Abstract
We recently cloned the zebrafish neuronal enolase-2 gene and showed that a 12kb eno2 promoter element was sufficient to drive transgene expression widely in CNS neurons in vivo from 48 hours post-fertilization through adulthood. The aim of the present study was to establish the expression pattern of the 12kb eno2 promoter element in the zebrafish visual system. Endogenous eno2 mRNA was detected in the developing retina from 2 days post-fertilization (dpf), and by 12dpf was localized to the retinal ganglion cell, inner and outer nuclear layers. Similar to endogenous eno2, GFP expression in the retina of Tg(eno2:egfp)Pt404 larvae was first evident at 2dpf, and by 12dpf intense GFP expression was seen in the retinal ganglion cell and photoreceptor layers, with weaker expression in the inner nuclear layer. We identified cell types expressing the eno2 promoter element by using two complementary strategies: (i) double label immunofluorescence analysis of Tg(eno2:egfp)Pt404 zebrafish, and (ii) generation of double transgenic zebrafish expressing red fluorescent protein under transcriptional control of the 12kb eno2 promoter and GFP under a rod- or cone-specific promoter. The 12kb eno2 promoter was expressed in retinal ganglion cells, amacrine cells, including a subset that co-expressed tyrosine hydroxylase, and rod photoreceptors. These data suggest that abnormalities of vision should be sought in transgenic models of diseases generated using this promoter. Owing to the specific expression of fluorescent reporters in neuronal subpopulations, Tg(eno2:egfp)Pt404 and Tg(eno2:RFP) zebrafish may be useful for studies of retinal lamination, neuronal differentiation and synapse formation in the visual system.
Introduction
The overall aim of our studies is to understand mechanisms underlying neurodegenerative disorders and to develop effective treatments for these diseases, by exploiting high-throughput screening [1, 11, 31] and imaging [15, 17] methodologies that can be deployed uniquely in the zebrafish as a model vertebrate organism. As a first step towards generating transgenic models of neurodegenerative diseases, we cloned a promoter element allowing expression of human neurodegeneration-associated genes within the zebrafish brain [2]. In mammals, three enolase isoenzymes are encoded by separate genes that show distinct expression patterns; the ENO2 gene encoding γ-enolase is expressed in neurons throughout the neuraxis [21]. We cloned zebrafish orthologs of the mammalian enolase genes and showed that zebrafish eno2 is highly conserved with respect to human ENO2, and is expressed widely in differentiated neurons [2]. A 12kb genomic fragment encompassing the 5′ flanking region, transcriptional start sites and exon 1 through exon 2 of the zebrafish eno2 gene was sufficient to direct neuronal transgene expression in multiple independent lines of stable Tg(eno2:egfp)Pt404 zebrafish. The 12kb eno2 promoter element showed properties to suggest that it may be useful for constructing transgenic zebrafish models of human neurodegenerative diseases, including robust transgene expression from late in development through adulthood and widespread activity in neuronal populations relevant to neurodegenerative diseases. During our initial analysis of the 12kb eno2 promoter, we noted that GFP fluorescence was evident in the eyes of Tg(eno2:egfp)Pt404 larvae, in addition to brain and peripheral nerves [2]. Since the retina is derived from the neural tube and contains numerous classes of neurons, we hypothesized that GFP is expressed in retinal neurons in Tg(eno2:egfp)Pt404 zebrafish. Characterization of retinal expression of the 12kb eno2 promoter fragment is important because transgenic models generated with this construct might show behavioral phenotypes that are caused or modulated by defects in visual function. Furthermore, interesting physiological or biochemical abnormalities might be detected in the retinas of transgenic animals.
The zebrafish retina shows similar lamination and cellular specialization to other vertebrate retinas [5, 13]. Ganglion cells residing in the innermost retinal layer send axonal projections to the brain. Photoreceptor cells located in the external nuclear layer consist of rods and four classes of cones (red-, green-, blue- and UV-sensitive). The inner nuclear layer is composed of interneurons, which include bipolar, amacrine, interplexiform, and horizontal cells [10]. In distinction to many other vertebrates, zebrafish embryos develop externally and are optically transparent, allowing direct microscopic observation of ocular and retinal developmental anatomy [20]. Consequently the zebrafish visual system has proven a powerful model in which to examine the development of retinal patterning and architecture [8, 19], axon guidance [24], cellular differentiation [9, 18], synapse formation [28] and visually-dependent behavior [7]. The application of fluorescent reporters expressed under control of specific promoter elements has allowed powerful in vivo analysis of the transcriptional regulation of cell-restricted and context-dependent genes, such as specific opsins in photoreceptor cells [27, 30] and genes induced during retinal and optic nerve repair [14, 15, 23], in addition to lineage-tracing studies of cell types involved in dynamic retinal regeneration [4].
In this study, we showed that endogenous eno2 and the 12kb eno2 promoter are expressed in the retina in similar patterns. Expression of the eno2:egfp transgene was detected at high levels in retinal ganglion cells and rod photoreceptors and at intermediate levels in some amacrine cells, including those that express tyrosine hydroxylase.
Materials and methods
Zebrafish, microscopy and RNA in situ hybridization
Experiments were carried out in accordance with Institutional Animal Use and Care Committee regulations and approvals. Zebrafish maintenance and microscopy, and RNA in situ hybridization were carried out as previously described [2, 3].
Transgenic zebrafish
An mRFP fragment was amplified using primers F 5′-atggcttcctccgaggacgtcatc-3′ and R 5′-ccactagttaaaaaacctcccacact-3′, inserted into the StuI/SpeI sites of pBS-I-Sce1-eno2, and a 12kb fragment of the eno2 gene was captured from BAC zC51M24 using gap repair recombination as previously described [2]. Transgenic lines were established as previously described [2] and genotyped by PCR using the mRFP primers shown above.
Immunohistology and antibodies
Immunohistochemistry and immunofluorescence were carried out as previously described [2, 3], using primary antibodies diluted: 1:250 (monoclonal GFP, Chemicon, Temecula, CA); 1:500 (polyclonal GFP, Invitrogen, Carlsbad, CA); 1:500 (RFP, Clontech, Mountain View, CA); 1:50 (Zn8, Developmental Studies Hybridoma Bank, University of Iowa) [26]; 1:300 (Zpr1, ZFIN); 1:400 (Lin7) [29]; or 1:500 (TH; Chemicon).
Results
Endogenous eno2 mRNA is expressed in the retina
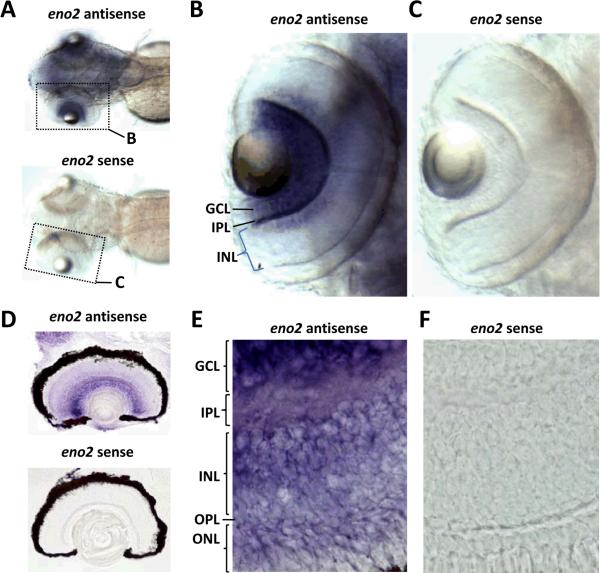
We examined the expression pattern of eno2 in the eye by using RNA in situ hybridization, employing a previously characterized eno2-specific cRNA probe that detects a single 3.5kb eno2 transcript on northern blot and labels neurons in whole-mount samples and cryosections [2]. The endogenous eno2 transcript was first detected in the developing eye of whole-mount preparations at 48 hours post-fertilization (hpf), and by 72hpf (at which point robust eno2 expression was seen in the brain) abundant eno2 mRNA was detected in the retina (figure 1A–C). Expression at this time point was seen in the inner segments of the retina, in the region of the developing retinal ganglion cell and inner nuclear layers. Retinal sections were subjected to RNA in situ hybridization at 12dpf (figure 1D–F). At this time point, abundant eno2 expression was visible in retinal ganglion cells. In addition, weaker signal was apparent in cells in the deep lamina of the inner nuclear layer and in some photoreceptors whose nuclear topography with respect to the outer plexiform layer was suggestive of rod photoreceptors. No reaction product was seen when whole mount samples or cryosections were hybridized with a sense control probe at any time point, confirming the specificity of the observed hybridization patterns.
Figure 1. Expression of endogenous eno2 in the retina.
RNA in situ hybridization was employed in order to analyze endogenous eno2 mRNA expression in the retina at 72 hpf (A–C) and 12 dpf (D–F)
A–C: Panel A shows a low power superior view of two whole mount 72hpf larvae (rostral left) that were hybridized with a cRNA probe specific to eno2 (upper larva) or a sense control probe (lower larva). Hybridized probe was detected using a histochemical reaction with a purple/blue reaction product. The region of each image encompassing the left eye is shown at higher magnification in panels B and C, as indicated by the dotted outline.
D–F: Panel D shows a low power view of two 15μm cryosections of eyes from 12dpf larvae that were hybridized with cRNA probe specific to eno2 (upper sample) or a control sense probe (lower sample). Hybridized probe was detected with a histochemical reaction producing a purple/blue product. Representative high magnification fields from similar sections are shown in panels E and F. Key: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer
GFP expression in the retina of Tg(eno2:egfp) zebrafish
Similar to endogenous eno2 mRNA, GFP expression was first detected in the eyes of intact Tg(eno2:egfp)Pt404 zebrafish by epifluorescence microscopy at 48hpf; the strength of the fluorescent signal increased thereafter, and by 5dpf bright GFP fluorescence was visible within the eye, in addition to the brain and peripheral nerve axons previously characterized (figure 2A). Confocal imaging in live larvae confirmed that ocular GFP expression was localized to the retina (figure 2B). In order to establish the developmental pattern of retinal GFP expression in Tg(eno2:egfp)Pt404 zebrafish, we examined sections between 48hpf and 12dpf by indirect immunofluorescence and confocal microscopy (figure 2 C–F). At 48hpf, expression was seen in the region surrounding the lens, containing developing retinal ganglion cells. Between 3 and 12 dpf, more distinct GFP-expressing cell profiles became visible in the developing retinal ganglion cell and inner plexiform layers, and weaker fluorescence became visible in the developing photoreceptor layer and inner nuclear layer. By 12 days post-fertilization, bright GFP expression was detected in the ganglion cell and photoreceptor layers, while inner nuclear and inner plexiform layers continued to show weak GFP expression. These data show that endogenous eno2 mRNA and the 12kb eno2:egfp transgene show similar laminar retinal expression patterns.
Figure 2. GFP expression in the retina of Tg(eno2:egfp)Pt404 zebrafish larvae.
A: The image shows a Z-plane projection of multiple confocal images of the head region of a live 5dpf Tg(eno2:egfp)Pt404 larva (rostral left, dorsal up) demonstrating GFP expression.
B: Single plane confocal image of GFP expression in the retina of a live 72hpf Tg(eno2:egfp)Pt404 larva.
C – F: Indirect immunofluorescence and confocal microscopy were employed to detect GFP expression in fixed 20μm thick retinal cryosections derived from Tg(eno2:egfp)Pt404 zebrafish at (C) 2dpf; (D) 3dpf; (E) 5dpf; and (F) 12dpf.
Key: R, retina; TeO, optic tectum; HB, hindbrain ON, optic nerve; L, lens; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer.
The 12kb eno2 promoter is expressed in retinal ganglion and amacrine cells
We next delineated cell types expressing the eno2:egfp transgene in the ganglion cell and inner nuclear layers by double label immunofluorescence. Examination of sections encompassing the retina and central visual pathways showed that GFP fluorescence co-localized with Zn8 immunoreactivity in retinal ganglion cell axons within the optic nerves and optic tracts, and their terminals in the superficial optic tectum, where particularly prominent GFP expression was visible (figure 3). GFP expression in the inner nuclear layer was localized to the region adjacent to the inner plexiform layer, suggesting that expression in this region may be restricted to amacrine cells. We tested this possibility by double immunofluorescence using an anti-lin7 antibody, which strongly labels bipolar cells [29] (figure 4A). The eno2:egfp transgene was not expressed in lin7-positive bipolar cells. In view of the importance of dopaminergic neurons in neurodegenerative diseases, we investigated whether the eno2:egfp transgene was expressed in the subset of amacrine cells that use dopamine as a neurotransmitter (figure 4B). The 12kb eno2 promoter was expressed at intermediate levels in a population of amacrine cells that were immunoreactive for tyrosine hydroxylase. There were other subpopulations of amacrine cells that either did not express the eno2:egfp transgene, or expressed the transgene at high levels.
Figure 3. Expression of GFP in retinal ganglion cells and their central projections in a Tg(eno2:egfp)Pt404 zebrafish larva.
A single plane confocal micrograph is shown of a cranial oblique coronal cryosection derived from a Tg(eno2:egfp)Pt404 zebrafish at 5dpf. Double indirect immunofluorescence was employed to detect GFP expression (green) and Zn8 immunoreactivity (red) simultaneously. Key: OT, optic tract; otherwise same as figures 1 and 2.
Figure 4. Expression of GFP in the inner nuclear layer of Tg(eno2:egfp)Pt404 retina.
Double Indirect immunofluorescence and confocal microscopy were employed to detect GFP expression and cell markers of interest in cryosections derived from 12dpf Tg(eno2:egfp)Pt404 zebrafish.
A: Lin7 (red; bipolar cells; left panel); eno2:egfp (green, center panel); overlay, right panel. Key same as figure 2.
B: Tyrosine hydroxylase (red; subset of amacrine cells; upper left panel); eno2:egfp (green, middle left panel); overlay (lower panel). Arrows mark TH-immunoreactive cells. A lower magnification image is shown on the right, indicating the region from which the images on the left were taken. Key same as figure 2.
Generation of Tg(eno2:mrfp)Pt407 zebrafish
We generated Tg(eno2:mrfp)Pt407 zebrafish in which monomeric red fluorescent protein (a kind gift from Dr. Roger Tsien, University of California, San Diego) was expressed under the transcriptional control of the 12kb eno2 promoter, allowing simultaneous detection of eno2 promoter activity and GFP expression in double transgenic fish. The eno2:mrfp transgene was constructed in an identical manner to the eno2:egfp construct that we previously described, except for the replacement of the cDNA encoding GFP with that encoding mRFP (figure 5A). A stable transgenic Tg(eno2:mrfp) line, which has been assigned allele Pt407, was established by the I-Sce1 meganuclease technique (figure 5B). Expression of mRFP was visible throughout the neuraxis of Tg(eno2:mrfp)Pt407 by 48hpf, the intensity of the fluorescent signal increasing thereafter (figure 5C). Within the retina, mRFP expression was evident in the GCL, INL and PRL. These data show that the expression pattern of mRFP in Tg(eno2:mrfp)Pt407 zebrafish is identical to the expression pattern seen in other transgenic lines constructed with this promoter, including multiple Tg(eno2:egfp)Pt404 lines, and is therefore determined by the 12kb eno2 promoter fragment rather than by the locus of transgene insertion or other artifact.
Figure 5. Generation of Tg(mRFP) zebrafish.
A: The schematic illustration shows the genomic organization of the eno2 gene and the DNA construct used to generate the transgenic Tg(eno2:mrfp)Pt407 zebrafish shown in panels B and C B: The 12kb eno2:mrfp construct was microinjected into single-cell zebrafish embryos and stable transgenic lines were derived as described in the text. The genotype of transgenic founders was confirmed by genomic PCR using eno2:mrfp-specific primers. The picture shows an ethidium bromide stained agarose gel. Lane 1, genomic DNA from wild-type strain AB* zebrafish spiked with mRFP plasmid; lane 2 genomic DNA from wild-type zebrafish; lane 3 genomic DNA from Tg(eno2:mrfp)Pt407 zebrafish.
C: mRFP expression in living stable Tg(eno2:mrfp)Pt407 zebrafish larva by epifluorescence microscopy. An oblique lateral view of a Tg(eno2:mrfp)Pt407 larva is shown at 5 days post-fertilization (rostral left, dorsal up). The yolk sac, Y, is autofluorescent. The major GFP-expressing divisions of the nervous system are labelled; Key: Tel, telencephalon; TeO, optic tectum; HB, hindbrain; SC, spinal cord; R, retina.
The 12kb eno2 promoter is expressed in rod photoreceptors
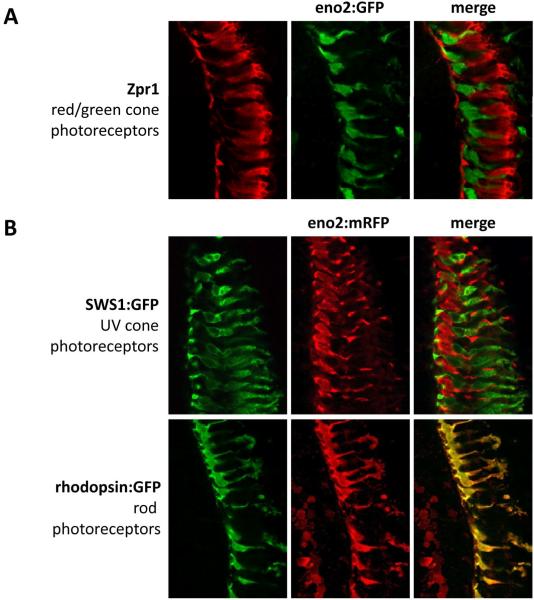
Finally, we sought to establish which population of photoreceptors expresses the 12kb eno2 promoter. First, we carried out double label immunofluorescence using an antibody to Zpr1, a marker of red/green photoreceptors (figure 6A). We saw no overlap in the expression pattern of Zpr1 and the eno2:egfp transgene, strongly suggesting that the 12kb eno2 promoter is not active in red/green cones. We next exploited the ability to examine GFP and mRFP expression simultaneously in double transgenic zebrafish, by crossing Tg(eno2:mrfp)Pt407 zebrafish with published zebrafish lines expressing GFP in either UV cone [25] or rod [16] photoreceptors (the lines were a kind gift from Dr. Shoji Kawamura, University of Tokyo). In double transgenic Tg(sws1:egfp)/Tg(eno2:mrfp)Pt407 retina, we did not see co-expression of GFP and mRFP in UV cones, suggesting that the eno2 promoter is not expressed in these cells (figure 6B). However, identical patterns of GFP and mRFP expression were seen in the photoreceptor layer of double transgenic Tg(rhodopsin:egfp)/Tg(eno2:mrfp)Pt407 zebrafish. These data show that the 12kb eno2 promoter is expressed in rod photoreceptors (figure 6B).
Figure 6. Expression of the 12kb eno2 promoter in photoreceptors of transgenic zebrafish.
A: Double indirect immunofluorescence and confocal microscopy were employed to detect zpr1 immunoreactivity (a marker of red and green-sensitive cone photoreceptors; red; left panel) and eno2:egfp expression (green; middle panel) in cryosections derived from 12dpf Tg(eno2:egfp)Pt404 zebrafish.
B, C: Tg(eno2:mrfp)Pt407 zebrafish were crossed with Tg(sws1:egfp) or Tg(rhodopsin:egfp) zebrafish that express GFP in either UV cone or rod photoreceptors respectively. GFP and mRFP were detected by double indirect immunofluorescence and confocal microscopy in larvae at 12dpf. Each set of images shows GFP (left, green); mRFP (middle, red); overlay (right).
Discussion
Our previous analysis suggested that the 12kb eno2 promoter fragment lacks elements involved in the full spatial expression pattern of eno2 in the adult brain; for example, the endogenous eno2 transcript, but not the eno2:egfp transgene, was expressed in cerebellar granule cells [2]. It is unclear whether the 12kb eno2 promoter exactly mirrors expression of the endogenous eno2 gene in the retina. During retinal development from 2 – 12 dpf, the temporal and spatial expression patterns of endogenous eno2 mRNA and the eno2:egfp transgene were very similar, although it was not possible to examine cells by double labeling in the absence of a specific antibody to zebrafish γ-enolase. A point of interest is that endogenous murine γ-enolase expression is restricted to cone photoreceptors in the mouse photoreceptor layer [22], whereas the 12kb eno2:egfp construct is exclusively expressed in rods in the zebrafish photoreceptor layer and RNA in situ hybridization analysis suggested that endogenous zebrafish eno2 is also expressed in rod photoreceptors. This implies that there may be differences in the retinal regulation of zebrafish eno2 and murine ENO2, although it will be necessary to develop specific zebrafish γ-enolase antibodies to exclude the possibility that a restricted subset of cis-acting signals within the 12kb construct gives rise to a different retinal expression pattern from the endogenous gene. The 12kb eno2 promoter element is expressed at different levels between different types of neurons. For example, the intermediate GFP expression level that we saw in TH-immunoreactive amacrine cells from Tg(eno2:egfp)Pt404 zebrafish was similar to the pattern that we saw in dopaminergic pre-tectal neurons in the brain, and the robust GFP expression we observed in retinal ganglion cells was similar to our previous findings in motor and reticulospinal neurons. It is possible that features common to particular cell types, for example a shared neurochemical phenotype or the metabolic demands of supporting a long axonal projection, reflect similarities in transcriptional regulation that are mechanistically related to those dictating expression from the enolase-2 transgene construct. These could result in similar GFP expression levels in related cell types.
The eno2 promoter element is currently being used to construct transgenic models of neurological diseases by expressing human proteins associated with neuronal dysfunction and loss, in the zebrafish nervous system. The data presented here suggest that it will be important to examine the visual system of resulting transgenic animals for signs of abnormal function or histopathology. For example, apparent behavioral deficits in models of neurodegenerative disease might result from visual abnormalities rather than motor problems. There are well-characterized tests of visual function in the zebrafish [6, 7, 12], and it might be possible to deploy these tests for the dual purpose of full characterization of phenotypic abnormalities in transgenic models, and development of assays for downstream applications. The high GFP expression level seen in rod photoreceptors and retinal ganglion cells in Tg(eno2:egfp)Pt404 zebrafish suggest that pathological examination of the retina might also be desirable in models constructed using this promoter. This may be especially true in instances where characteristic protein deposition is an expected histopathological abnormality.
The pattern of fluorescent reporter gene expression in the retinas of Tg(eno2:egfp)Pt404 and Tg(eno2:mrfp)Pt407 zebrafish encompasses several different cell types residing in different layers of the retina. Consequently, these transgenic lines might be useful for studies aiming to understand mechanisms of retinal lamination or neural connectivity, since multiple neuronal populations can be simultaneously visualized in relation to one another. The generation of double transgenic animals expressing mRFP and GFP may allow examination of the development of connectivity between adjacent or functionally related neuronal populations, for example by using Tg(eno2:mrfp)Pt407 zebrafish to label ganglion cells and other transgenic lines expressing GFP to delineate bipolar or amacrine cell subgroups.
Acknowledgements
We are grateful to the following colleagues for generously sharing reagents: Dr. Shoji Kawamura (University of Tokyo; Tg(sws1:egfp) and Tg(rhodopsin:egfp) lines); Dr. Neal Copeland (NCI, Frederick, MA; DY380 cells); Dr. Roger Tsien (University of California, San Diego; mRFP gene). This work was supported by research grants from: the University of Pittsburgh Competitive Medical Research Fund and the Pittsburgh Institute for Neurodegenerative Diseases.
References
- [1].Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bai Q, Garver JA, Hukriede NA, Burton EA. Generation of a transgenic zebrafish model of Tauopathy using a novel promoter element derived from the zebrafish eno2 gene. Nucleic Acids Res. 2007;35:6501–6516. doi: 10.1093/nar/gkm608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bai Q, Mullett SJ, Garver JA, Hinkle DA, Burton EA. Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 2006;1113:33–44. doi: 10.1016/j.brainres.2006.07.057. [DOI] [PubMed] [Google Scholar]
- [4].Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bilotta J, Saszik S. The zebrafish as a model visual system. Int J Dev Neurosci. 2001;19:621–629. doi: 10.1016/s0736-5748(01)00050-8. [DOI] [PubMed] [Google Scholar]
- [6].Branchek T. The development of photoreceptors in the zebrafish, brachydanio rerio. II. Function. J Comp Neurol. 1984;224:116–122. doi: 10.1002/cne.902240110. [DOI] [PubMed] [Google Scholar]
- [7].Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, Dowling JE. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc Natl Acad Sci U S A. 1995;92:10545–10549. doi: 10.1073/pnas.92.23.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Catalano AE, Raymond PA, Goldman D, Wei X. Zebrafish dou yan mutation causes patterning defects and extensive cell death in the retina. Dev Dyn. 2007;236:1295–1306. doi: 10.1002/dvdy.21148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Doerre G, Malicki J. Genetic analysis of photoreceptor cell development in the zebrafish retina. Mech Dev. 2002;110:125–138. doi: 10.1016/s0925-4773(01)00571-8. [DOI] [PubMed] [Google Scholar]
- [10].Dowling JE. Organization of vertebrate retinas. Invest Ophthalmol. 1970;9:655–680. [PubMed] [Google Scholar]
- [11].Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- [12].Easter SS, Jr., Nicola GN. The development of vision in the zebrafish (Danio rerio) Dev Biol. 1996;180:646–663. doi: 10.1006/dbio.1996.0335. [DOI] [PubMed] [Google Scholar]
- [13].Fadool JM, Dowling JE. Zebrafish: a model system for the study of eye genetics. Prog Retin Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goldman D, Hankin M, Li Z, Dai X, Ding J. Transgenic zebrafish for studying nervous system development and regeneration. Transgenic Res. 2001;10:21–33. doi: 10.1023/a:1008998832552. [DOI] [PubMed] [Google Scholar]
- [16].Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S. Visualization of rod photoreceptor development using GFP-transgenic zebrafish. Genesis. 2002;34:215–220. doi: 10.1002/gene.10155. [DOI] [PubMed] [Google Scholar]
- [17].Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- [18].Link BA, Fadool JM, Malicki J, Dowling JE. The zebrafish young mutation acts non-cell-autonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127:2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- [19].Malicki J, Driever W. oko meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell-cell interactions of the retinal neuroepithelial sheet. Development. 1999;126:1235–1246. doi: 10.1242/dev.126.6.1235. [DOI] [PubMed] [Google Scholar]
- [20].Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- [21].Marangos PJ, Schmechel DE. Neuron specific enolase, a clinically useful marker for neurons and neuroendocrine cells. Annu Rev Neurosci. 1987;10:269–295. doi: 10.1146/annurev.ne.10.030187.001413. [DOI] [PubMed] [Google Scholar]
- [22].Rich KA, Zhan Y, Blanks JC. Migration and synaptogenesis of cone photoreceptors in the developing mouse retina. J Comp Neurol. 1997;388:47–63. [PubMed] [Google Scholar]
- [23].Senut MC, Gulati-Leekha A, Goldman D. An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J Neurosci. 2004;24:7663–7673. doi: 10.1523/JNEUROSCI.2281-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Seth A, Culverwell J, Walkowicz M, Toro S, Rick JM, Neuhauss SC, Varga ZM, Karlstrom RO. belladonna/(Ihx2) is required for neural patterning and midline axon guidance in the zebrafish forebrain. Development. 2006;133:725–735. doi: 10.1242/dev.02244. [DOI] [PubMed] [Google Scholar]
- [25].Takechi M, Hamaoka T, Kawamura S. Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 2003;553:90–94. doi: 10.1016/s0014-5793(03)00977-3. [DOI] [PubMed] [Google Scholar]
- [26].Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- [27].Tsujimura T, Chinen A, Kawamura S. Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc Natl Acad Sci U S A. 2007;104:12813–12818. doi: 10.1073/pnas.0704061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Epps HA, Hayashi M, Lucast L, Stearns GW, Hurley JB, De Camilli P, Brockerhoff SE. The zebrafish nrc mutant reveals a role for the polyphosphoinositide phosphatase synaptojanin 1 in cone photoreceptor ribbon anchoring. J Neurosci. 2004;24:8641–8650. doi: 10.1523/JNEUROSCI.2892-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wei X, Luo Y, Hyde DR. Molecular cloning of three zebrafish lin7 genes and their expression patterns in the retina. Exp Eye Res. 2006;82:122–131. doi: 10.1016/j.exer.2005.05.009. [DOI] [PubMed] [Google Scholar]
- [30].Yu CJ, Gao Y, Willis CL, Li P, Tiano JP, Nakamura PA, Hyde DR, Li L. Mitogen-associated protein kinase- and protein kinase A-dependent regulation of rhodopsin promoter expression in zebrafish rod photoreceptor cells. J Neurosci Res. 2007;85:488–496. doi: 10.1002/jnr.21157. [DOI] [PubMed] [Google Scholar]
- [31].Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]