Abstract
MicroRNAs regulate pathways contributing to oncogenesis, and thus the mechanisms causing dysregulation of microRNA expression in cancer are of significant interest. Mature mir-29b levels are decreased in malignant cells, and this alteration promotes the malignant phenotype, including apoptosis resistance. However, the mechanism responsible for mir-29b suppression is unknown. Here, we examined mir-29 expression from chromosome 7q32 using cholangiocarcinoma cells as a model for mir-29b downregulation. Using 5′ rapid amplification of cDNA ends, the transcriptional start site was identified for this microRNA locus. Computational analysis revealed the presence of two putative E-box (Myc-binding) sites, a Gli-binding site, and four NF–κB-binding sites in the region flanking the transcriptional start site. Promoter activity in cholangiocarcinoma cells was repressed by transfection with c-Myc, consistent with reports in other cell types. Treatment with the hedgehog inhibitor cyclopamine, which blocks smoothened signaling, increased the activity of the promoter and expression of mature mir-29b. Mutagenesis analysis and gel shift data are consistent with a direct binding of Gli to the mir-29 promoter. Finally, activation of NF–κB signaling, via ligation of Toll-like receptors, also repressed mir-29b expression and promoter function. Of note, activation of hedgehog, Toll-like receptor, and c-Myc signaling protected cholangiocytes from TRAIL-induced apoptosis. Thus, in addition to c-Myc, mir-29 expression can be suppressed by hedgehog signaling and inflammatory pathways, both commonly activated in the genesis of human malignancies.
Keywords: Cholangiocellular carcinoma, FRA7H, Inflammation, miRNA, Sonic, Toll-like receptor
INTRODUCTION
MicroRNAs are small regulatory RNAs that affect protein expression post-transcriptionally, including suppressing translation or causing mRNA degradation. Altered expression levels of microRNAs have been described in many cancers and result in aberrant expression of proteins that influence malignant behavior, such as resistance to apoptosis, proliferation, and metastasis. Mechanisms of control of microRNA expression include altered transcriptional activity by transcription factor binding to enhancer and repressor elements [Chang et al., 2007; Chang et al., 2008; O’Donnell et al., 2005; Raver-Shapira et al., 2007; Tarasov et al., 2007] and regulated processing [Davis et al., 2008] or transport [Lee et al., 2008] of microRNA precursors.
Decreased expression of mir-29 family members has been described in multiple cancers, including cholangiocarcinoma, non-small cell lung cancer, nasopharyngeal cancer, and acute myeloid leukemia [Fabbri et al., 2007; Garzon et al., 2009b; Mott et al., 2007; Sengupta et al., 2008]. Potential targets of mir-29 contributing to the malignant phenotype include the anti-apoptotic protein Mcl-1 [Mott et al., 2007], phosphatidyl inositol 3 kinase [Park et al., 2009], DNA methyl transferase 3 [Fabbri et al., 2007], extracellular matrix proteins including collagens potentially involved in metastasis [Sengupta et al., 2008], and cell cycle regulators [Garzon et al., 2009a]. Expression of mir-29 is not universally decreased in cancer, as it has been found to be increased in metastatic breast cancer samples [Gebeshuber et al., 2009], illustrating the importance of the physiological context. The mechanisms regulating expression of mir-29 in normal and cancer cells are thus of interest because restoring mir-29 expression may alleviate multiple oncogenic signals, including tumor-suppressor promoter methylation, extracellular matrix remodeling, and antiapoptotic signaling. Recently, c-Myc was shown to contribute to mir-29 repression [Chang et al., 2008], and identification of additional mechanisms of suppression is of interest.
Inflammation-related cancers often utilize activation of NF–κB signaling to sustain the malignant phenotype [Karin, 2006; Pikarsky et al., 2004]. NF–κB has been described to stimulate expression of mir-143 in the hepatitis-B virus associated hepatocellular carcinoma [Zhang et al., 2009] through binding to the upstream DNA of the mir-143 locus. Alternatively, NF–κB activation represses let-7i expression in parasite-infected cholangiocarcinoma cells [O’Hara et al., 2009] through toll-like receptor (TLR) activation in cooperation with the transcription factor C/EBP-alpha. During myogenesis, NF–κB acts through the transcription factor Yin Yang-1 to repress expression of mir-29b-2/mir-29c from chromosome 1, and decreased Yin Yang-1 during differentiation permits increased mir-29b-2/mir-29c expression. This situation is reversed in rhabdomyosarcoma where mir-29b-2/mir-29c levels are decreased while Yin Yang-1 levels are increased [Wang et al., 2008]. Thus, NF–κB is a potential modulator of mir-29 expression.
Reactivation of the developmental hedgehog signaling pathway is a feature of many cancers [Berman et al., 2003; Jiang and Hui, 2008]. Hedgehog contributes to cancer cell survival through the activity of hedgehog-responsive Gli transcription factors that activate or repress target gene expression [Kasper et al., 2006]. Gli proteins are in turn inhibited by the cytoplasmic protein suppressor of fused. Recently mir-17-92 expression was described to be overexpressed in a subset of medulloblastomas that have activated hedgehog signaling [Northcott et al., 2009; Uziel et al., 2009]. During somitogenesis, where hedgehog signaling drives specification of muscle cell fates, mir-214 repressed suppressor of fused, increasing both the activator and repressor effects of Gli signaling, dependent upon hedgehog levels [Flynt et al., 2007]. Hedgehog signaling in malignant cells is a potential candidate for regulating microRNA expression.
Here, using cholangiocarcinoma cells as a model for mir-29 regulation, we report the cloning of a functional mir-29b-1/mir-29a promoter region. This promoter sequence is regulated by c-Myc, NF–κB, and hedgehog signaling pathways. These data provide mechanistic insight into the pathways contributing to mir-29 downregulation in human malignancies. Inhibition of these pathways with restoration of mir-29 expression is a potential therapeutic approach for cancers such as cholangiocarcinoma.
MATERIALS AND METHODS
Cell lines
H69 non-malignant cholangiocytes were cultured as described [Grubman et al., 1994]. KMCH and HuCCT-1 malignant cholangiocarcinoma cells were cultured in DMEM supplemented with 10,000 U/L penicillin, 10 mg/L streptomycin, 50 mg/L geneticin, and 10% (v/v) heat-inactivated fetal calf serum at 37°C in 5% CO2.
RNA isolation
Total RNA, including small RNA species, was isolated using the mirVana isolation kit (Ambion, Austin, TX). Cells were washed with PBS and lysed directly in the culture dish by addition of Lysis/Homogenization Buffer, followed by scraping. The resulting lysate was processed as instructed by the manufacturer.
RT-PCR
Expression of primary and precursor RNA for mir-29a, -29b-1, -29b-2, and -29c was assessed by RT-PCR using the following primers: precursor mir-29a, forward 5′ CTGATTTCTTTTGGTGTTCAG, reverse 5′ AACCGATTTCAGATGGTGC; precursor mir-29b-1, forward 5′ CATATGGTGGTTTAGATTT, reverse 5′ AACACTGATTTCAAATGGTG; precursor mir-29b-2, forward 5′ GCTGGTTTCACATGGTGGC, reverse 5′ AACACTGATTTCAAATGGTG; precursor mir-29c, forward 5′ CGATTTCTCCTGGTGTTCA, reverse 5′ ACCGATTTCAAATGGTGC; primary mir-29a, forward 5′ CAGAGACTTGAGCATCTGTG, reverse 5′ AACCGATTTCAGATGGTGC. Products were separated by 1% (w/v) agarose gel electrophoresis and visualized by ethidium bromide fluorescence. Bands were excised and processed for sequencing using the QiaQuick procedure (Qiagen, Valencia, CA). Mature mir-29b and the housekeeping RNA Z30 were quantified using realtime PCR with hydrolysis probe technology (Applied Biosystems, Foster City, CA).
5′ Rapid Amplification of cDNA ends
A 5′RACE kit (Invitrogen, Carlsbad, CA) was employed. Briefly, for 5′RACE total RNA isolated from H69 cells was reverse transcribed using a gene-specific primer (GSP 1 or GSP 3), and subsequently tailed with terminal deoxynucleotidyl transferase and dCTP. cDNA was then PCR amplified using the 5′ RACE Abridged Anchor Primer and a second, upstream gene-specific primer (GSP 2 or GSP 4). 5′ RACE employing gene-specific primers annealing between mir-29b-1 and mir-29a (GSP 1: 5′ GACCTGACTGCCATTTGTGAT; GSP 2: 5′ GACCTGACTGCCATTTGTGAT) demonstrated the expected cleavage product from Drosha processing (not shown). Thus, a pair of gene-specific primers located 3 nucleotides (GSP 3) and 32 nucleotides (GSP 4) upstream of the inferred 5′ Drosha cleavage site for mir-29b-1 was designed: GSP 3: 5′ CCTGAAGAAGCTTTATGATC, and GSP 4: 5′ AGAATTCGCTAGCTTCATAATGCTCTCTTACA. GSP 4 contained EcoRI and NheI restriction sites, underlined.
Enhancer element prediction
Sequences predicted to serve as transcription factor binding sites were defined by Enhancer Element Locator analysis (http://www.cs.helsinki.fi/u/kpalin/EEL/) using the human and mouse sequences as input.
Promoter constructs
The bacterial artificial chromosome RP11-36B6 (Invitrogen) was confirmed to contain the mir-29b-1/mir-29a sequence from chromosome 7 by PCR (not shown). The region flanking the transcription start site (determined by 5′RACE) was amplified by long-PCR using the PfuI polymerase and primers containing restriction sites (MluI or BglII) followed by sequence complementary to position −1530 (5′ TACCACGCGTCCTTTCAGAATTTCACATCC) and +165 (5′ GATCAGATCTGTAGTTAGCGACCTCTGCT) of the putative promoter. This amplicon was cloned into pGL3 in the MluI and BglII sites within the multiple cloning site adjacent to the firefly luciferase gene. The correct sequence was confirmed by automated sequencing. Site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA) was employed to target the putative Gli binding site. Transfections were performed using FuGene HD (Roche, Indianapolis, IN) and renilla luciferase from pCMV-RL was included to normalize expression of firefly luciferase, and data are expressed as luminescence from firefly luciferase divided by luminescence from renilla luciferase, detected using a Turner Designs TD-20/20 luminometer and the dual luciferase assay (Promega, Madison, WI).
Myc Expression
Enforced expression of c-Myc was performed by transfection of a c-Myc expression vector derived from pCDNA3 (Addgene plasmid 16011)[Ricci et al., 2004]. Control cells were transfected with pCDNA3.1.
NF–κB activation
The TLR ligands, including TLR-4 ligand lipopolysaccharide (LPS) and TLR-5 ligand Flagellin, were purchased from Axxora (San Diego, CA) as part of a TLR ligand set. LPS was used at 1 μg/mL and Flagellin at 100 ng/mL. Cells were treated for 30 minutes for immunofluorescence or 24 hours for RNA isolation. Immunofluorescence for p65 and p50 was performed on cells grown on collagen-coated coverslips and fixed in 3% (v/v) paraformaldehyde, 10 mM HEPES, 3 mM magnesium chloride, and 1 mM EGTA in PBS. Fixed cells were permeabilized in 0.0125% CHAPS in PBS, washed in PBS, blocked in PBS with 5% glycerol (w/v), 5% goat serum (v/v), 0.01% sodium azide (w/v) and incubated overnight at 4°C in 1:1000 rabbit anti-p65 primary antibody (C-20, Santa Cruz Biotechnology, Santa Cruz, CA) or 1:50 mouse anti-p50 primary antibody (E-10, Santa Cruz Biotechnology) in blocking buffer. AlexaFluor-488 conjugated secondary antibodies (Goat anti-rabbit or goat anti-mouse, respectively; Molecular Probes, Eugene OR) were used for visualization using a Zeiss 480 epi-fluorescent confocal microscope.
Electrophoretic Mobility Shift Assay
Double stranded 30-mer oligonucleotides labeled with cy5.5 at the 5′ terminus of one strand were synthesized (Mayo Clinic Advanced Genomics Technology Core, Rochester, MN), incubated with nuclear extract (2.5 μg protein) for 30 min at room temperature. Gel shift assays utilizing the Gli binding site probe (5′ TGCTACCGCAGCCCGCCCAGACGGATCTGC) were done in 140 mM KCl, 5 mM NaCl, 1 mM K2HPO4, 2 mM MgSO4, 20 mM HEPES (pH 7.05), 0.1 mM EGTA, 1 μM ZnSO4 supplemented with poly-dIdC (1:40), 1 μg salmon sperm DNA, 0.5% (w/v) non-fat dry milk, 1% (w/v) triton-X100. For NF–κB, the binding reaction was performed in 75 mM KCl, 2 mM HEPES (pH 7.5), 2.5 mM DTT, 5% glycerol (w/v), poly-dIdC (1:40), and 1 mg/mL bovine serum albumin. NF–κB probes were, −561: 5′ TCCTCCCCGAGTGGCTTTCCTCCCCACAAT; −110: 5′ ATGAACGTTGTGAAATCCCTCCTTTATAAT; +134: GCCAGGAGCTGGTGATTTCCTAAGCAGAGG. The reaction mix was separated on a 5% acrylamide-TBE gel that had been pre-run for 30 min in 0.5X TBE. The label was imaged using a Li-Cor Odyssey scanner.
Apoptosis assay
H69 cells were treated with 1 μg/mL LPS or 10 μg/mL sonic hedgehog ligand overnight, followed by TRAIL treatment (4 ng/mL) for 8 hours. Alternatively, cells were transfected with pCDNA/myc and pEGFP at 4:1 ratio at 48 and again 24 hours prior to apoptosis measurements. Transfected cells were treated with TRAIL (1 ng/mL) for 8 hours. Nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, 1000X stock = 5 mg/mL in water; Sigma, St. Louis MO) and visualized by fluorescence microscopy using excitation and emission wavelengths of 380 and 430 nm. Apoptosis was quantified by counting characteristic pyknotic nuclei that were strongly DAPI-positive.
Reagents
Sonic hedgehog signaling was inhibited using cyclopamine (LC Laboratories, Woburn, MA) at 5 μM, or the antagonistic antibody 5E1 (Developmental Studies Hybridoma Bank, Iowa City, IA) at 10 μg/mL. Silencer siRNA Construction kit from Ambion (Austin, TX) was utilized to generate siRNA to inhibit Gli-3 expression with the template sequence 5′-AAGGCATAATGTTGTCACAGACCTGTCTC (partial T7-promoter sequence is bold).
Statistical analysis
Unless otherwise indicated, all experiments were performed in triplicate. Statistical significance where multiple comparisons were possible was determined by one-way ANOVA with Bonferroni correction. Otherwise, unpaired two-tailed Student’s T-test was performed. A p value of less than 0.05 was considered significant. All data are means +/−SEM.
RESULTS
Identification and cloning of a functional mir-29b-1/mir-29a promoter
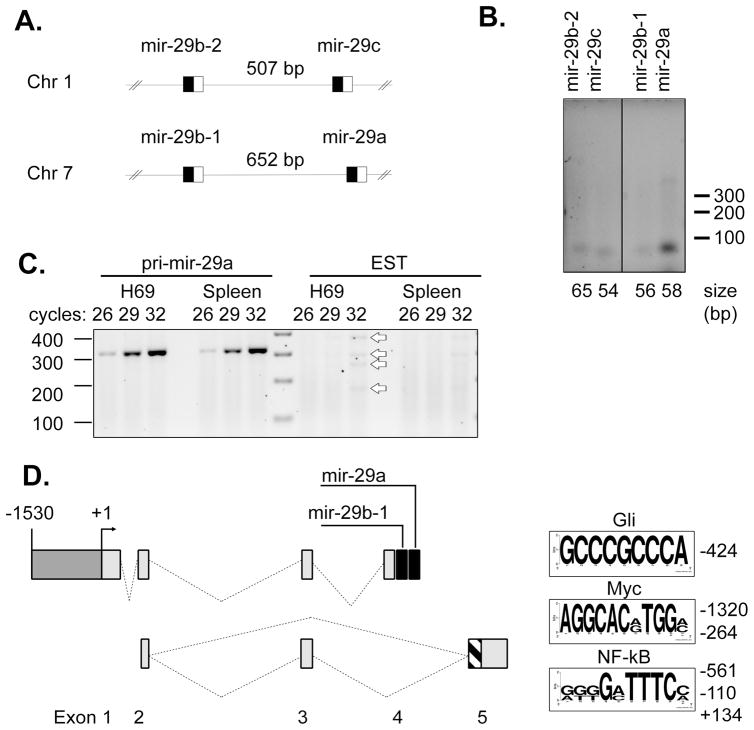
Family members of mir-29 include mir-29a, -29b-1, -29b-2, and -29c. These are arranged at two genomic sites in pairs, such that mir-29b-1 and mir-29a are separated by 652 bases on chromosome 7q32.3 and mir-29b-2 and mir-29c are separated by 507 bases on chromosome 1q32.2 (Figure 1A). To assess which of these loci express mir-29 in cholangiocytes, RT-PCR for the precursor microRNAs was performed. Amplification of pre-mir-29a yielded a strong product, while pre-mir-29b-1, -29b-2, and -29c had detectible, but less intense expression (Figure 1B). At the mir-29b-1/mir-29a locus on chromosome 7, a spliced expressed sequence tag (EST; BI768447) has been described from splenic RNA. To determine if mir-29b-1/mir-29a was expressed from the intron of this EST, RT-PCR was performed on RNA from H69 cholangiocyte cells and from total splenic RNA (Ambion). Primers positioned within the exons of the EST yielded only a faintly detectible series of products after 32 cycles of amplification in either sample, while RT-PCR for pri-mir-29a yielded a strong band in both spleen and cholangiocyte cell RNA (Figure 1C). The identity of each band was confirmed by automated sequencing of the gel purified band after subcloning into TOPO-pCR2.1. While faint, there were four distinct products of RT-PCR from the EST in both splenic and cholangiocyte RNA which were determined to be previously undescribed splice variants and were deposited in GenBank (accession numbers GU321463-321467).
Figure 1. mir-29b-1/mir-29a transcript from chromosome 7.
(A) Schematic diagram of mir-29 family members, which exist at paired loci on chromosome 1 (mir-29b-2 and mir-29c) and chromosome 7 (mir-29b-1 and mir-29a). The boxed regions indicate the position of the precursor microRNA sequence, with the black box representing the passenger strand (mir-29*) and the open box the mature microRNA. (B) RT-PCR for precursor mir-29 family members from total RNA isolated from the H69 cholangiocyte cell line. Primers were positioned on each side of the predicted stem-loop precursor and amplification was carried out for 30 cycles. The size of expected amplicons is noted below the electrophoretogram; size markers are indicated to the right. (C) RT-PCR for the primary mir-29a transcript (Left) or using primers complementary to exons described for the expressed sequence tag AI768447 demonstrated robust expression of the mir-29a transcript but low expression of the EST, even in spleen where the sequence was discovered. RT-PCR for the EST did yield the expected 172 bp product, as well as products of 252, 291, and 371 bp (open arrows). (D) Sequencing of cloned RT-PCR products from Figure 1C revealed alternative spice forms from the same locus composed of exons 2 and 5b (172 bp), exons 2, 3, and 5b (252 bp), exons 2 and 5a (291 bp), and exons 2, 3, and 5a (371 bp). These gene products are illustrated below the mir-29b-1/mir-29a locus. Exon 5 includes alternative 3′ acceptor sites, resulting in exon 5a (267 bp, hatched plus shaded bars) and a shorter exon 5b (148 bp, shaded bar). Sequence analysis of the putative promoter revealed predicted binding sites for Gli, Myc, and NF–κB, which are indicated as sequence logos created from manually aligned binding sites using weblogo. The height of each letter indicates the conservation at that position between the sites analyzed. There is a single Gli site, so all bases are shown at full height.
Thus mir-29b-1/mir-29a is likely expressed independently of the EST, suggesting an alternate transcription start site. 5′ RACE was performed on total RNA extracted from H69 cells that had been transfected with Drosha siRNA (Oligo #2, [Lee et al., 2003]) 72 hours previously to maintain un-processed primary mir-29 transcript. The resulting clone was sequenced and demonstrated a spliced transcript with 3 upstream exons and a 3′ splice acceptor site immediately upstream (59 bp from precursor) of mir-29b-1 consistent with mir-29b-1 and mir-29a residing on exon 4 (GenBank accession number GU321462). The identified 3′ splice acceptor exhibits the expected features of a canonical splice acceptor, including a polypyrimidine tract upstream of the TAG/G acceptor site [Black, 2003]. The 5′ terminus of the RACE product was taken as the transcription start site (+1) and flanking DNA was considered for promoter analysis (Figure 1D). Of note, Chang et al. recently described a 5′ RACE product from this locus which shares the upstream exons but has a transcription start 131 nucleotides upstream of the site we observed (GenBank accession EU154353).
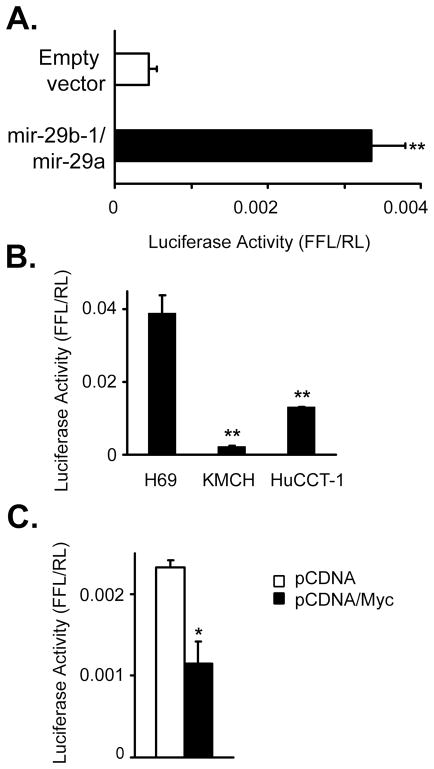
Initially, approximately 10 kbp of genomic DNA sequence from the human and mouse genomes were queried for predicted transcription factor/enhancer binding sites. Manual curation for transcription factors with cancer relevance revealed binding sites for hedgehog-responsive Gli, Myc and N-Myc, NF–κB, and p53. Both Gli and NF–κB binding sites were observed more than 5 times. A smaller fragment from −1530 to +165 relative to the transcription start site included binding sites for Gli, Myc, and NF–κB (Figure 1D) and was incorporated into a luciferase-based reporter plasmid. This putative promoter increased luciferase expression over the empty vector in KMCH cells, consistent with the presence of promoter activity (Figure 2A).
Figure 2. mir-29b-1/mir-29a promoter construct activity.
(A) The DNA sequence (−1530 to +165) flanking mir-29 transcriptional start site (+1) was ligated into the pGL3 reporter. This DNA sequence transfected into H69 cells displays promoter activity greater than the empty vector. (B) Comparison of luciferase activity in non-malignant H69 cells compared to malignant cholangiocarcinoma cell lines KMCH and HuCCT-1 cells 24 hours after transfection with the promoter reporter construct. (C) Enforced c-Myc expression reduced promoter activity, as determined luciferase assay following cotransfection of the reporter plus a c-Myc expression construct, or empty pCDNA3.1 control. All data are mean +/− SEM, and are representative of 3 separate experiments. * p < 0.05; ** p < 0.01.
Regulation of the mir-29b-1/mir-29a promoter region by c-Myc
To determine if the putative promoter demonstrates activity consistent with known expression of mir-29, we compared activity of the promoter in H69 cells to malignant cholangiocarcinoma cells KMCH and HuCCT-1. The promoter was significantly less active in malignant cells (Figure 2B), consistent with decreased expression of mir-29b in KMCH cells [Mott et al., 2007]. The mir-29b-1/mir-29a promoter region contains a canonical E-box Myc binding site (5′ CACGTG located at −261 from the transcription start site), as well as a close match (5′ CACATG; −1317) that were both predicted Myc binding sites. The site at −261 is perfectly conserved between human, rhesus monkey, mouse, and dog sequences, consistent with a functional site, while the −1317 site is conserved between human and rhesus. Indeed, repression of mir-29 expression by c-Myc expression has been demonstrated in B cell lymphoma [Chang et al., 2008]. Consistent with Myc-dependent repression, enforced expression of c-Myc decreased promoter activity by 50% (Figure 2C). Collectively, these data confirm suppression of mir-29 expression by c-Myc and provide support that the region upstream of the mir-29b-1/mir-29a transcriptional start site is indeed a functional promoter region.
Hedgehog signaling regulates the mir-29b-1/mir-29a promoter
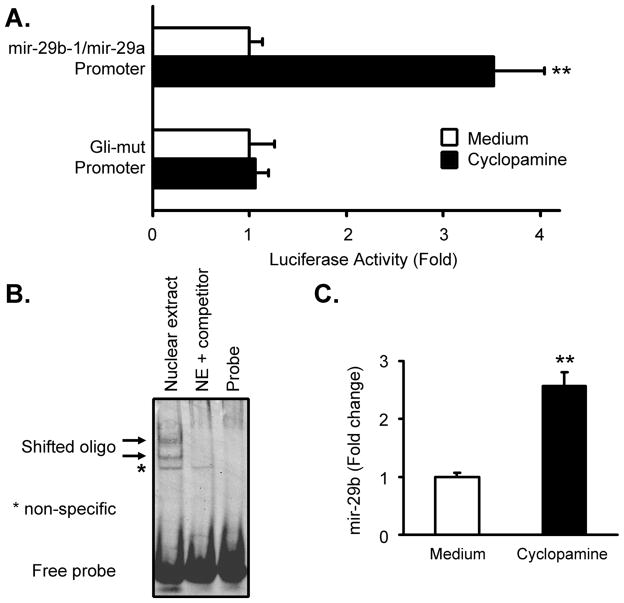
We have recently confirmed the expression of mRNA for hedgehog signaling components in KMCH cells (Kurita et al., unpublished) including the ligand Sonic hedgehog, the receptor patched-1, the 7-transmembrane signaling component smoothened, and Gli-1, -2, and -3. Computational analysis demonstrated a putative Gli-binding site at −424 (5′ GCCCGCCCA) in the human mir-29b-1/mir-29a promoter sequence. To assess whether hedgehog signaling affected the activity of the mir-29b-1/mir-29a promoter, reporter activity was measured in cells treated with cyclopamine, an inhibitor of smoothened. Luciferase activity increased significantly when cells were treated overnight with cyclopamine, but not when the putative Gli-binding site was disrupted (Gli-mut) by site-directed mutagenesis (Figure 3A). The loss of responsiveness to cyclopamine of the Gli-mut promoter suggested that Gli transcription factors directly bind this site in the mir-29 promoter. To further address this, a double-stranded DNA probe was designed based on the putative Gli-binding site for gel shift analysis. Nuclear extract from KMCH cells treated with cyclopamine exhibited binding activity to this probe, and excess cold probe competed for binding, demonstrating sequence specificity (Figure 3B). Thus the putative Gli-binding site is necessary for increased promoter function upon cyclopamine treatment and is sufficient to elicit the binding of nuclear proteins.
Figure 3. Expression of mir-29b-1/mir-29a is regulated by hedgehog signaling.
(A) Cyclopamine treatment (5 μM, 16 hours) of KMCH cells transfected with the mir-29b-1/mir-29a promoter reporter increased promoter activity 3–4 fold compared to medium. The same promoter construct except with a mutated Gli binding site (5′ GCCCGCCCA converted to 5′ GCCGGGCCA) however was not responsive to cyclopamine. Mean +/− SEM, ** p < 0.01. (B) Gel shift analysis of nuclear extract from cells treated overnight with cyclopamine demonstrates two shifted bands that can be competed by 200X molar excess of cold probe (arrows) and a non-specific band (*) that was not competed. (C) Mature mir-29b was measured by RT-PCR using a hydrolysis probe assay (Applied Biosystems) from cells treated with medium or cyclopamine (5 μM, 16 hours) revealing 2–3 fold increased levels of mature mir-29b. All data are normalized by dividing the mir-29b signal by that of the housekeeping RNA Z30 as an internal standard. Mean +/− SEM, ** p < 0.01.
To determine if hedgehog inhibition resulted in increased mature mir-29b expression, KMCH cells were treated with cyclopamine followed by RNA isolation and qRT-PCR for mir-29b. Expression of mir-29b increased 2.5-fold after treatment with cyclopamine (Figure 3C). When anti-Sonic hedgehog antiserum (5E1) or siRNA to Gli-3 was used to inhibit Sonic hedgehog, stimulation of expression of mir-29b was also observed to 46% and 43% over controls, respectively, after 12 hours (5E1) or 48 hours (siRNA) of treatment (not shown). In agreement with previous studies [Berman et al., 2003], the increased mir-29b expression suggested constitutive hedgehog signaling in cholangiocarcinoma cells, and we demonstrate here that this pathway repressed mir-29b expression.
Regulation of the mir-29b-1/mir-29a promoter by NF–κB
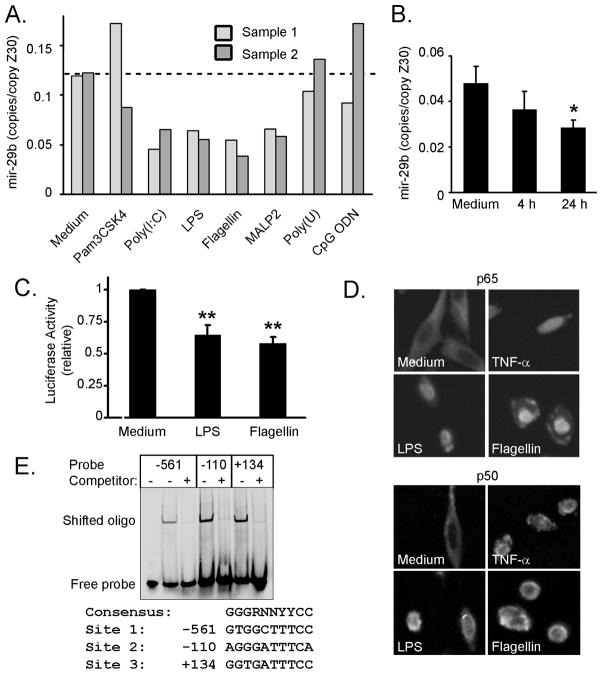
Three NF–κB sites were predicted within the −1530/+165 promoter so the effect of NF–κB activation on mir-29 expression was determined. Multiple TLRs are expressed in cholangiocytes and activate NF–κB upon receptor ligation [Chen et al., 2005; Harada et al., 2003]. Initially, we employed a panel of ligands for TLRs and determined mir-29b expression by RT-PCR in duplicate H69 cell samples. Cells treated with ligands for TLR-3 (Poly(I:C)), TLR-4 (LPS), TLR-5 (Flagellin), and TLR-6 (MALP-2) all showed similarly decreased mir-29b expression compared to medium-treated cells (Figure 4A). Ligands for TLR-1/2 (Pam3CSK4), TLR-7/8 (Poly(U)), and TLR-9 (CpG ODN) did not consistently decrease mir-29b expression (Figure 4A). The decreased mir-29b expression began after 4 hours of LPS treatment and increased by 24 hours (Figure 4B). Both LPS and Flagellin also repressed mir-29b-1/mir-29a promoter activity, consistent with a transcriptional mechanism of action (Figure 4C).
Figure 4. TLR ligands activate NF –κ B and decrease mir-29b expression.
(A) H69 cells were treated for 24 hours with ligands for TLR-1/2 (Pam3CSK4, 100 ng/mL), TLR-3 (Poly-I:C, 100 μg/mL), TLR-4 (LPS, 1 μg/mL), TLR-5 (Flagellin, 100 ng/mL), TLR-6 (MALP-2, 100 ng/mL), TLR-7/8 (Poly(U), 10μg/mL), or TLR-9 (CpG ODN, 10 μg/mL). RT-PCR for mir-29b was then performed on total RNA and normalized to Z30 expression. Two samples each were treated and analyzed, indicated as sample one and two, with generally good agreement between duplicates. (B) Time course analysis of mir-29b expression measured by RT-PCR after stimulation of H69 cells with LPS (1 μg/mL). Mean +/− SEM, * p < 0.05. (C) Ligation of TLR-4 (LPS) or TLR-5 (flagellin) reduced mir-29b-1/mir-29a promoter activity in H69 cells transfected with the luciferase reporter. Data are averaged from three independent experiments, expressed as luciferase activity compared to untreated cells. Mean, +/− SEM, ** p < 0.01. (D) Selected TLR ligands (LPS and flagellin) were chosen to verify activation of NF–κB by immunofluorescent staining of p65 (upper) and p50 (lower) in H69 cells treated for 30 minutes. Nuclear localization was observed in treated cells and the positive control TNF-α (28 ng/mL), while cytoplasmic staining was observed in untreated cells. (E) H69 cells were treated with LPS (1 μg/mL, 30 min) and nuclear proteins prepared for gel shift analysis using the three putative NF–κB binding sites identified. For each oligo, nuclear extract induced a shift that was specifically competed by 25–40 molar excess unlabeled duplex. The sequences of the binding sites are indicated, compared to the NF–κB consensus. R = purine (A or G); N = any nucleotide; Y = pyrimidine (C or T).
TLRs activate NF–κB and we confirmed that treatment with either LPS or Flagellin caused activation of NF–κB in H69 cholangiocytes. Indeed, treatment with either LPS, or Flagellin, induced nuclear localization of the p65 and p50 subunits of NF–κB, similar to treatment with TNF-α as a positive control (Figure 4D). We did not observe p65 or p50 nuclear localization in untreated cells, suggesting that NF–κB signaling is not constitutively active in cell culture. Double stranded oligonucleotide probes designed from each of the three predicted NF–κB sites, (−561, −110, and +134) exhibited a gel shift in the presence of nuclear extract from LPS-treated H69 cells (Figure 4E). Additionally, excess unlabeled oligonucleotides (25–40 fold molar excess) effectively competed for binding, demonstrating sequence specificity. Taken together, these data support the role of NF–κB activation through TLR signaling as a suppressor of mir-29b-1/mir-29a promoter function and expression of mature mir-29b through recognition of NF–κB binding sites.
Protection from apoptosis by hedgehog, TLR, and c-Myc signaling
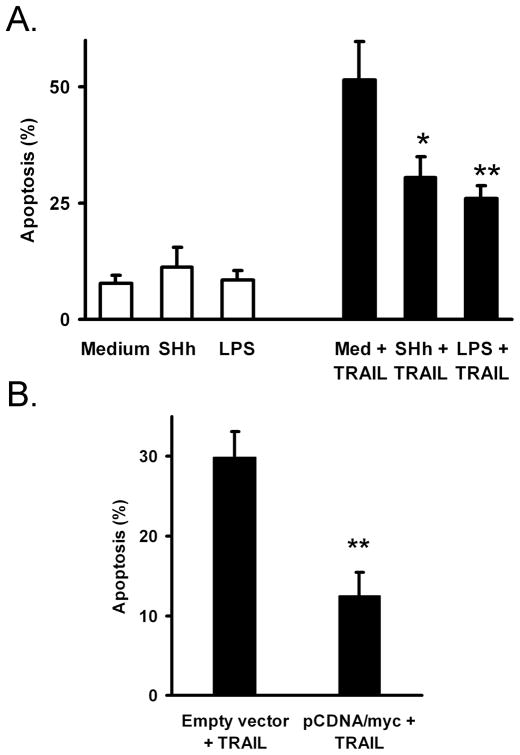
Functionally, mir-29 promotes apoptosis while repression of mir-29 levels is protective. Thus, we assessed the effect of hedgehog, TLR, and c-Myc activation on apoptosis in H69 cells after treatment with TRAIL. Non-malignant H69 cholangiocytes were sensitive to TRAIL-induced apoptosis, but treatment with recombinant sonic hedgehog or LPS protected cells from apoptosis (Figure 5A). Additionally, enforced c-Myc expression in transfected cells provided protection from TRAIL-induced apoptosis (Figure 5B). Thus, treatments that repress of mir-29 also provide protection from apoptosis, consistent with a role for mir-29 in cell death.
Figure 5. Cholangiocyte apoptosis is decreased when mir-29-suppressing pathways are activated.
(A.) H69 cells were pre-treated overnight with recombinant human Sonic Hedgehog ligand (10 μg/mL) or LPS (1 μg/mL) followed by TRAIL treatment (4 ng/mL) for 8 hours. Nuclei were stained with DAPI and nuclei with apoptotic morphology were counted. The percent of cells with apoptotic nuclei is indicated (mean, +/− SEM). ** p < 0.01. (B.) H69 cells were transfected with GFP plus either pCDNA3.1 (empty vector) or pCDNA/myc 48 hours and again 24 hours prior to apoptosis counting (1:4 GFP:pCDNA plasmid ratio). Cells were then treated with TRAIL (1 ng/mL) for 8 hours, and GFP-positive cells were then assayed for apoptosis by DAPI staining (mean, +/− SEM). ** p < 0.01. A lower concentration of TRAIL was employed compared to panel A to account for some stress due to transfection.
DISCUSSION
The principle findings of this study provide mechanistic insight regarding the transcriptional regulation of mir-29b-1/mir-29a in cholangiocarcinoma cells. Our results aid in understanding mir-29 expression by: 1) cloning of a functional mir-29b-1/mir-29a promoter; 2) demonstrating inhibition of promoter function and mir-29b expression by hedgehog signaling; and 3) demonstrating NF–κB activation through TLR activation represses mir-29b-1/mir-29a. These observations may allow for the design of rational treatment strategies to increase expression of mir-29 in an effort to counter the oncogenic effects of mir-29 targets. The results will be discussed in the context of the pathobiology of cholangiocarcinoma.
The proto-oncogene Myc is activated in hepatobiliary cancers [Tokumoto et al., 2005; Voravud et al., 1989] and can either activate or repress transcription of target genes. Notably, this transcription factor has been demonstrated to increase expression of the microRNA cluster mir-17-92 [O’Donnell et al., 2005] and also to decrease expression of more than 10 microRNAs, including the mir-29 family [Chang et al., 2008]. In the same manuscript, the authors used chromosomal immunoprecipitation analysis to show direct occupation by c-Myc of the putative promoter region [Chang et al., 2008]. The presumed promoter region amplified in that study corresponds to positions −122 to −72 relative to the transcriptional start site identified here, and is adjacent to the E-box identified in our analysis (−261). Our data extend these observations by identifying alternate transcripts from this locus, providing functional evidence of promoter activity, and demonstrating a direct effect on promoter activity of c-Myc overexpression.
Our 5′ RACE analysis shows similarities to the gene structure proposed by Chang et al. while our experiments were in progress [Chang et al., 2008], with differences noted at the transcriptional start site and whether mir-29b-1/mir-29a are intronic or exonic. The start site we observed is 131 base pairs downstream of that reported. Additionally, the 5′ RACE primers in our experiment were positioned immediately upstream of mir-29b-1 and demonstrated splicing to exon 3, implicating the mir-29b-1/mir-29a cluster is in exon 4, in contrast to the intronic location reported by Chang and colleagues (between exons 3 and 5). RT-PCR using primers in exons 2 and 5b in H69 cholangiocyte cells or total splenic RNA revealed, in addition to the expected 172 bp product, three additional alternatively spliced gene products. None of the cDNA clones generated by RT-PCR with primers in exons 2 and 5 contatined exon 4, suggesting that exons 4 and 5 serve as alternate (mutually exclusive) terminal exons. The dominant splice pattern likely depends on the cell type and other contextual cues, accounting for the intronic versus exonic expression. Notably, functional microRNA can be processed either from exonic or intronic locations [Kim and Kim, 2007].
Recent data have indicated that cholangiocarcinoma cells express signaling factors comprising the hedgehog pathway, and that inhibition by cyclopamine could limit cell survival and promote regression of xenografted cholangiocarcinoma cells [Berman et al., 2003]. Thus, the presence of a predicted Gli-binding site in the mir-29b-1/mir-29a promoter suggested that hedgehog signaling may affect mir-29 expression. Indeed, activity of the mir-29b-1/mir-29a promoter fragment (−1530/+165) was increased 3–4 fold in cells treated with cyclopamine, an effect that was prevented by mutagenesis of the Gli binding site. Additionally, cyclopamine treatment increased mir-29b expression 2–3 fold in these cells. Inhibition of the pathway by antagonistic antibody (5E1) or by siRNA to Gli-3 modestly increased mir-29b expression by 40–50%. This suggests either incomplete inhibition by the latter approaches or possibly that non-canonical hedgehog signaling contributes to mir-29 repression. Of interest, there are as yet no reports of other Gli-regulated microRNAs, though hedgehog can increase mir-17-92 expression indirectly [Northcott et al., 2009].
Classically, activation of the inflammation-sensitive NF–κB transcriptional complex promotes cell survival, including through increased expression of the prosurvival protein Bcl-2 [Grossmann et al., 2000] and related family members Bcl-xL [Lee et al., 1999; Tsukahara et al., 1999], A1 [Zong et al., 1999], and Mcl-1 [Ricci et al., 2007], which is particularly relevant to cholangiocarcinoma survival [Kobayashi et al., 2005; Taniai et al., 2004]. NF-κB is a transcription factor made up of different subunits, including p65 and p50 proteins, and is activated by TLR signaling. The finding that the mir-29b-1/mir-29a promoter on chromosome 7 contains multiple NF–κB binding sites suggested that in addition to a direct increase of Mcl-1 transcription, NF–κB may increase Mcl-1 protein expression indirectly by blocking expression of mir-29. Our data demonstrate that multiple TLR ligands of the innate immune system induce nuclear localization of NF–κB (p65 and p50) and decrease expression of the mature mir-29b. Repression of microRNA expression by NF–κB has been described for let-7i [O’Hara et al., 2009] as well as mir-29b-2/mir-29c, through Yin Yang-1 [Wang et al., 2008]. Alternatively, NF–κB drives expression of mir-143 in hepatocellular carcinoma [Zhang et al., 2009]. Thus NF–κB mediated microRNA regulation may be a frequent mechanism used to regulate cellular responses. In combination with LPS or Flagellin treatment, 24 hours of NF-κB inhibition (by SN50, MG-132, or Bay-11-7082) in these cells induced significant toxicity (not shown). Thus, while the observations of mir-29 repression and NF-κB activation are consistent with mir-29 repression by NF-κB, possibly through p65, p50, or other NF-κB subunits, we cannot exclude repression by alternative, non-NF-κB, TLR signaling pathways.
Functionally, mir-29 family members have been implicated in promoting apoptosis by us [Mott et al., 2007] and others [Park et al., 2009]. Thus, it is of interest that activation of Hedgehog signaling, TLR signaling, or c-Myc expression, each of which repress mir-29 expression in cholangiocytes, also act to protect H69 cells from apoptosis by TRAIL treatment. The effects of Hedgehog and NF-κB pathways on preventing apoptosis has been found to be multifactorial [Bigelow et al., 2004; Kucharczak et al., 2003; Kump et al., 2008] and we propose that mir-29 repression may contribute to this protective phenotype. Interestingly, c-Myc expression is classically considered to promote apoptosis [Evan et al., 1992], but can also have protective effects [D’Agnano et al., 2001; Gatti et al., 2009]. Thus, the pro- or anti-apoptotic effect of c-Myc may depend on the integration of competing signals, including mir-29 repression.
The results presented here are consistent with regulation of mir-29b-1/mir-29a expression by multiple signaling pathways relevant to cancer biology (Figure 6). The particular balance of these repressing factors may be different between cancers, but the finding that mir-29 expression is decreased in cancers of diverse origin [Fabbri et al., 2007; Garzon et al., 2009b; Mott et al., 2007; Pekarsky et al., 2006; Porkka et al., 2007; Sengupta et al., 2008; Yanaihara et al., 2006] is consistent with multiple potential repressive mechanisms. It remains possible, even likely, that additional cis acting regulatory elements contribute to the transcriptional control of mir-29b-1/mir-29a. As loss of mir-29 contributes to apoptosis, metastasis/invasion, and epigenetic signaling, approaches to increase endogenous mir-29b-1/mir-29a expression may have a beneficial effect on cancer treatment through multiple inter-related pathways. As small molecule inhibitors of Myc have not been developed, inhibition of hedgehog or NF–κB signaling may have the most promise in the near-term.
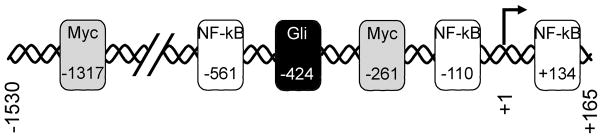
Figure 6. Transcription factor binding sites for mir-29b-1/mir-29a.
Schematic illustration of the functional mir-29b-1/mir-29a promoter used in this study. By computational analysis, two c-Myc binding sites (−1317 and −261) were identified (shaded boxes). Three NF-kB sites were predicted based on sequence and verified by gel shift analysis (−561, −110, +134; indicated by open boxes). A single Gli binding site was identified (at position −424; black box) by sequence analysis and confirmed by site-directed mutagenesis and gel shift assay.
Acknowledgments
The authors thank Erin Nystuen-Bungum for secretarial assistance.
Grants: This publication was supported by grants DK 79875 (JLM), CA 136526 (MEF-Z), and the Optical Microscopy Core of P30 (DK 84567) from the National Institute of Diabetes and Digestive and Kidney Diseases, as well as the Mayo Foundation Rochester, Minnesota, USA.
Abbreviations
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DMEM
Dulbecco’s modified eagle medium
- DTT
dithiothreitol
- EGTA
ethylene glycol tetraacetic acid
- EST
expressed sequence tag
- Gli-mut
Gli-binding site mutant promoter
- GSP
gene-specific primer
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LPS
lipopolysaccharide
- NF-κB
nuclear factor kappa B
- PBS
phosphate-buffered saline
- 5′ RACE
5′ rapid amplification of cDNA ends
- RT-PCR
reverse transcription-polymerase chain reaction
- SEM
standard error of the mean
- siRNA
short interfering RNA
- TBE
Tris-Borate-Ethylenediaminetetraactetic acid
- TLR
Toll-like receptor
- TNF-α
tumor necrosis factor-α
References
- Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, Toftgard R, McDonnell TJ. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–205. doi: 10.1074/jbc.M310589200. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, O’Hara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, Limper AH, LaRusso NF. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial defense responses to Cryptosporidium parvum via activation of NF-kappaB. J Immunol. 2005;175:7447–56. doi: 10.4049/jimmunol.175.11.7447. [DOI] [PubMed] [Google Scholar]
- D’Agnano I, Valentini A, Fornari C, Bucci B, Starace G, Felsani A, Citro G. Myc down-regulation induces apoptosis in M14 melanoma cells by increasing p27(kip1) levels. Oncogene. 2001;20:2814–25. doi: 10.1038/sj.onc.1204392. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–28. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–63. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, Andreeff M, Croce CM. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009a doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti G, Maresca G, Natoli M, Florenzano F, Nicolin A, Felsani A, D’Agnano I. MYC prevents apoptosis and enhances endoreduplication induced by paclitaxel. PLoS One. 2009;4:e5442. doi: 10.1371/journal.pone.0005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–5. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann M, O’Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. Embo J. 2000;19:6351–60. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol. 1994;266:G1060–70. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- Harada K, Ohira S, Isse K, Ozaki S, Zen Y, Sato Y, Nakanuma Y. Lipopolysaccharide activates nuclear factor-kappaB through toll-like receptors and related molecules in cultured biliary epithelial cells. Lab Invest. 2003;83:1657–67. doi: 10.1097/01.lab.0000097190.56734.fe. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–45. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. Embo J. 2007;26:775–83. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–65. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Kucharczak J, Simmons MJ, Fan Y, Gelinas C. To be, or not to be: NF-kappaB is the answer--role of Rel/NF-kappaB in the regulation of apoptosis. Oncogene. 2003;22:8961–82. doi: 10.1038/sj.onc.1207230. [DOI] [PubMed] [Google Scholar]
- Kump E, Ji J, Wernli M, Hausermann P, Erb P. Gli2 upregulates cFlip and renders basal cell carcinoma cells resistant to death ligand-mediated apoptosis. Oncogene. 2008;27:3856–64. doi: 10.1038/onc.2008.5. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. Rna. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Dadgostar H, Cheng Q, Shu J, Cheng G. NF-kappaB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc Natl Acad Sci U S A. 1999;96:9136–41. doi: 10.1073/pnas.96.16.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–55. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- O’Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, Chen XM, Larusso NF. NF{kappa}B p50-CCAAT-enhancer binding protein beta (C/EBP{beta})-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem. 2009 doi: 10.1074/jbc.M109.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Ricci MS, Jin Z, Dews M, Yu D, Thomas-Tikhonenko A, Dicker DT, El-Deiry WS. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol. 2004;24:8541–55. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MS, Kim SH, Ogi K, Plastaras JP, Ling J, Wang W, Jin Z, Liu YY, Dicker DT, Chiao PJ, Flaherty KT, Smith CD, El-Deiry WS. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007;12:66–80. doi: 10.1016/j.ccr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:5874–8. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, Kaufmann SH, Gores GJ. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–24. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tokumoto N, Ikeda S, Ishizaki Y, Kurihara T, Ozaki S, Iseki M, Shimizu Y, Itamoto T, Arihiro K, Okajima M, Asahara T. Immunohistochemical and mutational analyses of Wnt signaling components and target genes in intrahepatic cholangiocarcinomas. Int J Oncol. 2005;27:973–80. [PubMed] [Google Scholar]
- Tsukahara T, Kannagi M, Ohashi T, Kato H, Arai M, Nunez G, Iwanaga Y, Yamamoto N, Ohtani K, Nakamura M, Fujii M. Induction of Bcl-x(L) expression by human T-cell leukemia virus type 1 Tax through NF-kappaB in apoptosis-resistant T-cell transfectants with Tax. J Virol. 1999;73:7981–7. doi: 10.1128/jvi.73.10.7981-7987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–7. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voravud N, Foster CS, Gilbertson JA, Sikora K, Waxman J. Oncogene expression in cholangiocarcinoma and in normal hepatic development. Hum Pathol. 1989;20:1163–8. doi: 10.1016/s0046-8177(89)80006-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–9. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–7. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]