 2006;281:33030. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 40.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yand D, Wang S. Proc Natl Acad Sci U S A. 2008;105:3933. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K. J Med Chem. 2006;49:3432. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 42.Datta S. PhD Thesis. Purdue University; Aug, 2010. [Google Scholar]
- 43.Kalle K, Tam JP. Tetrahedron Lett. 1998;39:9327–9330. [Google Scholar]
- 44.Zhang L, Tam JP. J Am Chem Soc. 1999;121:3311. [Google Scholar]
- 45.Naumann TA, Savinov SN, Benkovic SJ. Biotechnol Bioeng. 2005;92:820. doi: 10.1002/bit.20644. [DOI] [PubMed] [Google Scholar]
- 46.Platt R, Drescher C, Park SK, Philips GJ. Plasmid. 2000;43:12. doi: 10.1006/plas.1999.1433. [DOI] [PubMed] [Google Scholar]
- 47.Kane SA, Fleener CA, Zhang YS, Davis LJ, Musselman AL, Huang PS. Anal Biochem. 2000;278:29. doi: 10.1006/abio.1999.4413. [DOI] [PubMed] [Google Scholar]
- 48.Villafane OC. PhD Thesis. Purdue University; Dec, 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.