Abstract
The size of an organ is largely determined by the number of cells it contains, which in turn is regulated by two opposing processes: cell proliferation and cell death, but it is generally not clear how cell proliferation and cell death are coordinated during development. Here we characterize the zebrafish dou yanmi234 mutation that results in a dramatic reduction of retinal size and a disruption of retinal differentiation and lamination. The retinal size reduction is caused by increased retinal cell death in a non-cell-autonomous manner during early development. The phenotypic defect in dou yanmi234 arises coincident with the onset of retinal neurogenesis and differentiation. Interestingly, unlike many other small eye mutations, the mutation does not increase the level of cell death in the brain, implying the brain and retina utilize different mechanisms to maintain cell survival. Identification and further study of the dou yan gene will enhance our understanding of the molecular mechanisms regulating retinal cellular homeostasis, i.e. the balance between cell proliferation and cell death.
Keywords: retina, dou yan, retinal lamination, cell death, small eye
INTRODUCTION
The development of multicellular organisms from a fertilized, one-cell egg requires constant and dynamic coordination between cell proliferation, cell differentiation, and cell death. Cell proliferation and differentiation provide the building blocks for ontogenesis, whereas programmed cell death eliminates inappropriate cells and plays an indispensable role in sculpting tissues and organs (Jacobson et al., 1997). The balance between cell proliferation and cell death determines the total number of cells in a given tissue or organ, such as the vertebrate retina, and consequently its size. The size of the vertebrate retina appears to be largely regulated by intrinsic factors, as demonstrated by the preservation of the small eye size of Ambystoma punctatum even when transplanted to a large and fast-growing Ambystoma tigrinum host, and vise versa (Stone, 1930). The intrinsic mechanisms that regulate the balance between cell proliferation and death during retinal development are unknown. Cell proliferation and death occur both before and during retinal neurogenesis in all vertebrates (Donovan and Dyer, 2005; Vecino et al., 2004; Linden et al., 1999; Robinson, 1988; Sengelaub et al., 1986; Potts et al., 1982; Biehlmaier et al., 2001; Cole and Ross, 2001), suggesting that early developmental stages are critical in regulating the size of the differentiated retina.
In the past, genetic and molecular analyses of animal models and human patients have revealed a number of genes that play a role in the control of retinal size. Some of these genes are involved in cell proliferation, such as Rb, Rab11-FIP4, Mcm5, and XOptx2 (Friend et al., 1987; Muto et al., 2006; Ryu et al., 2005; Zuber et al., 1999). Other genes affect cell differentiation, such as shh, pitx3, and dzd (Neumann and Nuesslein-Volhard, 2000; Wang et al., 2005; Shi et al., 2005; Perkins et al., 2005). Still others are necessary for the survival of retinal cells at early development stages, such as mikre oko, oval, and elipsa, or for the survival of retinal cells in adults, such as pde6b and TULPs (Doerre and Malicki, 2001, 2002; Reme et al., 1998; Pittler and Baehr 1991; Hagstrom et al., 1998). While the list of genetic disorders that affect retinal size continues to grow, it is unclear how the functions of these genes are coordinated to achieve retinal cellular homeostasis.
Here we report the isolation of the zebrafish dou yanmi234 mutant line. Phenotypic characterization of the line indicates that the mutation reduces eye size and disrupts retinal lamination during embryonic development. We show that the reduction in eye size is the result of extensive retinal cell death rather than reduced cell proliferation. Interestingly, the dou yan mutation does not increase cell death in the brain, even though the retina is originally derived from an anterior region of the brain neuroepithelium. We conclude that the product of the dou yan gene is essential specifically for retinal cell survival.
RESULTS
dou yanmi234 mutation causes a small eye phenotype
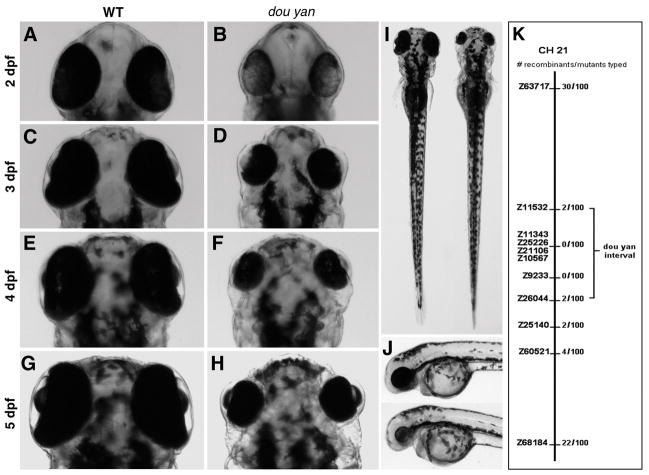
We isolated a recessive zebrafish mutant line that displays dramatic reduction in eye size in homozygous embryos. The genetic locus is named dou yan (meaning “beady eyes”) and the allele is mi234 (a serial number of the mutation isolated from a Michigan mutagenesis screen). A bulk segregant mapping analysis revealed that the dou yan gene is located between markers Z11532 and Z26044 on chromosome 21 (Fig. 1K). Prior to 28 hpf, the eye size phenotype is not distinguishable between wildtype and dou yanmi234 mutant embryos (data not shown). At 36 hpf, the eyes of the mutant embryos start to appear smaller than their wildtype siblings. However, genotyping by the size of the eyes is only about 80% accurate at this stage, likely due to developmental variations among individual embryos. By 48 hpf, the small eye phenotype becomes evident in 100% of the mutants. At this stage, the diameter of the mutant eyes along the anterior-posterior (AP) axis is about 70% of the diameter of wildtype eyes (Fig. 1A and B). The size difference between mutant and wildtype eyes increases over time. By 5 dpf, the mutant eyes are only about 50% of the diameter of the wildtype eyes along the AP axis (Fig. 1. A–H). Histological analysis indicates that the small eye phenotype can be primarily attributed to the small size of the mutant retinas (Fig. 2. A). In addition, we observed about 6% and 23% reduction in lens diameter at 48 hpf and 4 dpf, respectively (Fig. 2. B). Comparing to the 30% and 50% reduction in the eye size as a whole at the same stages, lens reduction is a secondary cause of the eye reduction. Because the retinal pigment epithelium (RPE) is only a single layer of flat cells, the RPE does not influence the diameter of the eye, even though it also shrinks and maintains a tight contact with the retina in the mutants (Fig. 2. A). Because retinal neurogenesis starts at about 28 hpf with the differentiation of the first retinal ganglion cells (Hu and Easter, 1999), the timing of the onset of the small eye phenotype suggests that the dou yanmi234 mutation does not affect the genes that are involved in specification of the eye field, formation of the optic primoridium, or early morphogenesis of the eye.
Figure 1.
The zebrafish dou yanmi234 mutation causes reduction in eye size. A–H. The small eye phenotype in the mutants is evident at 2 dpf. The mutant eyes remain smaller than the wildtype eyes at subsequent developmental stages. I. The dou yanmi234 mutation does not lead to reduction of body size in mutants. In a dorsal view at 4 dpf, the body of this mutant fish (right) is slightly longer than that of a wildtype sibling (left). J. A lateral view of a wildtype (top) and mutant fish (bottom) at 3 dpf shows no paracardiac edema in dou yan mutants. K. A bulk segregant mapping analysis revealed that the dou yan gene localizes to a genomic region that is flanked by markers Z11532 and Z26044 on chromosome 21.
Figure 2.
The dou yanmi234 mutation disrupts cytoarchitecture of the retina and reduces the retinal size more severely than it does to the lens. A. Histological analyses sections of wildtype and mutant retinas at 2, 3, 4 and 5 dpf reveals that retinal lamination is severely disrupted in the mutant eyes. Arrows indicate the darkly stained pyknotic nuclei in the mutant retina. Arrowheads indicate the disrupted plexiform layers. B. The isolated lenses of mutant embryos are 6% and 30% smaller than their counterparts of wildtype embryos at 48 hpf and 4 dpf, respectively. The scale bars represent 100 μm.
A reduction of organ size is apparent only in the eyes from an external examination. The body size of the dou yanmi234 mutants is comparable to that of wildtype, and on average, the mutants are actually 2–3% longer than their wildtype siblings at 4 dpf (Fig. 1. I.). In addition, unlike other retinal patterning mutants such as nok, where the body axis is curled up and a paracardiac edema develops as early as 36 hpf (Malicki et al., 1996), the body axis of the dou yanmi234 mutant is as straight as the wildtype fish (Fig. 1. I) with no development of paracardiac edema (Fig. 1. J). However, abdominal edema and paraocular edema start to develop at 5 dpf (data not shown). The dou yanmi234 mutant embryos are viable up to approximately ten days. It is uncertain what the cause of their eventual death is. Together, our observations suggest that the dou yan gene is essential for maintaining the proper size of the retina during early development.
dou yanmi234 mutation preferentially affects retinal structure
To determine if the dou yanmi234 mutation also affects the cellular architecture of the retina, we examined JB4 plastic sections of mutant and wildtype embryos between 2 dpf and 5 dpf and found that retinal lamination is disrupted by the mutation. In wildtype retinas, retinal lamination develops between 2 dpf and 5 dpf (Fig. 2, top panel). As they differentiate, retinal neurons are segregated into three distinct nuclear laminae according to their identity, and the plexiform layers containing neuronal processes separate the layers containing cell bodies. In the mutant retinas, the plexiform layers are segregated into islands of tangled neuronal processes (Fig. 2. arrowheads). In addition, many darkly stained pyknotic nuclei are observed in the mutant retinas, especially at 3 dpf (Fig. 2. arrows). In contrast, by examination of the isolated 4 dpf brains, we found that the size of the mutant brains is similar to that of the wildtype brains, except for an apparent reduction of the optic tectum (Fig. 3A–B). At 10 dpf, the mutant brain of surviving fish is similar to the mutant brain at 4 dpf, though slightly smaller (Fig. 3C–D). The histological analysis confirmed that the cellular architecture, such as the pattern of grey and white matter, of the forebrain and hindbrain of the mutant is similar to that of wildtype (Fig. 3E–F, I–J). The cross sections of 4 dpf wildtype and mutant through the midbrain region confirm that the optic tectum is almost missing in the mutant embryos (Fig. 3G–H). However, in the same cross sections, other brain structures, such as the posterior commissure, posterior tuberculum, and the dorsal thalamus are present and appear to be healthy in the mutant embryos (Fig. 3G–H). We then analyzed the midbrain region at an early stage, 3 dpf. It revealed that the cells in the potential optic tectum region do not organize properly as in wildtype (Fig. 3. K–L). We infer that the overall function of the nervous system, except for the visual system, is not dramatically disrupted in the mutants. This inference is supported by the observation of a normal touch response in mutant embryos (data not shown). Thus, we conclude that the dou yanmi234 mutation specifically disrupts the development of the retina and the optic tectum, or at least the effect is much more profound in the visual system than elsewhere in the nervous system.
Figure 3.
The dou yanmi234 mutation preferentially affects the development of the optic tectum more than it does to the other regions of the brain. A–B. Lateral views of the isolated wildtype (A) and mutant brains (B) at 4 dpf confirm that most regions of the mutant brain are only slightly smaller than that of the wildtype brain. However, the optic tectum is drastically reduced in the mutant embryos (arrows). The scale bars in A–B represent 100 μm. C–D. The arrows indicate the optic tectum regions of brains isolated from wildtype (C) and surviving mutants (D) at 10 dpf. The scale bars in C–G represent 100 μm. E–F. Cross-sections of the heads of wildtype (E) and mutant (F) embryos at the optic nerve region indicate similar forebrain size at 4 dpf, with the dorsal-ventral diameter of the mutant forebrains (arrows) being about 94% of that of the wildtype brains. The overall patterning of the gray (cell bodies and nuclei) and white (neuronal processes) matter is similar between the mutant and the wildtype embryos. Arrowheads indicate the optic nerves, which serve as a reference point for comparable section planes. Scale bars represent 50 μm. G–H. The cross sections of 4 dpf wildtype (G) and mutant (H) heads through the midbrain region reveal that the optic tectum (OT) is almost missing in the comparable region in the mutant embryos (arrows, H). However, the posterior commissure (POC), posterior tuberculum (PT), and the dorsal thalamus (DT) are comparable between mutant and wildtype embryos. I–J. The cellular patterning of the hindbrain mutant (J) is similar to that of the wildtype brain (I) at 4 dpf. Arrows indicate the hindbrains. The otic capsules (OC) were selected as a reference point for comparable section planes. K–L. At 3 dpf, cross sections through the midbrain revealed the optic tectum in wildtype embryos (K). However, no optic tectum-like structure is evident in the mutant embryos (L) at the corresponding region (arrow). The posterior commissure (POC) is selected as a reference point for matching comparison. Scale bars in G–L represent 50 μm.
dou yanmi234 mutation causes extensive cell death in the retina but not in the brain
Because pyknotic nuclei are a hallmark of dead cells, the observation of pyknotic nuclei in the mutant retinas suggests that many retinal cells are dying prematurely during the early stages of retinal neurogenesis and differentiation (Fig. 2. arrows). To confirm this prediction, we performed acridine orange vital staining of living fish at 3 dpf. Acridine orange preferentially stains the cell nuclei of dying and dead cells (McCall and Peterson, 2004). Numerous round and condensed nuclei showing strong acridine orange staining were found in the mutant retinas (Fig. 4. D–F, arrows) but not in the wildtype retinas (Fig. 4. A). These pyknotic nuclei are concentrated in central regions of retina and are scattered throughout the thickness of the retinal tissue. However, in the proliferative germinal zone at the ciliary margin of the mutant retinas, most cell nuclei display a normal, elongated morphology (Fig. 4D–F, arrowheads). At 3 dpf this region contains undifferentiated, proliferating cells, while the central retina predominately consists of differentiating cells, suggesting that the extensive retinal cell death caused by dou yanmi234 primarily affects post-mitotic cells. This inference is consistent with the observation that retinal size is not affected and cell death is not observed in dou yan mutants until after 28 hpf, when retinal cells begin to differentiate (Hu and Easter, 1999). In contrast, the brains of mutant fish display the same level of cell death found in the wildtype brains (Fig. 4G–H). We also did not observe an elevation in cell death in the mutant lenses by acridine orange nuclear staining (Fig. 4A–F). In the mutant embryos, the RPE tightly juxtaposes the retina, and also displays a reduction in size. However, due to the monolayer structure and the low density of the cell nuclei in the RPE, our observation of acridine orange staining could not yield conclusive evidence regarding whether the level of cell death in the RPE is increased by the mutation. We conclude that the dou yan mutation causes extensive cell death in the retina but not in the brain or lens.
Figure 4.
The dou yanmi234 mutation causes extensive cell death in the retina but not in the brain or lens. A–F. Staining with acridine orange (AO, green), which preferentially stains the nuclei of dead or dying cells, indicates extensive cell death in the central regions of the mutant retinas (D–F, arrows) compared with the wildtype retinas (A–C). The cell nuclei are stained with TO-PRO-3 (red). Cell nuclei with an elongated shape are found at the marginal zone of the mutant retina (D–F, arrowheads), similar to the germinal zone of proliferating cells found at the retinal margin in wildtype retinas at this stage (A–C). In the lenses (asterisks), no increase in cell death was observed in dou yan. However, non-specific acridine orange staining is sometimes observed in non-nuclear regions in the lens. Scale bars represent 20 μm. G and H. Cell nuclei that are heavily stained with acridine orange are mainly found in the mutant retina (arrows) but not in the brain (asterisks). The scale bars represent 50 μm.
dou yanmi234 mutation causes retinal cell death in a non-cell-autonomous manner
Cell death can result either from signals within the cell or from external environmental triggers. To determine whether the increased cell death observed in dou yanmi234 mutant retinas is cell-autonomous or non-cell-autonomous, we created genetic mosaic embryos by performing transplantation analysis with biotinylated dextran-labeled donor cells. In living cells, the dextran label diffuses throughout the cytoplasm of donor cells and highlights their morphology (Fig. 5B, arrows). However, when donor cells die, the dextran molecules become concentrated in cell debris and appear as small, brightly stained foci (Fig. 5E, arrowheads). Consequently, the staining pattern reflects donor cell viability in the host retinas. To quantify viability of wildtype donor cells following transplantation, we counted the numbers of living and dead clones in both the brains and the retinas of wildtype and mutant hosts. The results indicate that wildtype donor cells transplanted into mutant retinas die at significantly higher rates than wildtype cells transplanted into wildtype retinas (Fig. 5. G, H). We cannot exclude the possibility that some closely spaced dead clones might have been mistakenly scored as one clone, so our counts of dead clones might actually underestimate the actual numbers. Interestingly, the dou yanmi234 mutation does not increase the level of cell death in the brain as much as in the retina (Fig. 5. G, H). Furthermore, the reciprocal experiment of transplanting mutant cells into wildtype embryos revealed that the wildtype retinal environment promotes the survival of mutant cells (Fig. 5. H–N). We thus conclude that the dou yanmi234 mutation causes retinal cell death in a non-cell-autonomous manner. These observations indicate that the mutation specifically alters the microenvironment in the retina and reduces retinal cell survival.
Figure 5.
The dou yanmi234 mutation causes non-cell-autonomous retinal cell death. A–C. Clones of dextran-labeled wildtype donor cells (red) in the wildtype retina form vertical columns of cells with a differentiated morphology that span all three retinal nuclear layers (arrows). The cell nuclei were stained with YO-PRO-1 (green); C is the merged image of A and B. D–F. Extensive punctate staining of donor cells (red, arrowheads in E) was observed in the mutant retinas, indicating that many wildtype donor cells were already dead at the time of fixation. Some donor cells appeared intact in the mutant retina, but they had an elongated morphology typical of undifferentiated retinal progenitors (E, arrow). Pyknotic nuclei heavily stained with YO-PRO-1 are indicated with arrows in D. G. A histogram illustrates the percentages of dead wildtype donor clones in four host conditions: in wildtype brains, in wildtype retinas, in mutant brains, and in mutant retinas. The total number of clones counted in each condition is indicated in parentheses. Comparison of the percentages of dead donor cell clones with the Chi-Square test indicates that the increase in dead wildtype donor clones in the mutant retina, but not the brain, is statistically significant. H–J. Healthy-appearing dou yan mutant donor cells (red, arrows) develop properly in the wildtype retina and can localize to all three retinal cellular layers. The cell nuclei are counterstained with YO-PRO-1 (green). K–M. Large amount of punctate staining foci derived from mutant donor cells (arrowheads) were observed in mutant host retinas, indicating that many of the donor cells were already dead at the time of fixation. N. A histogram illustrates the percentages of dead clones of mutant donor cells in four host conditions: in wildtype brains, in wildtype retinas, in mutant brains, and in mutant retinas. The total numbers of clones counted in each condition are indicated in parentheses. Comparison of the percentages of dead donor cell clones with the Chi-Square test indicates that statistically more mutant donor cells survived in the wildtype retinas than in the mutant retinas at the time of examination. The scale bars represent 10 μm.
dou yanmi234 mutation does not inhibit cell proliferation in the retina
To determine whether or not cell proliferation is also affected by the dou yanmi234 mutation and consequently contributes to the small eye phenotype, we analyzed the level of active cell proliferation. We stained the retinas with anti-phospho-Histone H3 (pH3) antibody, which specifically stains cell nuclei in M-phase (Fig. 6A–F). We calculated the average number of positive cells per 3,600 μm2 of retinal section, and we found no significant difference between the relative numbers of pH3-positive cells in mutant retinas vs. the wildtype retinas at 40, 48, and 72 hpf (Fig. 6. G, with P values of 0.7129, 0.3729, and 0.6734, respectively). BrdU labeling at 49 to 51.5 hpf verified the existence of active DNA replication in mutant and wildtype retinas (Fig. 6. H and I). Thus, the dou yanmi234 mutation appears not to grossly affect cell proliferation dynamics in the zebrafish retina. This conclusion is consistent with the observation that the mutant retinas do not appear smaller before 28 hpf when the retinal primordium consists entirely of proliferating progenitor cells.
Figure 6.
The dou yanmi234 mutation does not disrupt cell proliferation in the retina. A–F. M-phase nuclei, visualized by anti-phospho-Histone 3 antibody (PH3, red), are observed in both wildtype (A–C) and mutant retinas (D–F) at 3 dpf. All nuclei are stained with YO-PRO-1 iodide (YO, green). C is the merged image of A and B. F is the merged image of D and E. G. The average number of PH3-positive cells per 3,600 μm2 in mutant retinas is similar to wildtype retinas at 40 hpf, 48 hpf, and 72 hpf. The error bars denote the standard error of the mean. H. BrdU labeling of the wildtype retina between 49 hpf and 51.5 hpf reveals proliferating cells in most regions of the retina. Arrowheads indicate BrdU-negative basal regions in the wildtype retina. I. BrdU labeling of dou yan mutants between 49 hpf and 51.5 hpf firms that many retinal cells are actively synthesizing DNA. The scale bars represent 20 μm.
Distinct classes of retinal cells are specified in dou yan
We next investigated whether or not the dou yanmi234 mutation affects retinal cell specification. We performed immunohistochemical analysis with cell type-specific markers (Wei et al., 2006) to determine whether red/green double cones, interplexiform cells, rods, Muller cells, ganglion cells, and lin7-positive bipolar cells differentiated in the mutant retinas. We found that all of these cell–specific markers appeared in dou yan mutant retinas although the labeled cells were greatly reduced in number compared with wildtype retinas at the same developmental stages (Fig. 7A–T). We did not find any cells that simultaneously expressed two markers that are normally expressed in different retinal cell classes, but the morphologies of the labeled cells were markedly different from their wildtype counterparts. For example, photoreceptors in the mutants did not have the typical elongated shape (Fig. 7. B and F), and Müller cells did not extend processes to the apical and basal ends of the retina (Fig. 7. K and O). We next performed a similar analysis with embryos at 78 hpf. We found that a considerable number of ganglion cells are specified in the mutants at 78 hpf. However many late-appearing cell types such as photoreceptors are not specified in most of the embryos at this stage (Fig. 8). Interestingly, the Lin7 is distributed throughout the mutant retinas, with an elevated expression level in the ganglion cell patch (Fig. 8. F). This broader expression pattern of Lin7 resembles its expression at around 48 hpf in wildtype (Wei et al., 2006a). These results suggest that the cell differentiation process is delayed in the mutant retinas. Taken together, we conclude that although cell specification apparently occurs in dou yan mutant retinas, the level of cell specification is reduced and cell differentiation is abnormal. Further investigation is needed to determine to what extent the extensive retinal cell death and delayed cell differentiation process contribute to the reduction in the amount of cells for each distinct retinal cell type.
Figure 7.
Distinct types of retinal cells are specified in dou yanmi234 mutants, but they fail to differentiate properly. A–H. At 5 dpf, green/red double cones (G/R, B and F) and Lin7-positive bipolar cells (Lin7, C and G) are observed in both mutant (E–H) and wildtype retinas (A–D). YO-PRO staining was used to visualize the entire population of cell nuclei (nuclei, A and E). D and H are merged images. I–P. Interplexiform cells (IPC, J and N) and Müller cells (Muller, K and O) are specified in both wildtype (I–L) and mutant retinas (M–P) at 5 dpf. Cell nuclei were visualized by YO-PRO staining (I and M). L and P are merged images. Q–T. Zn8 (red) expressing ganglion cells are specified in both wildtype (Q–R) and mutant retinas (S–T). Cell nuclei were visualized with YO-PRO staining (green). Scale bars represent 20 μm.
Figure 8.
Retinal cell specification is delayed by the dou yan mutation. A–D. At 78 hpf in wildtype retinas, Lin7 expression (blue) is concentrated in the bipolar cell body region and the plexiform layers. The ganglion cells stained with zn8 antibody (red) localize to the basal region of the retinas. The actin distribution as revealed by phalloidin staining highlights the plexiform layers. E–G. In dou yan mutant retina at 78 hpf, Lin7 (blue) is expressed in the entire retina with increased accumulation in the basal retinal region. This expression pattern is similar to Lin7 expression in wildtype retinas at 48 hpf (Wei et al., 2006a). A patch of retinal cells that express zn8 antigen is noticeable at the basal region of the retina in the mutants. I–L. Muller cells (blue) and green/red double cones (G/R, red) are properly differentiated in the wildtype retinas at 78hpf. M–P. In most of the mutant retinas at 78 hpf, only a few cells display positive expression of Muller cell marker carbonic anhydrase (arrows, blue) and double cone marker Zpr1 was not detectable. Q–T. Interplexiform layer cells (IPC, red) are present in wildtype retinas (Q and R) but not in the mutant retina (S and T) at 78 hpf. Scale bars represent 20 μm.
dou yanmi234 mutation does not affect retinal epithelial polarity
A number of retinal patterning mutations with phenotypes similar to dou yan (e.g., lack of retinal lamination) have been identified in earlier studies (Wei and Malicki, 2002; Jensen and Westerfield, 2004; Horne-Badovinac et al., 2001; Malicki et al., 2003), and all of these mutations disrupt epithelial polarity of the retina at early stages of development. To determine if retinal epithelial polarity is also affected by the dou yanmi234 mutation, we compared the distribution pattern of the apical polarity markers ZO-1, adherens junction-associating actin bundles, and M-phase nuclei in wildtype and mutant retinas. Because the mutant embryos cannot be phenotyped prior to 28 hpf, we first examined 48 hpf embryos. As shown in Figure 9, wildtype and mutant retinas do not differ in terms of the localization of these apical markers. To examine apical epithelial polarity at earlier stages, we randomly chose 20 embryos at 27 hpf from an incross of dou yan heterozygous parents (from which we would expect five, or one quarter, of the embryos to be homozygous mutant), and we found no differences in the localization or distribution of apical markers among the 20 embryos (data not shown). Thus the dou yanmi234 mutation does not appear to affect the polarity of the retinal or brain epithelia at early stages of development.
Figure 9.
The dou yanmi234 mutation does not affect apical epithelial polarity. A–C. Apical localization of adherens junctions in the retina (arrows) and brain (arrowheads) of the 48 hpf wildtype embryos is visualized by the staining patterns of ZO-1 (red) and adherens junction-associated actin bundles (A, green, arrows). D–F. ZO-1 (red) and adherens junction-associated actin bundles (green) localize properly to the apical surface of the retina (arrows) and brain (arrowheads) in dou yanmi234 embryos at 48 hpf. G–L. In 48 hpf retinas, the M-phase nuclei as visualized with anti-phosphorylated-Histone H3 antibody (PH3, green) localize to the apical regions of the retinas of mutant (J–L) and wildtype (G–I) embryos. Cell nuclei were labeled with TO-PRO (red). Scale bars represent 20 μm.
DISCUSSION
We have isolated and characterized a zebrafish mutation, dou yanmi234, whose primary phenotypic characteristic is failure of the eyes to grow after the onset of retinal neurogenesis. The mutation also disrupts retinal lamination and causes extensive retinal cell death.
A small eye phenotype can emerge at a variety of developmental stages depending on the genes that are affected. Loss-of-function of transcription factors that are required for early specification and morphogenesis of the eye, such as Pax6, Six3, Rx1, Lhx2, Tll and Optx2, can lead to early onset of eyeless or small eye phenotypes that appear before retinal neurogenesis begins (Perron et al., 1998; Zuber et al., 2003; Hill et al., 1991). On the other hand, defects in genes that are required for the survival of differentiated retinal cells would be predicted to interfere with retinal growth and may even cause shrinkage of the eye. In zebrafish, early morphogenesis of the eyecup is completed before 24 hpf and the first differentiated retinal ganglion cells appear at approximately 28 hpf (Schmitt and Dowling, 1994; Hu and Easter, 1999). Because the dou yanmi234 small eye phenotype becomes evident only after the onset of retinal differentiation, we suggest that the product of the dou yan gene is required for the differentiation and/or survival of retinal cells. This is consistent with the finding that cell proliferation in the undifferentiated retina is not affected in dou yan mutants, and the germinal zone at the ciliary margin of the mutant embryonic retina appears normal and contains proliferating retinal progenitor cells at least up to 3 dpf.
Retinal cell death might target either the entire retinal cell population or affect only certain classes of retinal cells. In human retinitis pigmentosa patients, the genetic defects initially affect only photoreceptors and lead to their degeneration (Wang et al., 2001). In zebrafish, the mikre oko, oval, pob, and fleer mutations cause cell death in photoreceptors (Doerre and Malicki, 2001; Taylor et al., 2005; Doerre and Malicki, 2002) and the chiorny mutation causes death in certain subtypes of amacrine cells (Avanesov et al., 2005). Unlike these mutations, the dou yan mutation leads to massive cell death throughout the central retina suggesting that the survival of all retinal cell classes is affected. We also showed that the cell survival defect in the dou yan mutation is non-cell-autonomous, so the mutation likely disrupts an aspect of the microenvironment in the retina that promotes cell-survival, perhaps a secreted neurotrophic factor.
Previous mutagenesis screens and reverse genetic approaches have identified several genes that are important in controlling the size of zebrafish eye: Rab11-FIP4, Mcm5, PITX3, small heart, no tectal neuron, and dazed (Muto et al., 2006; Babb et al., 2005; Perkins et al., 2005; Shi et al., 2005; Matsuda and Mishina, 2004; Ryu et al., 2005; Yuan and Joseph, 2004). These genes map to the chromosome regions different form the one in which the dou yan gene resides. Furthermore, in addition to the small eye phenotype, malfunction of these gene products also causes other defects. For example, disruption of Rab11-FIP4 and Mcm5 genes leads to extensive cell death not only in the retina but also in the brain (Ryu et al., 2005; Muto et al., 2006); the small heart mutation causes malformation of the heart and a curled body axis (Yuan and Joseph, 2004); disruption of the PITX3 gene function, which is required for lens development, likely has an indirect effect on retinal development as a consequence of disrupted interactions between lens and retina (Shi et al., 2005; Yamamoto and Jeffery, 2000); in no tectal neuron, the small eye phenotype does not appear until after 48 hpf, much later than that in dou yanmi234 (Matsuda and Mishina, 2004); and the dazed small eye mutant has normal retinal lamination (Perkins et al., 2005). These phenotypic differences suggest that the dou yan gene regulates the size of the eyes through a pathway different from the pathways that concern other identified genes.
In addition to a small eye, the dou yanmi234 mutation also causes a defect in retinal lamination. Five other zebrafish genes that are necessary for retinal lamination have been identified: nok, moe, N-cad, has, and yng (Wei and Malicki, 2002; Jensen and Westerfield, 2004; Horne-Badovinac et al., 2001; Malicki et al., 2003; Gregg et al., 2003). The first four mutations affect four different epithelial apical-basal polarity genes, which are expressed in the retina and in other tissues as well. Consequently, developmental defects in these mutants are not restricted to the retina, as manifested by a curled body axis, circulation defects and malformation of the digestive system (Horne-Badovinac et al., 2003). While the yng mutation causes retinal patterning defects, it does not disrupt retinal epithelial polarity (Link et al., 2000). In contrast, the dou yanmi234 mutant phenotype is restricted to the visual system: morphogenesis of the body is unaffected and embryos exhibit a normal touch response. Furthermore, the extensive retinal cell death observed in dou yanmi234 is not seen in nok, moe, N-cad, has, and yng mutants. And finally, unlike these other retinal patterning mutations, the dou yanmi234 mutation does not affect the apical-basal polarity of the retinal and brain epithelia. These phenotypic differences indicate that the loss of retinal lamination in dou yanmi234 mutants is mechanistically distinct from an apical-basal epithelial polarity-related patterning defect.
The retinal patterning defect in dou yanmi234 might be explained by extensive retinal cell death during early development, which could affect retinal lamination in one or more ways. First, cell death may constantly change the cellular environment in the retina, perhaps resulting in inconsistent migration and/or differentiation signals to the newly born retinal cells, leading them to aberrant destinations. Second, premature death of retinal cells in dou yan mutants might affect the normal cell-cell interactions required for the stabilization of proper retinal laminar structure. Third, lamination defect may occur because too few retinal neurons produce processes to form contiguous plexiform layers. These mechanisms are not mutually exclusive, and further work is needed to determine to what extent these or other factors contribute to the lamination defects observed in dou yanmi234.
Finally, it is noteworthy that a disruption of retinal lamination is not observed in all zebrafish mutants that have a small eye phenotype. Some mutations such as archie, round eye, out of sight, niezerka, and out of bounds, produce a downsizing of the retina and cause no retinal lamination defects (Fardool et al., 1997; Malicki et al., 1996; Vihtelic and Hyde; 2002). These observations indicate that the retinal size control and cellular pattern formation can be regulated independently. However, the dou yan mutation apparently affects both processes. Like dou yanmi234, another small eye mutation, barely started, also affects retinal pattern formation (Vihtelic and Hyde, 2002). However, in barely started the nuclei of retinal cells are enlarged and irregular, which is not observed in dou yan. Further molecular characterization of these mutations will help us better understand the complex mechanisms by which retinal size and lamination is generated and maintained.
EXPERIMENTAL PROCEDURES
Mutagenesis screen and fish care
Adult male zebrafish from an outbred wildtype colony (Mi fish line) were mutagenized with N-ethyl-N-nitrosurea (ENU, Sigma) according to established procedures (Mullins et al., 1994) followed by a three-generation breeding scheme to identify zygotic recessive mutations. The mutagenesis and screening was performed at the University of Michigan. The dou yan mi234 mutation was identified morphologically by the small size of the eyes in F3 mutant embryos at 3 days post-fertilization (dpf). Zebrafish embryos were raised at 28.5 °C in E3 embryo rearing medium (Westerfield, 2000) until the desired developmental stages and used for histological analysis. All procedures conformed to the University of Michigan and University of Pittsburgh standards for use and care of animals in research.
Mapping analysis of the dou yan gene
To identify the chromosome region in which the dou yan gene resides, we utilized the Mutant Mapping Facility at the University of Louisville for a bulk segregant mapping analysis. Heterozygous map fish were obtained by crossing Mi heterozygous fish with wildtype TU fish. One hundred mutant embryos from crosses between a pair of heterozygous map fish were collected. DNA isolated from these mutant embryos and their parent fin clips were used for the analysis, following the standard procedure at the Mapping Facility in Louisville (http://www.biochemistry.louisville.edu/zfmapping/index.html).
Histology and immunohistochemistry
To determine the cellular pattern of the dou yan mutant retina, we fixed embryos in 4% paraformaldehyde in 1× phosphate buffered saline (PBS) at RT overnight then dehydrated and embedded the tissues in JB4 resin (Polysciences, Inc.) following the manufacturer’s protocol. Embryos were sectioned at 4 μm thickness and stained with 1% methylene blue/1% azure II and observed and photographed with an AH2 Olympus microscope.
To determine if retinal cells are specified normally in dou yan mutants, we used immunohistochemical analysis with cell specific markers as described previously (Wei et al., 2006b). The following cell type-specific markers were used: zpr1 antibody for the Zpr1 antigen, which is specifically expressed in red/green double cones (ZFIN, 1:150 dilution); rabbit anti-Lin7 antibodies for Lin7-expressing cells (Wei et al., 2006a, 1:400 dilution); zn8 antibody against the Zn8 antigen that is expressed in retinal ganglion cells (ZFIN, 1:100 dilution); rabbit anti-carbonic anhydrase for Muller glia (gift from Dr. Paul Linser; 1:400 dilution); anti-phosphorylated-Histone H3 antibody for M-phase nuclei (Upstate, Cat.# 06-570); anti-tyrosine hydroxylase for interplexiform cells (Chemicon; 1:100 dilution). The nuclei were stained with TO-PRO-3 iodide or YO-PRO-1 iodide (Molecular Probes, Cat.# T-3605 and Cat.# Y-3603; at 1:500 dilution). Actin was visualized with Alexa Fluor 488-conjugated phalloidin (Molecular Probes, Cat. # A-12397, 1:500 dilution). ZO-1 was visualized with monoclonal mouse anti-ZO-1 antibody (Zymed, 1:200 dilution).
Acridine orange staining
To analyze cell death in the dou yan mutant retinas, we used acridine orange staining. Briefly, live fish embryos of both wildtype and mutant were stained with acridine orange (50 μg/ml in E3 embryo rearing medium) at room temperature for 30 minutes. Stained embryos were washed three times with PBS at room temperature for 30 minutes each. Embryos were then fixed overnight with 4% paraformaldehyde at RT. Fixed embryos were infiltrated with 40% sucrose in 1× PBS and cryosectioned at 30 μm, mounted in slow-fade mounting medium (Vector Laboratory) and observed under a BioRadMRC1024 laser scanning confocal microscope.
BrdU labeling of the retinas
The wildtype and mutant embryos were pulse labeled with 10 mM BrdU in E3 with 15% DMSO at 28 °C for 2.5 hours between 49 and 51.5 hours postfertilization (hpf). The embryos were then fixed with 4% paraformaldehyde at RT for two hours, followed by a DNA denaturation treatment with 2N HCl at 37 °C for 1 hour. The embryos were then processed for immunostaining as described above, with anti-BrdU antibody (Sigma Cat#: B2531).
Quantification of mitotic cells
Phospho-Histone H3 (pH3) is a marker of M-phase mitotic cells (Adams RR et al., 2001). Because the wildtype retinas are larger and contain more total cells than the mutant retinas, we normalized the number of pH3-positive cells to retinal area in wildtype and mutant retinas. In brief, we counted pH3-positive retinal cells in five sections from five embryos each at 40, 48, and 72 hpf. The retinal regions counted were contoured and measured with MetaMorph v6.1, and we calculated the average number of pH3-positive cells per 3,600 μm2 of retinal area. Statistical significance of the differences between mutant and wildtype was assessed with Student’s T-test.
Blastomere transplantation and mosaic analysis
For genetic mosaic analysis, we labeled wildtype donor embryos by injecting 5% purified lysine fixable biotin-conjugated dextran (Molecular Probes; Cat: BDA-10,000) at the 1- and 2-cell stages. The biotinylated dextran solution (in 0.2M KCl) was purified by filtration with Microcon Centrifugal Filters (Millipore, Cat #: 42420). Labeled blastomeres were transplanted to wildtype and mutant embryos as described (Wei & Malicki, 2002). The host embryos were raised to 3 dpf and fixed in 4% paraformaldehyde at room temperature for two hours. Fixed embryos were cryosectioned at 30 μm and stained with Cy3-conjugated avidin (Jackson InnunoResearch) for donor cells and with YO-PRO-1 for the cell nuclei. The distribution and morphology of the donor cells in wildtype and dou yan mutant embryos was examined and photographed with a BioRadMRC1024 laser scanning confocal microscope. The numbers of dead and live clones were counted according to the following criteria: 1) a group of vertically arranged, intact cells labeled with biotinylated dextran was counted as one live retinal clone; 2) in the brain, a group of spatially contiguous, intact labeled cells with no other labeled cells in the vicinity was counted as one clone; 3) a cluster of punctate biotinylated dextran staining, in which staining particles were less than one cell width apart was counted as one dead clone. Statistical comparisons were assessed with the Chi-Square test.
Acknowledgments
The authors thank Cindy Stone for performing JB4 plastic sections. The work was supported by an NIH core grant (5P30EY008098-17; PI, Robert Hendricks), NIH R01 EY015509 (P.A.R.), and the following funds to X.W.: NIH R01EY016099, University of Pittsburgh School of Medicine startup fund, Research to Prevent Blindness Career Development Award, and a UPMC Health System Competitive Medical Research Grant. DG was supported by a Michigan Life Sciences Corridor Grant, MEDC-38. Anne Catalano is also an undergraduate student in the Department of Biological Sciences at Carnegie Mellon University.
References
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–80. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanesov A, Dahm R, Sewell WF, Malicki JJ. Mutations that affect the survival of selected amacrine cell subpopulations define a new class of genetic defects in the vertebrate retina. Dev Biol. 2005;285:138–155. doi: 10.1016/j.ydbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Babb SG, Kotradi SM, Shah B, Chiappini-Williamson C, Bell LN, Schmeiser G, Chen E, Liu Q, Marrs JA. Zebrafish R-cadherin (Cdh4) controls visual system development and differentiation. Dev Dyn. 2005;233:930–945. doi: 10.1002/dvdy.20431. [DOI] [PubMed] [Google Scholar]
- Biehlmaier O, Neuhauss SC, Kohler K. Onset and time course of apoptosis in the developing zebrafish retina. Cell Tissue Res. 2001;306:199–207. doi: 10.1007/s004410100447. [DOI] [PubMed] [Google Scholar]
- Cole LK, Ross LS. Apoptosis in the developing zebrafish embryo. Dev Biol. 2001;240:123–142. doi: 10.1006/dbio.2001.0432. [DOI] [PubMed] [Google Scholar]
- Doerre G, Malicki J. A mutation of early photoreceptor development, mikre oko, reveals cell-cell interactions involved in the survival and differentiation of zebrafish photoreceptors. J Neurosci. 2001;21:6745–6757. doi: 10.1523/JNEUROSCI.21-17-06745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerre G, Malicki J. Genetic analysis of photoreceptor cell development in the zebrafish retina. Mech Dev. 2002;110:125–138. doi: 10.1016/s0925-4773(01)00571-8. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Brockerhoff SE, Hyatt GA, Dowling JE. Mutations affecting eye morphology in the developing zebrafish (Danio rerio) Dev Genet. 1997;20:288–295. doi: 10.1002/(SICI)1520-6408(1997)20:3<288::AID-DVG11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Friend SH, Horowitz JM, Gerber MR, Wang XF, Bogenmann E, Li FP, Weinberg RA. Deletions of a DNA sequence in retinoblastomas and mesenchymal tumors: organization of the sequence and its encoded protein. Proc Natl Acad Sci U S A. 1987;84:9059–9063. doi: 10.1073/pnas.84.24.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg RG, Willer GB, Fadool JM, Dowling JE, Link BA. Positional cloning of the young mutation identifies an essential role for the Brahma chromatin remodeling complex in mediating retinal cell differentiation. Proc Natl Acad Sci USA. 2003;100:6535–40. doi: 10.1073/pnas.0631813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom SA, North MA, Nishina PL, Berson EL, Dryja TP. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Rebagliati M, Stainier DY. A cellular framework for gut-looping morphogenesis in zebrafish. Science. 2003;302:662–665. doi: 10.1126/science.1085397. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Developmental Biology. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jensen AM, Westerfield M. Zebrafish mosaic eyes is a novel FERM protein required for retinal lamination and retinal pigmented epithelial tight junction formation. Curr Biol. 2004;14:711–717. doi: 10.1016/j.cub.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Linden R, Rehen SK, Chiarini LB. Apoptosis in developing retinal tissue. Prog Retin Eye Res. 1999;18:133–165. doi: 10.1016/s1350-9462(98)00020-2. [DOI] [PubMed] [Google Scholar]
- Link B, Fadool1 J, Malicki J, Dowling J. The zebrafish young mutation acts non-cell-autonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127:2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Malicki J, Jo H, Pujic Z. Zebrafish N-cadherin, encoded by the glass onion locus, plays an essential role in retinal patterning. Dev Biol. 2003;259:95–108. doi: 10.1016/s0012-1606(03)00181-7. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Mishina M. Identification of chaperonin CCT gamma subunit as a determinant of retinotectal development by whole-genome subtraction cloning from zebrafish no tectal neuron mutant. Development. 2004;131:1913–1925. doi: 10.1242/dev.01085. [DOI] [PubMed] [Google Scholar]
- McCall K, Peterson JS. Detection of apoptosis in Drosophila. Methods Mol Biol. 2004;282:191–205. doi: 10.1385/1-59259-812-9:191. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Muto A, Arai KI, Watanabe S. Rab11-FIP4 is predominantly expressed in neural tissues and involved in proliferation as well as in differentiation during zebrafish retinal development. Dev Biol. 2006 doi: 10.1016/j.ydbio.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Perkins BD, Nicholas CS, Baye LM, Link BA, Dowling JE. dazed gene is necessary for late cell type development and retinal cell maintenance in the zebrafish retina. Dev Dyn. 2005;233:680–694. doi: 10.1002/dvdy.20375. [DOI] [PubMed] [Google Scholar]
- Perron M, Kanekar S, Vetter ML, Harris WA. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199:185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci U S A. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts RA, Dreher B, Bennett MR. The loss of ganglion cells in the developing retina of the rat. Brain Res. 1982;255:481–486. doi: 10.1016/0165-3806(82)90013-x. [DOI] [PubMed] [Google Scholar]
- Reme CE, Grimm C, Hafezi F, Marti A, Wenzel A. Apoptotic cell death in retinal degenerations. Prog Retin Eye Res. 1998;17:443–464. doi: 10.1016/s1350-9462(98)00009-3. [DOI] [PubMed] [Google Scholar]
- Robinson SR. Cell death in the inner and outer nuclear layers of the developing cat retina. J Comp Neurol. 1988;267:507–515. doi: 10.1002/cne.902670405. [DOI] [PubMed] [Google Scholar]
- Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci U S A. 2005;102:18467–18472. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Dowling J. Early eye morphogenesis in thezebrafish, Brachydanio rerio. Journal of Comparative Neurology. 1994;344:532–542. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- Sengelaub DR, Dolan RP, Finlay BL. Cell generation, death, and retinal growth in the development of the hamster retinal ganglion cell layer. J Comp Neurol. 1986;246:527–543. doi: 10.1002/cne.902460409. [DOI] [PubMed] [Google Scholar]
- Shi X, Bosenko DV, Zinkevich NS, Foley S, Hyde DR, Semina EV, Vihtelic TS. Zebrafish pitx3 is necessary for normal lens and retinal development. Mech Dev. 2005;122:513–527. doi: 10.1016/j.mod.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Stone LS. Heteroplastic transplantation of eyes between larvae of two species of Amblystoma. J Exp Zool. 1930;55:193–261. [Google Scholar]
- Taylor MR, Kikkawa S, Diez-Juan A, Ramamurthy V, Kawakami K, Carmeliet P, Brockerhoff SE. The zebrafish pob gene encodes a novel protein required for survival of red cone photoreceptor cells. Genetics. 2005;170:263–73. doi: 10.1534/genetics.104.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecino E, Hernandez M, Garcia M. Cell death in the developing vertebrate retina. Int J Dev Biol. 2004;48:965–974. doi: 10.1387/ijdb.041891ev. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Zebrafish mutagenesis yields eye morphological mutants with retinal and lens defects. Vision Res. 2002;42:535–540. doi: 10.1016/s0042-6989(01)00261-9. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen Q, Zhao K, Wang L, Traboulsi EI. Update on the molecular genetics of retinitis pigmentosa. Ophthalmic Genet. 2001;22:133–154. doi: 10.1076/opge.22.3.133.2224. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–5113. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Wei X, Luo Y, Hyde DR. Molecular cloning of three zebrafish lin7 genes and their expression patterns in the retina. Exp Eye Res. 2006a;82:122–131. doi: 10.1016/j.exer.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wei X, Zou J, Takechi M, Kawamura S, Li L. Nok plays an essential role in maintaining the integrity of the outer nuclear layer in the zebrafish retina. Exp Eye Res. 2006b doi: 10.1016/j.exer.2005.10.030. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 4. Univ. of Oregon Press; Eugene: 2000. [Google Scholar]
- Yamamoto Y, Jeffery WR. Central role for the lens in cave fish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- Yuan S, Joseph EM. The small heart mutation reveals novel roles of Na+/K+-ATPase in maintaining ventricular cardiomyocyte morphology and viability in zebrafish. Circ Res. 2004;95:595–603. doi: 10.1161/01.RES.0000141529.48143.6e. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Perron M, Philpott A, Bang A, Harris WA. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]