Abstract
Recently, the oxidoreductase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), has become a subject of interest as more and more studies reveal a surfeit of diverse GAPDH functions, extending beyond traditional aerobic metabolism of glucose. As a result of multiple isoforms and cellular locales, GAPDH is able to come in contact with a variety of small molecules, proteins, membranes, etc. that play important roles in normal and pathologic cell function. Specifically, GAPDH has been shown to interact with neurodegenerative disease-associated proteins, including the β-amyloid precursor protein (AβPP). Studies from our laboratory have shown significant inhibition of GAPDH dehydrogenase activity in Alzheimer disease (AD) brain due to oxidative modification. Although, oxidative stress and damage is a common phenomenon in AD brain, it would seem that inhibition of glycolytic enzyme activity is merely one avenue in which AD pathology affects neuronal cell development and survival, as oxidative modification can also impart a toxic gain-of-function to many proteins, including GAPDH. In this review, we examine the many functions of GAPDH with respect to AD brain; in particular, GAPDH’s apparent role(s) in AD-related apoptotic cell death is emphasized.
Keywords: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-amyloid precursor protein (AβPP), amyloid β-peptide [Aβ(1-40)/(1-42)], Alzheimer disease (AD), hypometabolism, oxidative stress, apoptosis
Introduction
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzymes (EC 1.2.1.12) are a family of abundantly expressed oxidoreductases known for their role in glucose metabolism. Research on the role of mammalian GAPDH in various biological signaling pathways over the last couple of decades has increased interest in this classical glycolytic enzyme. Traditionally, GAPDH was used as a model, or control, in protein and gene structural- and catalytic mechanism-related studies, as well as a standard in Northern and Western blots because of its high degree of gene and protein sequence conservation across species [1–3]. However, recent research suggests that GAPDH possesses highly diverse, non-glycolytic functions, as its expression and activity are affected by multiple factors. In particular, GAPDH has been reported to bind DNA and RNA[1, 4–6], regulate transcription [7], possess kinase/phosphotransferase activity [8–15], catalyze microtubule formation and polymerization [16–22], facilitate vesicular transport [23], and bind integral membrane ion pumps associated with Ca2+ release [24, 25], as well as interact with a number of small molecules, including tumor necrosis factor (TNF)-α ribozymes [26], glutathione (GSH) [27, 28], p53 [29–31], and nitric oxide (NO) [32–34]. Additionally, since GAPDH also interacts with disease-associated proteins, like huntingtin [35] and the β-amyloid precursor protein (AβPP) [36]. GAPDH non-glycolytic activity is of great interest to neurodegenerative disease research, specifically in brains of subjects with Alzheimer disease (AD).
AD is the most common age-related neurodegenerative disorder affecting elderly populations over the age of 65. In familial cases, however, AD pathology can present as early in life, due to autosomal dominant mutations in the AβPP protein and presenilin genes 1 and 2 [37–40]. Characteristic symptoms of AD include progressive memory loss, declining cognition, impaired linguistic function, and dementia. Pathologically, the brain exhibits extensive synapse and neuronal cell loss, as well as the appearance of neurofibrillary tangles (NFT) and senile plaques, associated with widespread oxidative stress and damage [41–47]. Studies from our laboratory demonstrate that GAPDH is subject to many different types of oxidative modification in AD brain, which drastically affect its structure and function, including S-glutathionylation [28, 34, 48], S-nitrosylation [49–51], and direct or indirect reaction with reactive oxygen species (ROS) [52–55]. Moreover, a recent study by Petrak, et al. [56] found that the frequency with which GAPDH was shown differentially expressed in all 2D-gel electrophoresis (2-DE)-based experiments in human and rodent tissues was ~18%, securing it a spot on the top 15 list of most frequently reported differentially expressed proteins. Taking into consideration the multitude of functions GAPDH can carry out under normal conditions, in addition to a variety of subcellular locations, it is not surprising that this enzyme is so often affected by disease pathology. In this review we will discuss the different roles of GAPDH, and how those roles are affected by and/or contribute to neurodegenerative disease; our focus will be on understanding the function(s) of GAPDH in AD.
2.0 GAPDH Structure
GAPDH (EC 1.2.1.12) is a member of the dehydrogenase enzyme family, also known as oxidoreductases, and is essential to glucose metabolism. This glycolytic enzyme is ubiquitously expressed in both prokaryotes and eukaryotes, and comprises ~10–20% of the total cellular protein content [1]. All mammalian GAPDH genes, including human, have a complex genetic organization; gene expression studies reveal that human GAPDH has only one functional gene located on chromosome 12 (Gene ID: 2597), but around 150 or more pseudogenes with similar sequence identity [57–61]. Further investigation reveals the presence of a single GAPDH mRNA species in different tissues [57, 59–61].
GAPDH exists as a homologous tetramer (~150 kDa), monomer, and dimer, of which there is little information at present [8, 62]. The tetrameric form is located mainly in the cytoplasm and is comprised of four chemically identical subunits, O, P, Q, and R (Fig. 1), each approximately 37 kDa, with three asymmetric interfaces between subunits P, Q, and R [63, 64]. The monomeric form (~37 kDa) is localized to the nucleus, mainly during cell proliferation, and consists of 335 amino acids [65, 66]. Each GAPDH monomer contains two binding domains: an N-terminal NAD+-binding domain and a C-terminal catalytic, or glyceraldehyde-3-phosphate (G3P)-binding domain. The NAD+-binding domain contains amino acid residues 1-151, forming the main-chain, while the G3P-binding domain consists of residues 315-335, forming the C-terminal helix. The NAD+-binding domain, or coenzyme domain, also contains the well-known Rossmann fold structure necessary for binding dinucleotides. Interestingly, a number of active-site amino acids (human analogs, Asp-35, Cys-152, His-179, Thr-182, Thr-211, Arg-234, Tyr-314, Tyr-320) that are directly involved in binding of the G3P and NAD+ nicotinamide moiety and responsible for catalytic activity are part of a highly conserved sequence, that, on the phylogenic scale, maintains a highly conserved 3D-structure (Fig. 2) [2]. Conversely, the structure surrounding the adenine and phosphate binding sites vary across species [2, 3, 67, 68].
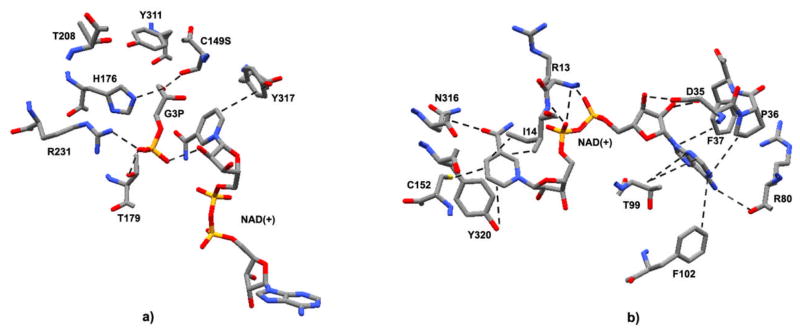
Figure 1. GAPDH Structure.
Ribbon view of the human placental GAPDH, subunits O, P, Q, and R. GAPDH active-sites clefts are indicated by arrows on each of the asymmetric subunits, P, Q, and R. Color codes: Green, subunit Q; Yellow, subunit O; Blue, subunit P; Red, subunit R; Gray, backbone carbon loops connecting secondary structure successions; Gray molecular structures, NAD(+). Data were obtained from the RCSB protein data bank as entry code 1u8f.pdb, DOI: 10.1107/S0907444905042289 [2]. This figure was drawn using the DeepView Swiss-PDB viewer program, version 4.0.
Figure 2. GAPDH active-site.
a) View of Bacillus stearothermophilus GAPDH active-site catalytic residues with a Cys149Ser substitution and occupied by D-G3P and NAD(+). Residues shown are those directly involved in G3P binding (human amino acid analogs discussed in Section 2.0). The structure was drawn in CPK format: Gray, carbons atoms; Blue, nitrogen atoms; Red, oxygen atoms; Orange, phosphorous atoms. Dotted lines represent potential electrostatic or non-polar interactions between G3P, NAD(+), and amino acids; residues T208 and Y311 are normally directly involved with binding of the C149 sulfate anion, however, the C149S substitution replaces the SH-group with a hydroxyl, as shown. Data were obtained from the RCSB protein data bank as entry code 1NQO.pdb, DOI: 10.1074/jbc.M211040200 [77]. This figure was drawn using the DeepView Swiss-PDB viewer program, version 4.0. b) View of human placental GAPDH active-site catalytic residues with no substitution and occupied only by NAD(+). Residues shown are those directly involved in NAD+ binding. The structure was drawn in CPK format: Gray, carbons atoms; Blue, nitrogen atoms; Red, oxygen atoms; Orange, phosphorous atoms; Yellow, sulfur atoms. Dotted lines represent potential electrostatic or non-polar interactions between NAD(+) and specified amino acids. Data were obtained from the RCSB protein data bank as entry code 1u8f.pdb, DOI: 10.1107/S0907444905042289 [2]. This figure was drawn using the DeepView Swiss-PDB viewer program, version 4.0.
Under normal conditions, the NAD+ molecule situates in a cleft formed by the coenzyme domain S-loops from adjacent subunits along the R-axis (Fig. 1); NAD+ binds to the C-terminal edge of a parallel β-sheet, flanking it between two α-helices. As the nicotinamide end of NAD+ faces the interior of the GAPDH tetramer, a cleft opens, revealing a spacious active-site pocket (Fig. 1) [2]. The catalytic domain, on the other hand, consists of twisted, eight-stranded parallel β-sheets connected by α-helices on one side, while the other side of this β-sheet forms extensive contacts with the β-sheet of an adjacent subunit (Fig. 1) [2, 69, 70]. In addition, the amino acid Val-240 (human analog) of each subunit falls in a disallowed region of Ramachandran plots, which is a characteristic of all GAPDH enzymes [70–74].
3.0 GAPDH Active-site Function
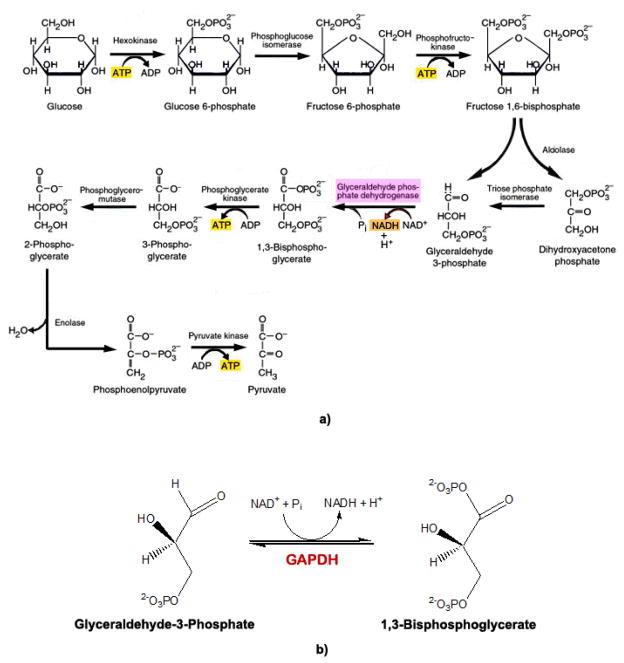
GAPDH is most well-known for its role in glycolysis, catalyzing the reversible phosphorylation of G3P to 1,3-bisphosphoglycerate (BPG) using NAD+ as a cofactor (Fig. 3). The first step of this mechanism involves nucleophilic attack by the sulfhydryl group of Cys-152 on the carbonyl of G3P, resulting in the formation of a hemiacetal (Fig. 4). The hemiacetal intermediate is then oxidized to a high energy thioester, by hydride transfer to NAD+, possibly facilitated by His-179 in the catalytic domain [75, 76]. This high energy thioester is then attacked by nucleophilic, inorganic phosphate (Pi) to form BPG (Fig. 4) [68, 69, 77]. Mutational studies in Bacillus stearothermophilus show replacement of Cys-149 (human analog Cys-152) with a Ser thiol significantly reduces GAPDH activity, while substitution with Ala completely inactivates the enzyme [77, 78].
Figure 3.
a) Glycolysis. Schematic representation of glucose metabolism, with reactions 5–10 completed twice (not shown) (adapted from [224]). b) Overall GAPDH reaction. The sixth reaction in glycolysis involves conversion of glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate (BPG) by GAPDH, using NAD+ as a cofactor.
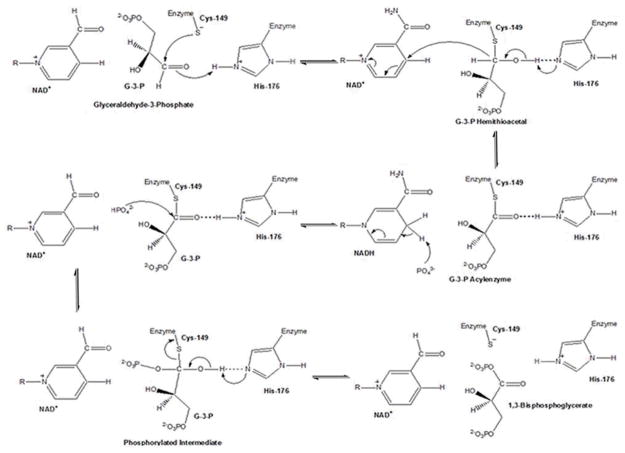
Figure 4. GAPDH Reaction.
The reaction catalyzed by GAPDH involves two processes: Oxidation of the glyceraldehyde-3-phosphate (G3P) aldehyde to a carboxylic acid by NAD+, and joining of the G3P carboxylic acid and the NAD+ orthophosphate to form 1,3-bisphosphoglycerate (BPG). The first step of this mechanism involves nucleophilic attack by the Cys-149 (human analog Cys-152) SH-group on the carbonyl of G3P, resulting in formation of a hemithioacetal. Acting as an acid catalyst, His-176 (human analog His-179) facilitates hemiacetal development by forming an ion pair with the Cys-149 SH-group, enhancing Cys-149 reactivity, and favoring the nucleophilic attack of GAPDH by the thiolate [76]. A hydride transfer to NAD+ oxidizes the hemiacetal intermediate to a high energy thioester, facilitated by His-176, now acting as a base catalyst during the oxidation step [75, 76]. His-176 stabilizes the hemithioacetal intermediate by hydrogen bonding the hemiacetal hydroxyl group to the Nε of its imidazole ring, while the acylenzyme intermediate, is stabilized by hydrogen bonding with the protonated imidazole Nε of His-176 [76]. Finally, the thioester undergoes nucleophilic attack by an inorganic phosphate (Pi), facilitated again by His-176 acting as an acid catalyst, to form BPG [75–77], which is then released from the ternary complex, along with NADH (adapted from [76]).
GAPDH has a very large active-site cavity in order to accommodate G3P, with its bulky phosphate ions, and the cofactor NAD+, located in a large cleft between the NAD+-and G3P-binding domains (Fig. 1). Structural analysis of bacterial and eukaryotic GAPDH reveals that there are two anion (i.e., phosphate) binding sites located in the catalytic domain that bind a Pi and the C3-phosphate of the substrate, G3P (Fig. 2b) [79]. Literature studies show that Arg-234 (bacterial analog Asp-231) of catalytic domain acts as a conformational switch that regulates G3P binding and release (Fig. 2a), as chemical modification of Arg-234 results in a 95% reduction in GAPDH activity [77, 79–81]. Moreover, in the catalytic domain, His-179 forms a hydrogen bond with human Cys-152 (Fig. 2a), which resides at the N-terminus of the first helix of the catalytic domain, where it acts as a base catalyst to facilitate hydride transfer [2]. In the NAD+-binding domain, nicotinamide carbonyls form hydrogen bonds with Asn-316 (human analog), while nicotinamide amines form intramolecular hydrogen bonds with its pyrophosphate groups in the N-terminal, glycine-rich, first helix loop of the Rossmann fold (Fig. 2b) [2]. Finally, a highly conserved Asp-35 residue forms two hydrogen bonds with the adenosine ribose moiety of NAD+, while a nonpolar interaction between NAD+ and Ile-14and Tyr-320 (human analogs) holds the nicotinamide ring in place (Fig. 2b) [2].
4.0 GAPDH Functional Diversity
Apart from a classical role in glycolysis, GAPDH also exhibits numerous non-glycolytic functions, in which the NAD+- and G3P-binding domains play a very important role. Analysis of such functions after proteolysis and competitive binding using NAD+ and polynucleotides indicates that the majority of GAPDH functional diversity is a direct result of the many different interactions that can occur within the NAD+-binding domain [1, 26, 82–85]. For example, the NAD+-binding domain Rossmann fold, essential to the dehydrogenase activity of GAPDH [2, 69, 70], is also important in the catalysis of aminoacyl tRNA synthesis by GAPDH [86], and may comprise the GAPDH nucleotide binding site for tubulin [87]. Moreover, GAPDH can act as a nuclear tRNA transport protein, whose activity is inhibited by NAD+ in a competitive manner [1, 88].
In addition, experimental evidence suggests that the catalytic G3P-binding domain is involved in regulation of GAPDH membrane binding and subcellular localization in mammalian cells. A study with human erythrocyte membranes shows that modification of G3P-binding site residues, Lys-191 and Lys-212, may determine GAPDH-membrane interactions [1, 89]. Moreover, amino acid sequence analysis of GAPDH reveals the presence of sequence motifs that are often used by mammalian cells to regulate the intracellular localization of a protein. One motif has the sequence KKVVK (residues 259-263), which is partially homologous to the nuclear localization signal (NLS), and the other is ALQNIJP (residues 202-208), which is partially homologous to a nuclear export domain [1, 90]. In contrast, other observations suggest GAPDH may not require such a NLS, since proteins as small as monomeric GAPDH do not require active transport [91–93]. Regardless of the mode of entry, monomeric GAPDH can be found in the nucleus of non-apoptotic cells [1, 65]. Based on this information, it can be suggested that the NAD+-binding domain is associated with GAPDH functional diversity, while the G3P-binding domain is responsible for GAPDH intracellular localization [1].
4.1 Nuclear/Perinuclear GAPDH Function
In general, non-glycolytic functions of GAPDH can be divided into nuclear, perinuclear, cytosolic, and membrane-related functions. As part of its nuclear and/or perinuclear activities, monomeric GAPDH interacts with DNA and RNA. Several studies report that GAPDH binds DNA [4] and exhibits DNA repair activity similar to a uracil DNA-glycosylase (UDG), a DNA repair enzyme that removes free uracils [1, 5, 6]. In vivo studies have also shown that suppression of nuclear UDG activity increases GAPDH mRNA levels and dehydrogenase activity [94]. GAPDH can also interact with the 5′-UTR and 3′-UTR regions of various mRNA sequences through its NAD+-binding domain [4]. A dose-response study with human parainfluenza virus shows that an increase in NAD+ decreases the ability of GAPDH to bind RNA by competitive inhibition [83]. Other nuclear/perinuclear functions of GAPDH include the ability to bind the Octomer binding protein -1 (Oct-1) involved in histone transcription [7], increase the activity of TNF-α ribozymes by interacting with their pyrimidine regions [26], and bind the dinucleoside phosphate, P1, P4-di(adenosine-5′) tetraphosphate (Ap4A) protein, which plays a significant role in DNA replication and repair (Section 5.1) [5, 95], among others (Table 1). Moreover, since the ratio of Ap3 to Ap4 changes in apoptosis, GAPDH is thought to modulate Ap4A protein functions at various stages of the cell-cycle [4, 5, 96].
Table 1.
GAPDH functional diversity
Cytosolic Functions:
|
Nuclear Functions:
|
Other Disease Involvement:
|
Interestingly, ROS has been shown to play an important role in modulating non-glycolytic nuclear/perinuclear functions of GAPDH. In HEK-293T cells expressed with human GAPDH, Hwang and coworkers [97] have shown that active-site cysteine residues (human analog Cys-152) of GAPDH are oxidized upon H2O2 treatment, forming cysteic acid and/or intramolecular disulfide bonds with Cys-156 depending on the extent of oxidative stress. Thus oxidized GAPDH interacts with heterodimeric RNA and DNA-binding proteins, 54kDa RNA-binding protein (p54nrb) and polypyrimidine tract-binding protein-associated splicing factor (PSF), enhancing topoisomerase-I activity. Involvement of GAPDH in modulation of topoisomerase-I activity during DNA replication and transcription, together with a multitude of other diverse nuclear/perinuclear GAPDH functions, strongly suggests GAPDH may also function as a transcription factor [97].
4.2 Cytosolic GAPDH Functions
As described above (Section 2.0), the most common GAPDH isoform found in the cytoplasm is tetrameric. In addition to its glycolytic activity, previous studies have confirmed numerous kinase and phosphotransferase activities elicited by cytosolic GAPDH, other than phosphorylation of G3P. First, dimeric GAPDH has been reported to associate with the tumor suppresser nucleoside diphosphate protein kinase, nm23 [8]. In order to elicit Ser/Thr phosphotransferase activity in human cells, nm23 must first complex with dimeric GAPDH, as nm23 alone shows no kinase activity [8]. Interestingly, the glycolytic activity of GAPDH was not inhibited by this interaction, suggesting that the NAD+- and/or G3P-binding domains are vital to GAPDH phosphotransferase/kinase function. In addition, mammalian GAPDH has been shown to phosphorylate the Hepatitis B virus (HBV) in a Ca2+-independent manner, opening new avenues investigating the role of GAPDH in viral pathogenesis [9].
Another facet of its phosphotransferase/kinase activity involves phosphorylation of GAPDH by various cellular kinases including, protein kinase C [10, 11], epidermal growth factor kinase [12], and Ca2+/calmodulin-dependant protein kinase II [13], among others (Table 1). The exact GAPDH amino acid modified by these kinases is unknown; however, preliminary studies suggest a Tyr as the site of phosphorylation [13, 98]. Furthermore, studies have shown that mammalian, tetrameric GAPDH was auto-phosphorylated in the presence of ATP and Mg2+ in a concentration-dependent manner and dephosphorylated in the presence of NAD+, NADH, and/or G3P [9, 14, 15]. Although, the role of GAPDH phosphorylation in normal cell function is unclear at present, it would appear that auto-phosphorylation is not necessarily a prerequisite to its other known phosphotransferase or kinase activities.
Beyond phosphorylation, cytosolic GAPDH also catalyzes microtubule formation and polymerization by binding the cytoskeletal protein tubulin [16–22], binds to the mitochondrial voltage-dependent anion channel protein (VDAC-1) [99], associates with small molecules like GSH (Section 5.1.1) [27, 28], p53 (Section 5.1.2) [29–31], NO (Section 5.1.3) [32–34], and interacts with proteins associated with neurodegenerative disease, such as the huntingtin protein in Huntington’s disease [35], the androgen receptor protein in spinocerebellar ataxia type-1 and spinobulbar muscular atrophy [100], and AβPP and amyloid β-peptide (Aβ) in AD (Table 1) [36, 101, 102], to be discussed in this review.
4.3 Membrane-associated GAPDH Activity
As part of its membrane-associated activities, earlier studies revealed that GAPDH binds to the human erythrocyte membrane [103] and is part of the phospholipid bilayer [1, 104]. This enzyme is also involved in vesicular transport from the Golgi apparatus to the endoplasmic reticulum (ER) [23], and has Ca2+-dependant fusogenic activity in human neutropil [105] and rat brain cytosol (Table 1) [106]. Previous studies demonstrated that fusogenic activity was dependent on Ser-234 [107], and inhibited by the substrate G3P. Interestingly, GAPDH fusogenic activity was inhibited by G3P, but not affected by the dehydrogenase inhibitor, koningic acid, as those GAPDH isoforms displayed fusogenic activity, but were void of dehydrogenase activity [106]. Furthermore, GAPDH has been shown to bind integral membrane proteins, such as the inositol-1,4,5-triphosphate receptor (IP3R) and sarcoplasmic reticulum Ca2+ (SERCA) pump (Table 1; Section 5.2) [24, 25], contributing to the regulation of intracellular Ca2+ levels.
5.0 GAPDH & Alzheimer Disease
GAPDH can undergo many different oxidative modifications, which influence its structure and activity. Active-site modifications include, GAPDH-transition metal complex formation [108], mono-ADP-ribosylation [50, 109], S-glutathionylation [28, 34, 48], S-nitrosylation by NO or other reactive nitrogen species (RNS) [49–51], and direct or indirect reaction with ROS, as measured by levels of protein carbonylation and modification by the lipid peroxidation product, 4-hydroxy-2-nonenal (HNE) [52–55]. In AD brain, such oxidative modifications are common, as numerous studies have shown oxidative stress and damage to be a major hallmark of AD pathology, precipitating the loss of neurons, synapses, and ultimately, normal brain function [41–47]. Studies conducted by our laboratory have revealed at least 42 proteins negatively affected by the shifting redox environment in AD brain, including GAPDH (Fig. 5) [48, 51, 52, 54, 55]. Furthermore, a study by Petrak, et al. [56] found that the frequency with which GAPDH was shown differentially expressed in all 2D-gel electrophoresis (2-DE)-based experiments in human and rodent tissues was ~18%, securing it a spot on the top 15 list of most frequently reported differentially expressed proteins. Considering the surplus of different GAPDH functions and locations within the cell, as well as the frequency with which it is differentially expressed, oxidative dysfunction of GAPDH may significantly contribute to loss of neuronal function and neurodegeneration in AD brain.
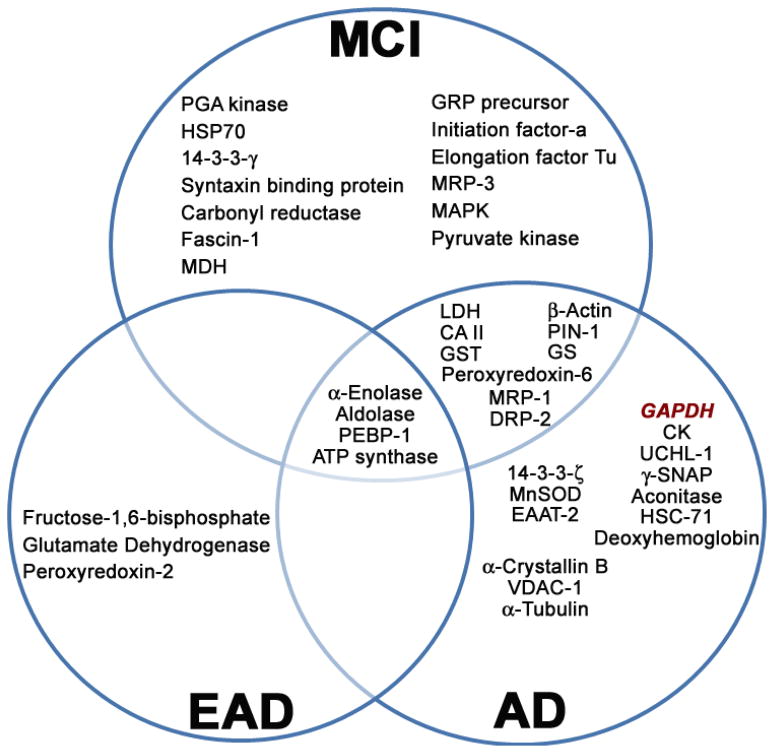
Figure 5. Proteins oxidatively modified in MCI, EAD, and AD brain identified by our laboratory [48, 51, 55, 225–231].
The inter-relationship of those proteins identified to be significantly modified by protein carbonylation, HNE- and 3-NT modification, as well as S-glutathionylation using redox proteomics on mild cognitive impairment (MCI), early AD (EAD), and AD human brain are shown. Abbreviations: PCG kinase, phosphoglycerate kinase; HSP-70, Heat-shock protein-70; MDH, Malate dehydrogenase; GRP precursor, Glucose-regulated protein precursor; MRP-1/MRP-3, Multidrug-resistant protein-1 or -3; MAPK, Mitogen-associated protein kinase; PEBP-1, phosphatidylethanolamine-binding protein-1; LDH, Lactate dehydrogenase; CAII, Carbonic anhydrase II; GST, Glutathione S-transferase; DRP-2, Dihydropyrimidinase-related protein-2; PIN-1, Peptidyl-prolyl cis/trans isomerase; GS, Glutamine synthetase; MnSOD, Manganese superoxide dismutase; EAAT-2, Excitatory amino acid transporter-2; VDAC-1, Voltage dependent anion channel-1; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; CK, Creatine kinase; UCHL-1, Ubiquitin carboxy-terminal hydrolase L-1; γ-SNAP, Soluble N-ethylmaleimide-sensitive factor attachment protein-γ; HSC-71, Heat-shock cognate-71.
5.1 GAPDH & Apoptosis
Though somewhat controversial, increasing evidence suggests apoptosis occurs in AD brain [110–116]. Apoptosis, involves a series of intracellular signaling events that ultimately lead to programmed cell death in response to relatively mild stimuli [117]. Tetrameric GAPDH stability is achieved by ionic interactions between positively charged NAD+ and negatively charged sulfate ions of active-site Cys residues. Modification of these cysteines decreases the tetramer’s stability, giving rise to monomers, dimers, and other denatured GAPDH products [62, 118], that individually elicit various levels of glycolytic function [119]. Interestingly, all GAPDH isozymes have been shown to be altered during apoptosis [120]. Numerous evidence supports the role of GAPDH in apoptosis, facilitating cell death induced by apoptotic stimuli and/or oxidative stress by nuclear translocation [30]. The first evidence of this relationship was observed in rat cerebellar granule and cortical neuronal cell cultures undergoing spontaneous apoptosis. In this study, it was found that monomeric GAPDH was over-expressed prior to apoptosis, while transfection with antisense GAPDH caused a significant reduction in apoptosis concomitant to reduction of GAPDH mRNA [121, 122]. However, even though these studies establish a role for GAPDH in apoptotic processes, how GAPDH participates is still a subject of much research.
While investigating mechanisms of GAPDH apoptotic function, studies have found that GAPDH acts as a pro-apoptotic protein after nuclear translocation. Analysis of cerebellar granule and cortical neuronal cell culture after cytosine arabinoside (AraC)-induced apoptosis showed increased levels of GAPDH in mitochondrial and crude nuclear fractions [121, 122]. Because treatment with antisense GAPDH completely blocked AraC-induced apoptosis, it is probable that GAPDH translocation functions as a potent initiator of apoptotic processes [121–123]. Interestingly, nuclear GAPDH accumulation was allied with a reduction in GAPDH UDG activity (Section 4.1), as glycolytic activity initially increased in response to AraC treatment and decreased with UDG activity [6, 120]. With respect to AD brain, these studies present two ways in which GAPDH could be involved in disease pathology: First, because GAPDH is a known transcription factor [7], nuclear translocation of cytosolic GAPDH could induce transcription of genes that mediate cell death. Second, suppression of GAPDH UDG activity after translocation would prevent DNA repair [5, 6, 94], thereby, increasing levels of damaged DNA, a common finding in apoptosis-related cell death. Nuclear translocation and accumulation of GAPDH also occurs with concomitant degradation of lamin B1, a nuclear membrane protein and caspase-3 substrate. Analysis of purified nuclei showed an increase in the levels of six GAPDH isoforms after AraC treatment, the greatest increase of which was found in more acidic isoforms [120]. Thus, the significance of acidic isozymes in neuronal apoptosis, in addition to the differential translocation behavior of different GAPDH isoforms, strongly suggests a key role for cytosolic GAPDH translocation in AD-related apoptotic processes.
A second way GAPDH is involved in apoptosis is through post-translational modification and small molecule/protein interactions, including Ap4A binding [5, 124], VDAC-1 binding [99], phosphorylation [8–15, 98], S-glutathionylation (Section 5.1.1), p53 binding (Section 5.1.2), S-nitrosylation (Section 5.1.3), and AβPP binding (Section 5.4). Ap4A is a part of the diadenosine oligophosphate (ApnA) signal-transduction family of molecules, which play a role in DNA replication and repair through binding cell membranes and the DNA polymerase-α complex [5, 95, 124]. Ap4A may act as a neurotransmitter (see review [125]). Previous reports have shown that Ap4A is neuroprotective against 6-hydroxydopamine-induced neurotoxicity in a rodent model of Parkinson’s disease [126]. Moreover, pre- and post-treatment with Ap4A was shown to protect neurons against hypoxic and ischemic brain injury [126, 127]. In other investigations, however, these diadenine nucleotides have also been linked to apoptosis. One study of human cell cultures reported that apoptosis was associated with decreased levels of Ap3A and increased levels of Ap4A [95, 128]. Furthermore, in HeLa cell studies, Mg2+-dependent GAPDH-Ap4A binding was observed using photo affinity probes, and later confirmed by gel filtration and SDS-PAGE analysis [4, 5]. Unfortunately, the role played by the association of GAPDH and Ap4A in apoptosis, let alone in AD, is unclear at present; however, its significance with respect to the role of Ap3A/Ap4A ratios in programmed cell death is a subject of ongoing investigation.
Interestingly, it has been reported that GAPDH can also be found in and bound to mitochondria, generally by association with the mitochondrial membrane protein, VDAC-1, a component of the mitochondrial permeability transition pore complex (MPTP) [99, 119]. Upon binding GAPDH, the MPTP opens, triggering inner mitochondrial membrane permeabilization, the loss of transmembrane potential, increased cytosolic Ca2+ uptake, matrix swelling, and the release of cytochrome C and apoptosis inducing factor (AIF) into the cytosol. In effect, this cascade is the beginning of an intrinsic apoptotic process initiated by GAPDH. It should be noted, however, that although GAPDH is present in mitochondria and enzymatically active under normal conditions [119, 129], overexpression of GAPDH and its import into mitochondria via VDAC-1 is required to trigger an apoptotic cascade through the MPTP [99]. In a study by Tarze, et al. [99], it was suggested that the GAPDH – VDAC-1 interaction occurred via inter- and/or intra-molecular disulfide bonding, as their association was inhibited by administration of the thiolating agent, dithiothreitol (DTT). Moreover, these researchers speculate that monomeric GAPDH isoforms were the most likely candidates involved in pro-apoptotic MPTP opening, as denaturing conditions did not interfere with GAPDH-VDAC-1 binding [99]. Likewise, it would also appear that enzymatic activity was not a prerequisite for VDAC-1 binding [99]. The opening of the MPTP in neurodegenerative disease is well know (reviewed in [130]), and favored by the overwhelmingly oxidative environment in AD brain. Although whether or not oxidized GAPDH binds VDAC-1 was not described, the aforementioned study by Tarze, et al. [99] strongly suggests the likelihood of their interaction, considering monomeric and/or other denatured isoforms of GAPDH readily associate with VDAC-1. Thus, it can be inferred that the pro-apoptotic GAPDH-VDAC-1 interaction also could lead to AD pathology.
Other post-translational modifications that may link GAPDH to apoptotic processes involve its phosphotransferase/kinase activity (Section 4.2). As a phosphotransferase, or kinase, GAPDH is able to substitute the hydrogen atom of a hydroxyl moiety on Ser, Thr, and Tyr residues with a highly negative phosphate group, causing significant conformational changes in the phosphorylated protein’s structure [1]; likewise, GAPDH structure is also dramatically altered when phosphorylated by other intracellular protein kinases (Section 4.2). Phosphorylation is one of the most common post-translational modifications the cell employs to complete a myriad of signaling cascades under normal conditions. However, a pathologic state can cause excessive phosphorylation of a variety of intracellular proteins, causing either complete inactivation, or yielding a toxic gain-of-function effect that could propagate apoptotic pathways. One deleterious example in AD brain is the hyperphosphorylation of the microtubule-associated protein, tau. Routine phosphorylation of tau is necessary to its function in the control of microtubule assembly and stability, as well as intracellular axonal transport [131–133]; however, hyperphosphorylation of tau results in the formation of toxic NFTs, intracellular deposits of hyperphosphorylated tau, involved in the pathogenesis of AD, as well as other tauopathies (also see Section 5.4) [134–138]. Therefore, phosphorylation of or by GAPDH during apoptosis could be either a protective mechanism or impart a toxic gain-of-function to, or elicited by, GAPDH that enhances apoptosis in AD brain. Unfortunately, the role(s) of GAPDH phosphotransferase/kinase activity in normal and/or pathological cell function has yet to be made clear.
A final way inhibition of GAPDH activity can contribute to apoptosis is through the glycolytic generation of toxic side-products. GAPDH can undergo many different oxidative modifications, influencing its structure and activity in a variety of neurodegenerative diseases, but especially in AD (Section 5.0). Normally, GAPDH catalyzes the reversible phosphorylation of G3P to BPG during glycolysis, while reducing its cofactor NAD+ to NADH. However, when GAPDH glycolytic activity is impeded, the triose phosphate isomerase and/or aldolase intermediates, dihydroxyacetone phosphate (DHAP) and G3P, begin to accumulate and give rise to the deleterious breakdown product, methylglyoxal (MG). MG is a highly reactive α-ketoaldehyde that readily oxidizes proteins, lipids, and other cellular components, leading to cytotoxicity [139]. Moreover, MG binds to Cys, Lys, and His residues by Michael addition at a faster kinetic rate than does the lipid peroxidation product, HNE [140], although MG, HNE, and acrolein can all alter protein conformation and function [141, 142]. A previous study of human red blood cells, in conjunction with type-1 diabetes, revealed the formation of MG after GAPDH inhibition, while in vivo administration of exogenous MG was shown to cause renal damage in mice [143]. Moreover, MG is also an isomer of the lipid peroxidation product malondialdehyde (MDA), another highly reactive aldehyde known to accumulate and promote oxidative stress and damage in mouse-models of accelerated aging, as well as AD brain [140, 144, 145]. Considering formation of MG has been shown to result from redox imbalance and perturbation of cellular NAD+/NADH ratios [146], it is reasonable to believe that MG would play a similar role in AD-related neurodegeneration as MDA.
In a study by Ahmed et al. [147] of cerebrospinal fluid (CSF) of 32 AD patients, it was found that the concentration of MG-derived hydroimidazolone (MG-H1) was increased 30-fold compared to age-matched controls. Correlative regression analysis indicated that increased amounts of free MG-H1 adducts were most likely due to oxidative inhibition of GAPDH glycolytic activity, as well as accumulation of triosephospahte in non-vascular compartments and neurons [147]. Moreover, a study by Kuhla et al., [148] examining the ability of reactive carbonyl compounds acrolein, glyoxal, MG, and MDA to induce and/or accelerate tau oligomerization, formation of thioflavin T-positive tau aggregates, and formation of paired helical filaments (PHF), further confirms MG involvement in AD pathology. In this study, MG was recognized as the second most reactive compound amongst the four analyzed, and was able to induce dimer, trimer, and tetramer formation (PHF-like) of synthetic tau filaments in a concentration-dependant manner. It was postulated that the mechanism of MG action on synthetic filaments was through carbon-carbon cross-linking of Lys, Arg, and Cys residues [148].
5.1.1 GAPDH & GSH
The interaction of GAPDH with GSH under normal and pathological conditions is better understood. GSH recognizes the Rossmann fold of GAPDH, and binds to active-site Cys residues (Sections 2.0 & 3.0), forming disulfide bonds [28]. This S-glutathionylation was first observed as a function of oxidative stress induced by H2O2 in human umbilical vein endothelial cells [34], and could be inhibited by both ATP and NAD+ [28]. S-glutathionylation by GSH changes the pI of GAPDH from 8.1 to 6.9, making the protein more acidic [120], a particular characteristic of those GAPDH isoforms that are often implicated in apoptotic processes (Section 5.1) [120]. Notably, GAPDH is found to be oxidized in AD brain (also see Section 5.0) [28, 34, 48–55, 149], and regional analysis shows that GAPDH is S-glutathionylated in the inferior parietal lobule (IPL) region of AD brain [48]. Though the role of S-glutathionylation is AD pathology is not yet completely understood, some studies suggest it is a mechanism to protect proteins, like GAPDH, against permanent damage resulting from oxidation of Cys residues to cysteic and cystinic acids by the oxidizing environment of the AD brain [150–152]. This protection helps maintain the redox status of the cellular environment [150–152].
Although, S-glutathionylation, among other oxidative modifications (Section 5.1.3), would inhibit glycolytic activity of GAPDH [48], it would also cause the cell to shift its dependence on glycolysis to the pentose phosphate shunt to augment NADPH production (also see Section 5.2). NADPH is a key cofactor used by GSH-reductase to reduce oxidized glutathione (GSSG), accelerating the cell’s antioxidant response elicited by recycling GSH. Therefore, it can be inferred that, under oxidative conditions, GAPDH can act as a switch to redirect glucose metabolism to more appropriate defensive pathways [30]. A momentary deviation from glucose metabolism in such cases would be beneficial, as this diversion provides a protective antioxidant response against ROS and RNS. Unfortunately, due to the unrelenting pathology of AD, the inhibition of glycolysis becomes permanent, and antioxidant defense mechanisms are ultimately overcome by the ever-increasing amount of ROS and RNS generated in AD brain, eliminating the possibility of cell survival [153].
5.1.2 GAPDH & p53
Like GAPDH, p53 is also a multifunctional protein, known largely for its role in DNA transcription and repair [154, 155], as well as for its various roles in programmed cell death [156, 157]. In particular, results from in vivo and in vitro studies have shown elevated levels of p53 in neurons related to AD [158–162]. Similarly, studies from our laboratory reveal that p53 levels are significantly higher within the IPL region of AD patients, of which, monomeric and dimeric p53 isoforms were the most abundant [163, 164]. Our studies show that p53 is a target of protein oxidation, nitration, and lipid peroxidation within advanced stages of AD neurodegeneration [163, 164]. Additionally, monomeric and dimeric p53 isoforms were found to be S-glutathionylated to a higher degree than tetrameric isoforms, which may also illustrate one neuronal defensive mechanism against oxidative stress-induced apoptosis in AD brain [165]. Therefore, the oxidative modification of p53 in AD brain can elicit a toxic gain-of-function to p53 activity that most likely mediates p53-induced apoptotic pathways in AD [163], that are usually associated with two B-cell lymphoma-2 (Bcl-2) family proteins, pro-apoptotic Bax and anti-apoptotic Bcl-2 [166].
During an investigation into the role of GAPDH in AraC-induced apoptosis, it was found that transfection of antisense-p53 mRNA into cell culture reduced p53, Bax, and GAPDH mRNA [31]. Moreover, neurons prepared from p53-deficient mice were resistant to AraC-induced apoptosis, in addition to suppressed GAPDH mRNA expression [31], while introducing p53 increased GAPDH expression and triggered apoptosis [29–31]. Other studies indicate that Bcl-2 may regulate the nuclear translocation of GAPDH, thus, protecting cells from apoptosis [29, 30, 167]. For example, in GT1-7 hypothalamic neurosecretory cells, GAPDH over-expression, nuclear translocation, and subsequent apoptosis were blocked by Bcl-2 over-expression alone [30, 31]. Interestingly, Bcl-2 had no effect on the translocation of a GAPDH-GFP conjugate, indicating a unique degree of complexity in the GAPDH translocation mechanism [168]. However, observations with neuroblastoma cells during 6-hydroxydopamine treatment revealed that GAPDH translocates to the Golgi prior to localizing within the nucleus [168]. Taking the above observations into account, it can be inferred that GAPDH is positively regulated by p53; we speculate that p53 can directly induce GAPDH over-expression and nuclear translocation, initiating apoptotic pathways that can be blocked by Bcl-2 in AD brain (Fig. 6).
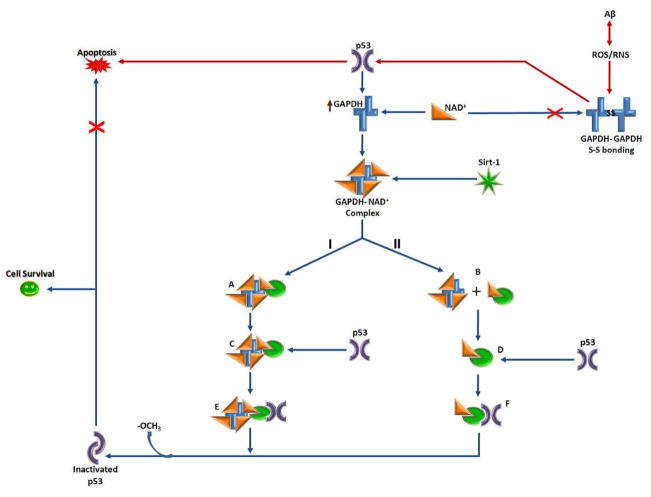
Figure 6. p53 and GAPDH interactions.
This diagram represents possible feedback-loop mechanisms of p53 modulation by GAPDH and the NAD+-dependant protein deacetylase, Sirt-1. Upon binding the GAPDH-NAD+ complex, Sirt-1 undergoes a conformational change that enables it to bind and remove acetyl groups (−OCH3) from C-terminal Lys residues of p53, inactivating it. Inactivated p53 is unable to induce apoptotic processes; thus, this pathway could lead to cell survival (path shown in blue). At present, the nature of the interaction between Sirt-1 and GAPDH unknown; therefore in this diagram, two possibilities are indicated: I, indicates formation of a GAPDH-NAD+-Sirt-1 complex (A), while II, indicates NAD+ transfer from GAPDH, leading to NAD+-Sirt-1 complex formation (B). In either case, the NAD+-Sirt-1 interaction likely changes the conformation of Sirt-1 (C and D), which may further facilitate Sirt-1 interaction with p53 to form complexes E and/or F. Additionaly, oxidative stress in AD causes GAPDH to undergo intermolecular disulfide bonding, inhibiting its activity and ability to bind NAD+, as well as further hindering Sirt-1-induced inactivation of pro-apoptotic p53 (path shown in red).
However, regulation of GAPDH by p53 may not always be to the detriment of the cell. Some studies speculate that induction of GAPDH over-expression by p53 may be an anti-apoptotic mechanism, wherein GAPDH provides an NAD+ to the p53 inhibitor protein, Sirt-1, in an effort to prevent apoptosis [169]. Sirt-1 is an NAD+-dependant histone deacetylase protein that removes acetyl groups from a Lys residue present on the C-terminal of p53 [170]. p53 inhibition can increase cellular resistance to stress stimuli, thereby improving cell survival rates [169]. Therefore, regulation of GAPDH expression by p53 could represent a feedback-loop, in which p53-induced over-expression of GAPDH increases the supply of NAD+ to Sirt-1 that, in turn, inhibits excessive p53-related apoptotic activity (Fig. 6). Moreover, the oxidative modification of GAPDH in AD would decrease its affinity for NAD+, which would preclude NAD+ transfer to Sirt-1, resulting in unchecked p53 activity and apoptosis [30, 169]. Given the potentially diverse results from interactions of p53 and GAPDH, additional studies of the roles and interactions of p53 and GAPDH in AD brain are warranted.
5.1.3 NO & GAPDH
NO is a gaseous signaling molecule that plays many critical roles in the central nervous system associated with cognitive function, synaptic plasticity, hormone secretion, and neurotransmission, among others [171, 172]. However, as with many signaling molecules, NO exhibits a duality of function, wherein excess NO can become cytotoxic [173–175]. Furthermore, NO becomes increasingly redox reactive in cells undergoing oxidative stress, a predominant feature in AD brain (Section 5.0), by forming toxic RNS and inducing widespread nitrosative stress [172, 174, 175]. There has been much research indicating a direct interaction between NO and GAPDH by reversible S-nitrosylation of critical active-site Cys residues (mainly Cys-149; Section 3.0), which inhibits GAPDH dehydrogenase activity [176–178]. Many studies have demonstrated that NO can induce a post-translational modification specific to GAPDH through non-enzymatic, covalent NAD+ modification [177, 179–181] not observed among other dehydrogenases [180]. Modification of NAD+ is achieved through auto-ADP-ribosylation of cytosolic GAPDH, in which the adenosine 5′-diphosphoribose (ADP-ribose) moiety of NAD+ is transferred to another protein [182]. The NO induced S-nitrosylation of active-site Cys-149 stimulates mono-ADP-ribosylation of tetrameric GAPDH [181, 183], by providing a critical SH-group in close proximity to the NAD+-binding site [179]. Unfortunately, the role(s) of ribosylated GAPDH in normal and/or pathological cell function have not been described beyond inhibition of enzymatic activity at this point in time. Nevertheless, GAPDH is considered a sensor for NO and nitrosative stress [173, 175].
Nitrosative stress is a common phenomenon in neurodegenerative disorders, especially in AD, in which 3-nitrotyrosine (3-NT) formation has been well-documented (Section 5.0). Under pathological conditions, NO is produced in excess by one of three NO synthase (NOS) isoforms in the brain, as a pro-inflammatory response [173, 174]. Interestingly, although S-nitrosylation by NO inhibits enzymatic activity of most proteins, such enzymatic inhibition is not always injurious to the cell. For example, NO can nitrosylate catalytic Cys residues of caspase-3, thereby preventing an apoptotic process [184, 185]. However, the consequences of GAPDH NO-modification are not as favorable. In addition to inhibiting its dehydrogenase activity, S-nitrosylation of GAPDH confers upon it the ability to bind the E3 ubiquitin ligase, Siah-1 [2, 32, 33]. Siah-1 is widely expressed in the brain [186, 187], and, by itself, can initiate apoptosis by translocating to the nucleus, acetylating, ubiquitinating, and degrading a variety of nuclear proteins, including p53, that mediate cell death [33, 188–190].
However, previous studies show that transfection of apoptotic HEK293 cells with GAPDH amplifies nuclear levels of Siah-1 [188], suggesting the stabilized GAPDH-Siah-1 complex is not readily degraded by the ubiquitin proteasome system, thus augmenting GAPDH-Siah-1 nuclear translocation and apoptotic cell death [188]. Moreover, our laboratory has shown a significant increase in GAPDH expression and nitrosylation in AD (Section 5.0) [51], suggesting that AD pathology creates a synergistic environment augmenting apoptosis induced, in part, by the NO/GAPDH/Siah-1 apoptotic cell death cascade. Consistent with these considerations, a recent study by Sen, et al. [190] demonstrated that inhibition of GAPDH S-nitrosylation prevented GAPDH-Siah-1 interactions, thereby preventing the initiation of apoptotic processes. These authors also describe a neuroprotective cytosolic protein, GOSPEL (~52 kDa), that competitively binds cytosolic GAPDH and prevents its nuclear translocation [190]. Interestingly, they found that S-nitrosylation of GOSPEL promotes GAPDH binding, thereby enhancing its neuroprotective capabilities [190]. Furthermore, this study indicated that GOSPEL may be a useful approach to modulate neurodegeneration in AD, Huntington’s disease, and other neurodegenerative conditions in which GAPDH may play a critical role.
5.2 Hypometabolism, GAPDH Membrane Binding, & Ca2+ Flux
A well-known aspect of AD pathology is extensive glucose hypometabolism in concert with hypoxia [191], as the oxidative modification and subsequent inactivation of glycolytic enzymes, such as GAPDH, results in a substantial decline in energy and oxygen bioavailability [191]. Interestingly, many studies have demonstrated that key glycolytic enzymes, such as GAPDH, are electrostatically bound to cell membranes, as well as endoplasmic and sarcoplasmic membranes, of many cell types [25, 192–195]. In fact, GAPDH glycolytic activity is directly affected by its interaction with these membranes [194, 196–198]. Studies by Galli, et al. [194], show that S-nitrosylation of active-site thiol groups (Section 5.1.3) precludes GAPDH membrane binding, in addition to inhibiting enzyme activity. Therefore, one way the cell regulates glucose metabolism is through GAPDH membrane binding, which prevents modification of active-site Cys residues that are essential to glycolytic activity [194]. A study by Brorson, et al. [199] involving NO and neuronal energy production demonstrated that an acute depletion of ATP production by high levels of NO was most likely due to inhibition of both mitochondrial respiration and glycolysis, a favored target being GAPDH. Their experiments also imply that the NO-threshold which inhibits glycolytic ATP production is greater than that which inhibits ATP generated by the electron transport chain (ETC) [199]. Inhibition of GAPDH would cause the cell to shift its reliance on glycolysis to the pentose phosphate shunt, which produces NADPH in lieu of NADH. This metabolic switch during oxidative stress can be beneficial as antioxidant enzymes such as glutaredoxin, thioredoxin, and GSH-reductase require NADPH (Section 5.1.1). However, since these anti-oxidant defense mechanisms are eventually overcome in the progression of AD, this switch could permanently uncouple the production of ATP and pyruvate from glycolysis [30, 200], thereby contributing to the growing anaerobic environment found in AD brain [194]. Therefore, these experiments imply, in part, a cellular preference for maintenance of glycolytic function over oxidative phosphorylation.
Even though neither glycolysis nor oxidative phosphorylation alone is capable of sustaining cellular energy deficits, the primary source of ATP energy for membrane ion pumps, such as the Na+/K+-ATPase and Ca2+-ATPase, is glycolytic [191, 195, 201]. Previous studies have shown that GAPDH binds to the IP3 receptor and SERCA pump [24, 25], thereby coupling glycolysis and ion pump activity [195, 202], which would account for the cell’s glycolytic ATP preference [195, 203]. In a study by Kahlert, et al. [202], glycolytic ATP was shown to be more important for maintaining ER Ca2+ stores and IP3-mediated Ca2+ signaling than the ETC, as the addition of iodoacetic acid (IA; non-specific SH-group modifier) together with exogenous pyruvate to cultured astrocytes could not restore intracellular ATP and basal Ca2+ levels, nor membrane potential [201, 202]. IP3R-bound GAPDH modulates Ca2+ release via NADH production [24]; therefore, NO modification (like IA) of critical GAPDH active-site Cys residues under pathological conditions would not only cause a metabolic shift from glycolysis to the pentose phosphate pathway, but also trigger the cytoplasmic release of ER Ca2+ stores, leading to Ca2+ excitotoxicity, predominant in AD pathology [204–206]. Therefore, maintenance of glycolytic GAPDH function is essential not only to ATP and pyruvate production, but also to the maintenance of intracellular Ca2+ levels and prevention of membrane depolarization that could lead to cell death [191], especially in AD brain.
5.3 GAPDH, AβPP, Aβ, & Tau
A systematic meta-analysis of AD genetic association study by Bertram et al. [207] showed that 13 genes play a significant role in development of AD, one of which, was GAPDH. Moreover, a study by Wang, et al. [208] reported the conversion of GAPDH to a detergent-insoluble state was coupled with AD progression. These analyses, in addition to others, serve to emphasize the critical role GAPDH plays in neurodegeneration, as the GAPDH gene (GAPD) and pseudogenes [57–61] have been suggested to be risk factors in late-onset AD [209, 210]. Initial investigations into the involvement of GAPDH in AD reported that a ~38 kDa protein, identified as GAPDH, was consistently increased during age-induced apoptosis [211]. Further studies demonstrated cross-reactivity between GAPDH and the monoclonal antibody Am-3, which recognizes AβPP but not Aβ, raised against amyloid plaques extracted from the brain of AD patients [212]. However, Tamaoka et al. [213] suggested that such cross-reactivity was not a result of similar homology between epitopes, since they would lie outside the Aβ(1-42) sequence region on AβPP [213–218], but were most likely due to conformational similarities between Aβ and GAPDH. Furthermore, a direct interaction between GAPDH and AβPP has been reported, in which rat brain monomeric GAPDH interacted with the cytoplasmic C-terminal domain of recombinant AβPP, while maintaining its glycolytic activity [36]. Later studies confirmed that brain-derived GAPDH also could bind a variety of Aβ isoforms, displaying greatest affinity for Aβ(1-42) [101, 102, 219].
Indeed, GAPDH is often identified as a major component of amyloid plaques and even NFTs in AD brain [102, 208]; however, some researchers have questioned whether or not its presence indicates a direct role in Aβ aggregation and/or NFT formation. Recent research suggests the manner in which GAPDH accumulates in amyloid plaques is not due to a high concentration in neurons, or the binding of GAPDH to pre-aggregated Aβ; rather, these studies demonstrate that only oxidized and denatured forms of GAPDH (i.e., monomeric, dimeric, or unfolded polypeptide chains) were able to form highly stable complexes with Aβ [102, 219]. Moreover, these non-native GAPDH isoforms only could bind soluble Aβ species, as opposed to pre-aggregated structures, indicating the direct involvement of GAPDH in amyloid aggregation. Interestingly, it also was suggested that Aβ could potentially accelerate [thermo] inactivation of native GAPDH, as the interaction between Aβ and partially unfolded GAPDH species would shift the equilibrium to favor denaturation [219]. Likewise, a study by Cumming and Schubert [220] showed that Aβ promotes GAPDH disulfide binding, suggesting that oxidative stress induced by Aβ neurotoxicity not only increases levels of denatured GAPDH, but also promotes its nuclear translocation and pro-apoptotic action (Sections 5.1.1 & 5.1.3).
Research on NFT formation in AD brain reports results similar to those of GAPDH and Aβ/AβPP. As mentioned above (Section 5.1), hyperphosphorylation of the microtubule-associated protein tau is rampant in AD brain, leading to formation of neurotoxic NFTs, of which, PHF-tau is a major component [136, 137, 221]. Previous LC-MS/MS analysis demonstrated that GAPDH co-localized with NFTs and immunoprecipitated with PHF-tau from the temporal cortex of AD brain [208, 222]. Further studies by Chen, et al. [223], showed that tau was able to bind and promote the denaturation and inactivation of GAPDH in vitro. However, their research also established that phosphorylated and pre-aggregated PHF-tau were unable to bind or affect GAPDH denaturation or activity [223], suggesting a direct involvement of GAPDH in tau aggregation and NFT formation in AD brain. Although, the above-mentioned research reveals the critical role GAPDH plays in both Aβ and tau aggregation, how these GAPDH-Aβ, -AβPP, and -tau complexes affect other cellular processes in AD brain remains to be determined. However, it is clear that maintenance of GAPDH form and function is integral to the prevention of protein aggregation pathology in AD brain.
6.0 Conclusion
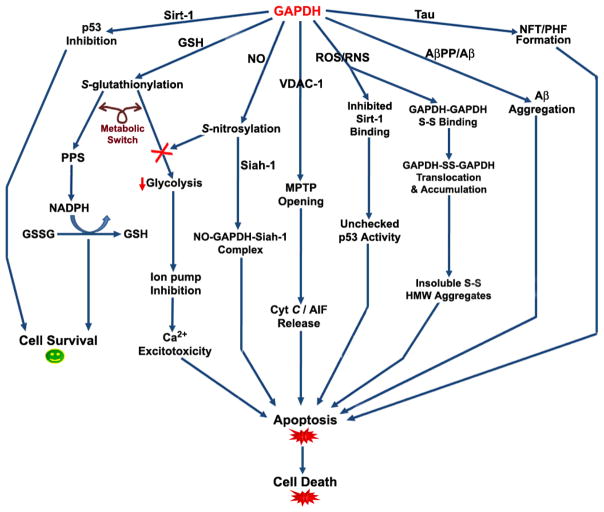
It is clear that GAPDH possesses a myriad of functions in addition to basic glycolysis, and the purpose of those functions in normal and pathologic cellular environments remains a topic of great interest. Based on the research described above, there is strong evidence for direct and indirect involvement of GAPDH in AD pathology (Fig. 7). Decreased GAPDH glycolytic activity, in addition to oxidative and post-translational modifications, like S-gluatathionylation and S-nitrosylation, are distinct markers of cellular stress in AD pathology that significantly impact intracellular homeostasis by promoting GAPDH-induced Ca2+ excitotoxicity through IP3R and SERCA binding, as well as interaction with pro-apoptotic proteins such as p53, Siah-1, and the MPTP through VDAC-1 binding. These indirect apoptotic insults contribute to a host of feedback-loop mechanisms, in which AD pathology is able to perpetuate an ongoing cycle of cell death that ultimately overcomes inherent anti-oxidant defenses. Although S-glutathionylation and S-nitrosylation of GAPDH may initially act as a metabolic switch to protect the cell, in AD brain these modifications eventually inhibit GAPDH activity and glucose metabolism indefinitely.
Figure 7. GAPDH functional diversity in AD brain.
This arrow diagram is a summary of the various indirect and direct signalling pathways, pro- and/or anti-apoptotic, that GAPDH triggers upon association with various proteins in AD brain. Abbreviations: GSH, gluthathione; GSSG, oxidized GSH; PPS, pentose phosphate shunt; NO, nitric oxide; MPTP, mitochondrial permeability transition pore; ROS, reactive oxygen species; RNS, reactive nitrogen species; S-S, disulfide bond; HMW, high molecular weight; NFT, neurofibrillary tangles; PHF, paired helical filaments. See text.
The direct involvement of GAPDH in AD is somewhat more intriguing, as its association with AβPP, Aβ(1-40)/Aβ(1-42), and tau demonstrates its direct participation in the aggregation of these three species into their insoluble counterparts. However, it should be noted that in order for GAPDH to partake in amyloid or tau aggregation, it must first succumb to denaturation and/or oxidative modification, demonstrating that GAPDH inhibition and pro-apoptotic function is a result of ongoing oxidative stress and damage that is itself but a secondary effect of an as of yet unknown primary scaffold of AD pathology. Although the exact mechanisms of many of the GAPDH interactions and processes described in this review are not yet clear, one can be certain that the structure and subcellular localization of GAPDH is vital to understanding the many roles it plays in normal and pathological cell function. Therefore, it seems that maintenance of GAPDH structure and activity may be a promising therapeutic target to slow or halt neurodegeneration in AD brain.
Acknowledgments
This work was supported in part by NIH grants to D.A.B. [AG-10836; AG-05119].
References
- 1.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins JL, Tanner JJ. High-resolution structure of human D-glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2006;62:290–301. doi: 10.1107/S0907444905042289. [DOI] [PubMed] [Google Scholar]
- 3.Branlant G, Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1985;150:61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- 4.Sirover MA. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J Cell Biochem. 1997;66:133–140. [PubMed] [Google Scholar]
- 5.Baxi MD, Vishwanatha JK. Uracil DNA-glycosylase/glyceraldehyde-3-phosphate dehydrogenase is an Ap4A binding protein. Biochemistry. 1995;34:9700–9707. doi: 10.1021/bi00030a007. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, deRiel JK, Sirover MA. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L, Roeder RG, Luo Y. S-phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 8.Engel M, Seifert M, Theisinger B, Seyfert U, Welter C. Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A. Two old enzymes combine for the novel Nm23 protein phosphotransferase function. J Biol Chem. 1998;273:20058–20065. doi: 10.1074/jbc.273.32.20058. [DOI] [PubMed] [Google Scholar]
- 9.Duclos-Vallee JC, Capel F, Mabit H, Petit MA. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J Gen Virol. 1998;79 (Pt 7):1665–1670. doi: 10.1099/0022-1317-79-7-1665. [DOI] [PubMed] [Google Scholar]
- 10.Reiss N, Hermon J, Oplatka A, Naor Z. Interaction of purified protein kinase C with key proteins of energy metabolism and cellular motility. Biochem Mol Biol Int. 1996;38:711–719. [PubMed] [Google Scholar]
- 11.Reiss N, Oplatka A, Hermon J, Naor Z. Phosphatidylserine directs differential phosphorylation of actin and glyceraldehyde-3-phosphate dehydrogenase by protein kinase C: possible implications for regulation of actin polymerization. Biochem Mol Biol Int. 1996;40:1191–1200. doi: 10.1080/15216549600201833. [DOI] [PubMed] [Google Scholar]
- 12.Reiss N, Kanety H, Schlessinger J. Five enzymes of the glycolytic pathway serve as substrates for purified epidermal-growth-factor-receptor kinase. Biochem J. 1986;239:691–697. doi: 10.1042/bj2390691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashmarina LI, Louzenko SE, Severin SE, Jr, Muronetz VI, Nagradova NK. Phosphorylation of D-glyceraldehyde-3-phosphate dehydrogenase by Ca2+/calmodulin-dependent protein kinase II. FEBS Lett. 1988;231:413–416. doi: 10.1016/0014-5793(88)80861-5. [DOI] [PubMed] [Google Scholar]
- 14.Kawamoto RM, Caswell AH. Autophosphorylation of glyceraldehyde-phosphate dehydrogenase and phosphorylation of protein from skeletal muscle microsomes. Biochemistry. 1986;25:657–661. doi: 10.1021/bi00351a022. [DOI] [PubMed] [Google Scholar]
- 15.Wu K, Aoki C, Elste A, Rogalski-Wilk AA, Siekevitz P. The synthesis of ATP by glycolytic enzymes in the postsynaptic density and the effect of endogenously generated nitric oxide. Proc Natl Acad Sci U S A. 1997;94:13273–13278. doi: 10.1073/pnas.94.24.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumagai H, Sakai H. A porcine brain protein (35 kDa protein) which bundles microtubules and its identification as glyceraldehyde-3-phosphate dehydrogenase. J Biochem. 1983;93:1259–1269. doi: 10.1093/oxfordjournals.jbchem.a134260. [DOI] [PubMed] [Google Scholar]
- 17.Huitorel P, Pantaloni D. Bundling of microtubules by glyceraldehyde-3-phosphate dehydrogenase and its modulation by ATP. Eur J Biochem. 1985;150:265–269. doi: 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 18.Caswell AH, Corbett AM. Interaction of glyceraldehyde-3-phosphate dehydrogenase with isolated microsomal subfractions of skeletal muscle. J Biol Chem. 1985;260:6892–6898. [PubMed] [Google Scholar]
- 19.Walsh JL, Keith TJ, Knull HR. Glycolytic enzyme interactions with tubulin and microtubules. Biochim Biophys Acta. 1989;999:64–70. doi: 10.1016/0167-4838(89)90031-9. [DOI] [PubMed] [Google Scholar]
- 20.Launay JF, Jellali A, Vanier MT. Glyceraldehyde-3-phosphate dehydrogenase is a microtubule binding protein in a human colon tumor cell line. Biochim Biophys Acta. 1989;996:103–109. doi: 10.1016/0167-4838(89)90101-5. [DOI] [PubMed] [Google Scholar]
- 21.Volker KW, Knull H. A glycolytic enzyme binding domain on tubulin. Arch Biochem Biophys. 1997;338:237–243. doi: 10.1006/abbi.1996.9819. [DOI] [PubMed] [Google Scholar]
- 22.Durrieu C, Bernier-Valentin F, Rousset B. Binding of glyceraldehyde 3-phosphate dehydrogenase to microtubules. Mol Cell Biochem. 1987;74:55–65. doi: 10.1007/BF00221912. [DOI] [PubMed] [Google Scholar]
- 23.Bryksin AV, Laktionov PP. Role of glyceraldehyde-3-phosphate dehydrogenase in vesicular transport from Golgi apparatus to endoplasmic reticulum. Biochemistry (Mosc) 2008;73:619–625. doi: 10.1134/s0006297908060011. [DOI] [PubMed] [Google Scholar]
- 24.Patterson RL, van Rossum DB, Kaplin AI, Barrow RK, Snyder SH. Inositol 1,4,5-trisphosphate receptor/GAPDH complex augments Ca2+ release via locally derived NADH. Proc Natl Acad Sci U S A. 2005;102:1357–1359. doi: 10.1073/pnas.0409657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu KY, Becker LC. Ultrastructural localization of glycolytic enzymes on sarcoplasmic reticulum vesticles. J Histochem Cytochem. 1998;46:419–427. doi: 10.1177/002215549804600401. [DOI] [PubMed] [Google Scholar]
- 26.Sioud M, Jespersen L. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1996;257:775–789. doi: 10.1006/jmbi.1996.0201. [DOI] [PubMed] [Google Scholar]
- 27.Lind C, Gerdes R, Schuppe-Koistinen I, Cotgreave IA. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem Biophys Res Commun. 1998;247:481–486. doi: 10.1006/bbrc.1998.8695. [DOI] [PubMed] [Google Scholar]
- 28.Puder M, Soberman RJ. Glutathione conjugates recognize the Rossmann fold of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1997;272:10936–10940. doi: 10.1074/jbc.272.16.10936. [DOI] [PubMed] [Google Scholar]
- 29.Chuang DM, Chen RW, Saunders PA, Ishitani R. Roles of GAPDH, p53, and Bax in neuronal apoptosis. Naunyn-Schmiedebergs Arch Pharmacol. 1998;358:R431. [Google Scholar]
- 30.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 31.Chen RW, Saunders PA, Wei H, Li Z, Seth P, Chuang DM. Involvement of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and p53 in neuronal apoptosis: evidence that GAPDH is upregulated by p53. J Neurosci. 1999;19:9654–9662. doi: 10.1523/JNEUROSCI.19-21-09654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuppe-Koistinen I, Moldeus P, Bergman T, Cotgreave IA. S-thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur J Biochem. 1994;221:1033–1037. doi: 10.1111/j.1432-1033.1994.tb18821.x. [DOI] [PubMed] [Google Scholar]
- 35.Burke JR, Enghild JJ, Martin ME, Jou YS, Myers RM, Roses AD, Vance JM, Strittmatter WJ. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 36.Schulze H, Schuler A, Stuber D, Dobeli H, Langen H, Huber G. Rat brain glyceraldehyde-3-phosphate dehydrogenase interacts with the recombinant cytoplasmic domain of Alzheimer’s β-amyloid precursor protein. J Neurochem. 1993;60:1915–1922. doi: 10.1111/j.1471-4159.1993.tb13420.x. [DOI] [PubMed] [Google Scholar]
- 37.Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ. Mutation of the β-amyloid precursor protein in familial Alzheimer’s disease increases β-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 38.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 39.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci U S A. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wisniewski T, Dowjat WK, Buxbaum JD, Khorkova O, Efthimiopoulos S, Kulczycki J, Lojkowska W, Wegiel J, Wisniewski HM, Frangione B. A novel Polish presenilin-1 mutation (P117L) is associated with familial Alzheimer’s disease and leads to death as early as the age of 28 years. Neuroreport. 1998;9:217–221. doi: 10.1097/00001756-199801260-00008. [DOI] [PubMed] [Google Scholar]
- 41.Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: potential causes and consequences involving amyloid β-peptide-associated free radical oxidative stress. Free Radic Biol Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 42.Butterfield DA, Stadtman ER. Protein oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- 43.Butterfield DA, Drake J, Pocernich C, Castegna A. Evidence of oxidative damage in Alzheimer’s disease brain: central role for amyloid β-peptide. Trends Mol Med. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- 44.Butterfield DA, Castegna A, Lauderback CM, Drake J. Evidence that amyloid β-peptide-induced lipid peroxidation and its sequelae in Alzheimer’s disease brain contribute to neuronal death. Neurobiol Aging. 2002;23:655–664. doi: 10.1016/s0197-4580(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 45.Butterfield DA. Amyloid β-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer’s disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 46.Markesbery WR. The role of oxidative stress in Alzheimer disease. Arch Neurol. 1999;56:1449–1452. doi: 10.1001/archneur.56.12.1449. [DOI] [PubMed] [Google Scholar]
- 47.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 48.Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, Klein JB, Turner DM, Butterfield DA. An increase in S-glutathionylated proteins in the Alzheimer’s disease inferior parietal lobule, a proteomics approach. J Neurosci Res. 2007;85:1506–1514. doi: 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- 49.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 50.Mohr S, Stamler JS, Brune B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 51.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer’s disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Boyd-Kimball D, Sultana R, Poon HF, Lynn BC, Casamenti F, Pepeu G, Klein JB, Butterfield DA. Proteomic identification of proteins specifically oxidized by intracerebral injection of amyloid β-peptide (1-42) into rat brain: implications for Alzheimer’s disease. Neuroscience. 2005;132:313–324. doi: 10.1016/j.neuroscience.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 53.Cahuana GM, Tejedo JR, Jimenez J, Ramirez R, Sobrino F, Bedoya FJ. Nitric oxide-induced carbonylation of Bcl-2, GAPDH and ANT precedes apoptotic events in insulin-secreting RINm5F cells. Exp Cell Res. 2004;293:22–30. doi: 10.1016/j.yexcr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Opii WO, Joshi G, Head E, Milgram NW, Muggenburg BA, Klein JB, Pierce WM, Cotman CW, Butterfield DA. Proteomic identification of brain proteins in the canine model of human aging following a long-term treatment with antioxidants and a program of behavioral enrichment: relevance to Alzheimer’s disease. Neurobiol Aging. 2008;29:51–70. doi: 10.1016/j.neurobiolaging.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perluigi M, Sultana R, Cenini G, Di Domenico F, Memo M, Pierce WM, Coccia R, Butterfield DA. Redox proteomics identification of HNE-modified brain proteins in Alzheimer’s disease: Role of lipid peroxidation in AD pathogenesis. Proteomics Clin Appli. 2009 doi: 10.1002/prca.200800161. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 57.Piechaczyk M, Blanchard JM, Riaad-El Sabouty S, Dani C, Marty L, Jeanteur P. Unusual abundance of vertebrate 3-phosphate dehydrogenase pseudogenes. Nature. 1984;312:469–471. doi: 10.1038/312469a0. [DOI] [PubMed] [Google Scholar]
- 58.Benham FJ, Hodgkinson S, Davies KE. A glyceraldehyde-3-phosphate dehydrogenase pseudogene on the short arm of the human X chromosomes defines a multigene family. EMBO J. 1984;3:2635–2640. doi: 10.1002/j.1460-2075.1984.tb02186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ercolani L, Florence B, Denaro M, Alexander M. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1988;263:15335–15341. [PubMed] [Google Scholar]
- 60.Hanauer A, Mandel JL. The glyceraldehyde-3-phosphate dehydrogenase gene family: structure of a human cDNA and of an X chromosome linked pseudogene; amazing complexity of the gene family in mouse. EMBO J. 1984;3:2627–2633. doi: 10.1002/j.1460-2075.1984.tb02185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tso JY, Sun XH, Kao TH, Reece KS, Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985;13:2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlile GW, Chalmers-Redman RM, Tatton NA, Pong A, Borden KE, Tatton WG. Reduced apoptosis after nerve growth factor and serum withdrawal: conversion of tetrameric glyceraldehyde-3-phosphate dehydrogenase to a dimer. Mol Pharmacol. 2000;57:2–12. [PubMed] [Google Scholar]
- 63.Buehner M, Ford GC, Moras D, Olsen KW, Rossmann MG. Structure determination of crystalline lobster D-glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1974;82:563–585. doi: 10.1016/0022-2836(74)90249-6. [DOI] [PubMed] [Google Scholar]
- 64.Roitel O, Vachette P, Azza S, Branlant G. P- but not R-axis interface is involved in cooperative binding of NAD on tetrameric phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. J Mol Biol. 2003;326:1513–1522. doi: 10.1016/s0022-2836(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 65.Cool BL, Sirover MA. Immunocytochemical localization of the base excision repair enzyme uracil DNA glycosylase in quiescent and proliferating normal human cells. Cancer Res. 1989;49:3029–3036. [PubMed] [Google Scholar]
- 66.Corbin IR, Gong Y, Zhang M, Minuk GY. Proliferative and nutritional dependent regulation of glyceraldehyde-3-phosphate dehydrogenase expression in the rat liver. Cell Prolif. 2002;35:173–182. doi: 10.1046/j.1365-2184.2002.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verlinde CL, Callens M, Van Calenbergh S, Van Aerschot A, Herdewijn P, Hannaert V, Michels PA, Opperdoes FR, Hol WG. Selective inhibition of trypanosomal glyceraldehyde-3-phosphate dehydrogenase by protein structure-based design: toward new drugs for the treatment of sleeping sickness. J Med Chem. 1994;37:3605–3613. doi: 10.1021/jm00047a017. [DOI] [PubMed] [Google Scholar]
- 68.Warizaya M, Kinoshita T, Kato A, Nakajima H, Fujii T. Cloning, expression, purification, crystallization and preliminary X-ray analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2004;60:567–568. doi: 10.1107/S0907444904000265. [DOI] [PubMed] [Google Scholar]
- 69.Ismail SA, Park HW. Structural analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005;61:1508–1513. doi: 10.1107/S0907444905026740. [DOI] [PubMed] [Google Scholar]
- 70.Skarzynski T, Moody PC, Wonacott AJ. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 A resolution. J Mol Biol. 1987;193:171–187. doi: 10.1016/0022-2836(87)90635-8. [DOI] [PubMed] [Google Scholar]
- 71.Aronov AM, Verlinde CL, Hol WG, Gelb MH. Selective tight binding inhibitors of trypanosomal glyceraldehyde-3-phosphate dehydrogenase via structure-based drug design. J Med Chem. 1998;41:4790–4799. doi: 10.1021/jm9802620. [DOI] [PubMed] [Google Scholar]
- 72.Cowan-Jacob SW, Kaufmann M, Anselmo AN, Stark W, Grutter MG. Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2003;59:2218–2227. doi: 10.1107/s0907444903020493. [DOI] [PubMed] [Google Scholar]
- 73.Shen YQ, Li J, Song SY, Lin ZJ. Structure of apo-glyceraldehyde-3-phosphate dehydrogenase from Palinurus versicolor. J Struct Biol. 2000;130:1–9. doi: 10.1006/jsbi.2000.4220. [DOI] [PubMed] [Google Scholar]
- 74.Yun M, Park CG, Kim JY, Park HW. Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes. Biochemistry. 2000;39:10702–10710. doi: 10.1021/bi9927080. [DOI] [PubMed] [Google Scholar]
- 75.Soukri A, Mougin A, Corbier C, Wonacott A, Branlant C, Branlant G. Role of the histidine 176 residue in glyceraldehyde-3-phosphate dehydrogenase as probed by site-directed mutagenesis. Biochemistry. 1989;28:2586–2592. doi: 10.1021/bi00432a036. [DOI] [PubMed] [Google Scholar]
- 76.Talfournier F, Colloc’h N, Mornon JP, Branlant G. Comparative study of the catalytic domain of phosphorylating glyceraldehyde-3-phosphate dehydrogenases from bacteria and archaea via essential cysteine probes and site-directed mutagenesis. Eur J Biochem. 1998;252:447–457. doi: 10.1046/j.1432-1327.1998.2520447.x. [DOI] [PubMed] [Google Scholar]
- 77.Didierjean C, Rahuel-Clermont S, Vitoux B, Dideberg O, Branlant G, Aubry A. A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+ J Mol Biol. 1997;268:739–759. doi: 10.1006/jmbi.1997.0998. [DOI] [PubMed] [Google Scholar]
- 78.Boschi-Muller S, Branlant G. The active site of phosphorylating glyceraldehyde-3-phosphate dehydrogenase is not designed to increase the nucleophilicity of a serine residue. Arch Biochem Biophys. 1999;363:259–266. doi: 10.1006/abbi.1998.1080. [DOI] [PubMed] [Google Scholar]
- 79.Moras D, Olsen KW, Sabesan MN, Buehner M, Ford GC, Rossmann MG. Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1975;250:9137–9162. doi: 10.2210/pdb1gpd/pdb. [DOI] [PubMed] [Google Scholar]
- 80.Nagradova NK. Study of the properties of phosphorylating D-glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Mosc) 2001;66:1067–1076. doi: 10.1023/a:1012472627801. [DOI] [PubMed] [Google Scholar]
- 81.Souza DH, Garratt RC, Araujo AP, Guimaraes BG, Jesus WD, Michels PA, Hannaert V, Oliva G. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase: structure, catalytic mechanism and targeted inhibitor design. FEBS Lett. 1998;424:131–135. doi: 10.1016/s0014-5793(98)00154-9. [DOI] [PubMed] [Google Scholar]
- 82.Carlile GW, Tatton WG, Borden KL. Demonstration of a RNA-dependent nuclear interaction between the promyelocytic leukaemia protein and glyceraldehyde-3-phosphate dehydrogenase. Biochem J. 1998;335 (Pt 3):691–696. doi: 10.1042/bj3350691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De BP, Gupta S, Zhao H, Drazba JA, Banerjee AK. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 84.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 85.Schultz DE, Hardin CC, Lemon SM. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 86.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 87.Nogales E, Downing KH, Amos LA, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 88.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 89.Eby D, Kirtley ME. Role of lysine residues in the binding of glyceraldehyde-3-phosphate dehydrogenase to human erythrocyte membranes. Biochem Biophys Res Commun. 1983;116:423–427. doi: 10.1016/0006-291x(83)90540-5. [DOI] [PubMed] [Google Scholar]
- 90.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 91.Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 92.Jans DA. Nuclear signaling pathways for polypeptide ligands and their membrane receptors? FASEB J. 1994;8:841–847. doi: 10.1096/fasebj.8.11.8070633. [DOI] [PubMed] [Google Scholar]
- 93.Yoneda Y. How proteins are transported from cytoplasm to the nucleus. J Biochem. 1997;121:811–817. doi: 10.1093/oxfordjournals.jbchem.a021657. [DOI] [PubMed] [Google Scholar]
- 94.McNulty SE, Toscano WA., Jr Transcriptional regulation of glyceraldehyde-3-phosphate dehydrogenase by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun. 1995;212:165–171. doi: 10.1006/bbrc.1995.1951. [DOI] [PubMed] [Google Scholar]
- 95.Kisselev LL, Justesen J, Wolfson AD, Frolova LY. Diadenosine oligophosphates (Ap(n)A), a novel class of signaling molecules? FEBS Lett. 1998;427:157–163. doi: 10.1016/s0014-5793(98)00420-7. [DOI] [PubMed] [Google Scholar]
- 96.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 97.Hwang NR, Yim SH, Kim YM, Jeong J, Song EJ, Lee Y, Lee JH, Choi S, Lee KJ. Oxidative modifications of glyceraldehyde-3-phosphate dehydrogenase play a key role in its multiple cellular functions. Biochemical Journal. 2009;423:253–264. doi: 10.1042/BJ20090854. [DOI] [PubMed] [Google Scholar]
- 98.Sergienko EA, Kharitonenkov AI, Bulargina TV, Muronetz VV, Nagradova NK. D-glyceraldehyde-3-phosphate dehydrogenase purified from rabbit muscle contains phosphotyrosine. FEBS Lett. 1992;304:21–23. doi: 10.1016/0014-5793(92)80580-a. [DOI] [PubMed] [Google Scholar]
- 99.Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N, Zamzami N, Jan G, Kroemer G, Brenner C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26:2606–2620. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- 100.Koshy B, Matilla T, Burright EN, Merry DE, Fischbeck KH, Orr HT, Zoghbi HY. Spinocerebellar ataxia type-1 and spinobulbar muscular atrophy gene products interact with glyceraldehyde-3-phosphate dehydrogenase. Hum Mol Genet. 1996;5:1311–1318. doi: 10.1093/hmg/5.9.1311. [DOI] [PubMed] [Google Scholar]
- 101.Oyama R, Yamamoto H, Titani K. Glutamine synthetase, hemoglobin α-chain, and macrophage migration inhibitory factor binding to amyloid β-protein: their identification in rat brain by a novel affinity chromatography and in Alzheimer’s disease brain by immunoprecipitation. Biochim Biophys Acta. 2000;1479:91–102. doi: 10.1016/s0167-4838(00)00057-1. [DOI] [PubMed] [Google Scholar]
- 102.Verdier Y, Huszar E, Penke B, Penke Z, Woffendin G, Scigelova M, Fulop L, Szucs M, Medzihradszky K, Janaky T. Identification of synaptic plasma membrane proteins co-precipitated with fibrillar β-amyloid peptide. J Neurochem. 2005;94:617–628. doi: 10.1111/j.1471-4159.2005.03158.x. [DOI] [PubMed] [Google Scholar]
- 103.Tanner MJ, Gray WR. The isolation and functional identification of a protein from the human erythrocyte ‘ghost’. Biochem J. 1971;125:1109–1117. doi: 10.1042/bj1251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wooster MS, Wrigglesworth JM. Adsorption of glyceraldehyde-3-phosphate dehydrogenase on condensed monolayers of phospholipid. Biochem J. 1976;153:93–100. doi: 10.1042/bj1530093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hessler RJ, Blackwood RA, Brock TG, Francis JW, Harsh DM, Smolen JE. Identification of glyceraldehyde-3-phosphate dehydrogenase as a Ca2+-dependent fusogen in human neutrophil cytosol. J Leukoc Biol. 1998;63:331–336. doi: 10.1002/jlb.63.3.331. [DOI] [PubMed] [Google Scholar]
- 106.Glaser PE, Gross RW. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- 107.Robbins AR, Ward RD, Oliver C. A mutation in glyceraldehyde-3-phosphate dehydrogenase alters endocytosis in CHO cells. J Cell Biol. 1995;130:1093–1104. doi: 10.1083/jcb.130.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krotkiewska B, Banas T. Interaction of Zn2+ and Cu2+ ions with glyceraldehyde-3-phosphate dehydrogenase from bovine heart and rabbit muscle. Int J Biochem. 1992;24:1501–1505. doi: 10.1016/0020-711x(92)90078-f. [DOI] [PubMed] [Google Scholar]
- 109.Pancholi V, Fischetti VA. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci U S A. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kitamura Y, Shimohama S, Kamoshima W, Ota T, Matsuoka Y, Nomura Y, Smith MA, Perry G, Whitehouse PJ, Taniguchi T. Alteration of proteins regulating apoptosis, Bcl-2, Bcl-x, Bax, Bak, Bad, ICH-1 and CPP32, in Alzheimer’s disease. Brain Res. 1998;780:260–269. doi: 10.1016/s0006-8993(97)01202-x. [DOI] [PubMed] [Google Scholar]
- 111.Bader Lange ML, Cenini G, Piroddi M, Abdul HM, Sultana R, Galli F, Memo M, Butterfield DA. Loss of phospholipid asymmetry and elevated brain apoptotic protein levels in subjects with amnestic mild cognitive impairment and Alzheimer disease. Neurobiol Dis. 2008;29:456–464. doi: 10.1016/j.nbd.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 113.Honig LS, Rosenberg RN. Apoptosis and neurologic disease. Am J Med. 2000;108:317–330. doi: 10.1016/s0002-9343(00)00291-6. [DOI] [PubMed] [Google Scholar]
- 114.Engidawork E, Gulesserian T, Seidl R, Cairns N, Lubec G. Expression of apoptosis related proteins in brains of patients with Alzheimer’s disease. Neurosci Lett. 2001;303:79–82. doi: 10.1016/s0304-3940(01)01618-4. [DOI] [PubMed] [Google Scholar]
- 115.Su JH, Anderson AJ, Cummings BJ, Cotman CW. Immunohistochemical evidence for apoptosis in Alzheimer’s disease. Neuroreport. 1994;5:2529–2533. doi: 10.1097/00001756-199412000-00031. [DOI] [PubMed] [Google Scholar]
- 116.Su JH, Zhao M, Anderson AJ, Srinivasan A, Cotman CW. Activated caspase-3 expression in Alzheimer’s and aged control brain: correlation with Alzheimer pathology. Brain Res. 2001;898:350–357. doi: 10.1016/s0006-8993(01)02018-2. [DOI] [PubMed] [Google Scholar]
- 117.Margolis RL, Chuang DM, Post RM. Programmed cell death: implications for neuropsychiatric disorders. Biol Psychiatry. 1994;35:946–956. doi: 10.1016/0006-3223(94)91241-6. [DOI] [PubMed] [Google Scholar]
- 118.Arutyunov DY, Muronetz VI. The activation of glycolysis performed by the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase in the model system. Biochem Biophys Res Commun. 2003;300:149–154. doi: 10.1016/s0006-291x(02)02802-4. [DOI] [PubMed] [Google Scholar]
- 119.Ryzlak MT, Pietruszko R. Heterogeneity of glyceraldehyde-3-phosphate dehydrogenase from human brain. Biochim Biophys Acta. 1988;954:309–324. doi: 10.1016/0167-4838(88)90086-6. [DOI] [PubMed] [Google Scholar]
- 120.Saunders PA, Chen RW, Chuang DM. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase isoforms during neuronal apoptosis. J Neurochem. 1999;72:925–932. doi: 10.1046/j.1471-4159.1999.0720925.x. [DOI] [PubMed] [Google Scholar]
- 121.Ishitani R, Chuang DM. Glyceraldehyde-3-phosphate dehydrogenase antisense oligodeoxynucleotides protect against cytosine arabinonucleoside-induced apoptosis in cultured cerebellar neurons. Proc Natl Acad Sci U S A. 1996;93:9937–9941. doi: 10.1073/pnas.93.18.9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ishitani R, Kimura M, Sunaga K, Katsube N, Tanaka M, Chuang DM. An antisense oligodeoxynucleotide to glyceraldehyde-3-phosphate dehydrogenase blocks age-induced apoptosis of mature cerebrocortical neurons in culture. J Pharmacol Exp Ther. 1996;278:447–454. [PubMed] [Google Scholar]
- 123.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and non-neuronal cell death. Proc Natl Acad Sci U S A. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vishwanatha JK, Wei Z. Diadenosine tetraphosphate binding protein from human HeLa cells: purification and characterization. Biochemistry. 1992;31:1631–1635. doi: 10.1021/bi00121a007. [DOI] [PubMed] [Google Scholar]
- 125.Baxi MD, Vishwanatha JK. Diadenosine polyphosphates: their biological and pharmacological significance. J Pharmacol Toxicol Methods. 1995;33:121–128. doi: 10.1016/1056-8719(94)00127-p. [DOI] [PubMed] [Google Scholar]
- 126.Wang Y, Chang CF, Morales M, Chiang YH, Harvey BK, Su TP, Tsao LI, Chen S, Thiemermann C. Diadenosine tetraphosphate protects against injuries induced by ischemia and 6-hydroxydopamine in rat brain. J Neurosci. 2003;23:7958–7965. doi: 10.1523/JNEUROSCI.23-21-07958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pintor J, Carracedo G, Alonso MC, Bautista A, Peral A. Presence of diadenosine polyphosphates in human tears. Pflugers Arch. 2002;443:432–436. doi: 10.1007/s004240100696. [DOI] [PubMed] [Google Scholar]
- 128.Vartanian A, Prudovsky I, Suzuki H, Dal Pra I, Kisselev L. Opposite effects of cell differentiation and apoptosis on Ap3A/Ap4A ratio in human cell cultures. FEBS Lett. 1997;415:160–162. doi: 10.1016/s0014-5793(97)01086-7. [DOI] [PubMed] [Google Scholar]
- 129.Mazzola JL, Sirover MA. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim Biophys Acta. 2003;1622:50–56. doi: 10.1016/s0304-4165(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 130.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 131.Cleveland DW, Hwo SY, Kirschner MW. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol. 1977;116:227–247. doi: 10.1016/0022-2836(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 132.Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer’s disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 133.Brandt R, Lee G. Functional organization of microtubule-associated protein tau. Identification of regions which affect microtubule growth, nucleation, and bundle formation in vitro. J Biol Chem. 1993;268:3414–3419. [PubMed] [Google Scholar]
- 134.Roder HM, Eden PA, Ingram VM. Brain protein kinase PK40erk converts tau into a PHF-like form as found in Alzheimer’s disease. Biochem Biophys Res Commun. 1993;193:639–647. doi: 10.1006/bbrc.1993.1672. [DOI] [PubMed] [Google Scholar]
- 135.Iqbal K, Alonso AD, Gondal JA, Gong CX, Haque N, Khatoon S, Sengupta A, Wang JZ, Grundke-Iqbal I. Mechanism of neurofibrillary degeneration and pharmacologic therapeutic approach. J Neural Transm Suppl. 2000;59:213–222. doi: 10.1007/978-3-7091-6781-6_22. [DOI] [PubMed] [Google Scholar]
- 136.Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 137.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Butterfield DA, Abdul HM, Opii W, Newman SF, Joshi G, Ansari MA, Sultana R. Pin1 in Alzheimer’s disease. J Neurochem. 2006;98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- 139.Beisswenger PJ, Howell SK, Smith K, Szwergold BS. Glyceraldehyde-3-phosphate dehydrogenase activity as an independent modifier of methylglyoxal levels in diabetes. Biochim Biophys Acta. 2003;1637:98–106. doi: 10.1016/s09254439(02)00219-3. [DOI] [PubMed] [Google Scholar]
- 140.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 141.Subramaniam R, Roediger F, Jordan B, Mattson MP, Keller JN, Waeg G, Butterfield DA. The lipid peroxidation product, 4-hydroxy-2-trans-nonenal, alters the conformation of cortical synaptosomal membrane proteins. J Neurochem. 1997;69:1161–1169. doi: 10.1046/j.1471-4159.1997.69031161.x. [DOI] [PubMed] [Google Scholar]
- 142.Lovell MA, Xie CS, Markesbery WR. Acrolein is increased in Alzheimer’s disease brain and is toxic to primary hippocampal cultures. Neurobiology of Aging. 2001;22:187–194. doi: 10.1016/s0197-4580(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 143.Golej J, Hoeger H, Radner W, Unfried G, Lubec G. Oral administration of methylglyoxal leads to kidney collagen accumulation in the mouse. Life Sci. 1998;63:801–807. doi: 10.1016/s0024-3205(98)00336-1. [DOI] [PubMed] [Google Scholar]
- 144.Palmer AM, Burns MA. Selective increase in lipid peroxidation in the inferior temporal cortex in Alzheimer’s disease. Brain Res. 1994;645:338–342. doi: 10.1016/0006-8993(94)91670-5. [DOI] [PubMed] [Google Scholar]
- 145.Butterfield DA, Howard BJ, Yatin S, Allen KL, Carney JM. Free radical oxidation of brain proteins in accelerated senescence and its modulation by N-tert-butyl-alpha-phenylnitrone. Proc Natl Acad Sci U S A. 1997;94:674–678. doi: 10.1073/pnas.94.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tilton WM, Seaman C, Carriero D, Piomelli S. Regulation of glycolysis in the erythrocyte: role of the lactate/pyruvate and NAD/NADH ratios. J Lab Clin Med. 1991;118:146–152. [PubMed] [Google Scholar]
- 147.Ahmed N, Ahmed U, Thornalley PJ, Hager K, Fleischer G, Munch G. Protein glycation, oxidation and nitration adduct residues and free adducts of cerebrospinal fluid in Alzheimer’s disease and link to cognitive impairment. Journal of Neurochemistry. 2005;92:255–263. doi: 10.1111/j.1471-4159.2004.02864.x. [DOI] [PubMed] [Google Scholar]
- 148.Kuhla Br, Haase C, Flach K, Lüth H-J, Arendt T, Münch G. Effect of Pseudophosphorylation and Cross-linking by Lipid Peroxidation and Advanced Glycation End Product Precursors on Tau Aggregation and Filament Formation. Journal of Biological Chemistry. 2007;282:6984–6991. doi: 10.1074/jbc.M609521200. [DOI] [PubMed] [Google Scholar]
- 149.Sultana R, Perluigi M, Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer’s disease: insights into mechanism of neurodegeneration from redox proteomics. Antioxid Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 150.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. Actin S-glutathionylation: evidence against a thiol-disulphide exchange mechanism. Free Radic Biol Med. 2003;35:1185–1193. doi: 10.1016/s0891-5849(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 151.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 152.Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 153.Sultana R, Perluigi M, Butterfield DA. Oxidatively modified proteins in Alzheimer’s disease (AD), mild cognitive impairment and animal models of AD: role of Abeta in pathogenesis. Acta Neuropathol. 2009;118:131–150. doi: 10.1007/s00401-009-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 155.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 156.Gottlieb TM, Oren M. p53 and apoptosis. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 157.Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 158.Alves da Costa C, Paitel E, Mattson MP, Amson R, Telerman A, Ancolio K, Checler F. Wild-type and mutated presenilin 2 trigger p53-dependent apoptosis and down-regulate presenilin 1 expression in HEK293 human cells and in murine neurons. Proc Natl Acad Sci U S A. 2002;99:4043–4048. doi: 10.1073/pnas.062059899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.de la Monte SM, Sohn YK, Wands JR. Correlates of p53- and Fas (CD95)-mediated apoptosis in Alzheimer’s disease. J Neurol Sci. 1997;152:73–83. doi: 10.1016/s0022-510x(97)00131-7. [DOI] [PubMed] [Google Scholar]
- 160.Gilman CP, Chan SL, Guo Z, Zhu X, Greig N, Mattson MP. p53 is present in synapses where it mediates mitochondrial dysfunction and synaptic degeneration in response to DNA damage, and oxidative and excitotoxic insults. Neuromolecular Med. 2003;3:159–172. doi: 10.1385/NMM:3:3:159. [DOI] [PubMed] [Google Scholar]
- 161.Kitamura Y, Shimohama S, Kamoshima W, Matsuoka Y, Nomura Y, Taniguchi T. Changes of p53 in the brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 1997;232:418–421. doi: 10.1006/bbrc.1997.6301. [DOI] [PubMed] [Google Scholar]
- 162.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 163.Cenini G, Sultana R, Memo M, Butterfield DA. Effects of oxidative and nitrosative stress in brain on p53 proapoptotic protein in amnestic mild cognitive impairment and Alzheimer disease. Free Radic Biol Med. 2008;45:81–85. doi: 10.1016/j.freeradbiomed.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cenini G, Sultana R, Memo M, Butterfield DA. Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE, in brain from subjects with amnestic mild cognitive impairment and Alzheimer’s disease. J Cell Mol Med. 2008;12:987–994. doi: 10.1111/j.1582-4934.2008.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Di Domenico F, Cenini G, Sultana R, Perluigi M, Uberti D, Memo M, Butterfield DA. Glutathionylation of the pro-apoptotic protein p53 in Alzheimer’s disease brain: implications for AD pathogenesis. Neurochem Res. 2009;34:727–733. doi: 10.1007/s11064-009-9924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pellegrini M, Strasser A. A portrait of the Bcl-2 protein family: life, death, and the whole picture. J Clin Immunol. 1999;19:365–377. doi: 10.1023/a:1020598632068. [DOI] [PubMed] [Google Scholar]
- 167.Maruyama W, Akao Y, Youdim MB, Davis BA, Naoi M. Transfection-enforced Bcl-2 overexpression and an anti-Parkinson drug, rasagiline, prevent nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase induced by an endogenous dopaminergic neurotoxin, N-methyl(R)salsolinol. J Neurochem. 2001;78:727–735. doi: 10.1046/j.1471-4159.2001.00448.x. [DOI] [PubMed] [Google Scholar]
- 168.Maruyama W, Oya-Ito T, Shamoto-Nagai M, Osawa T, Naoi M. Glyceraldehyde-3-phospate dehydrogenase is translocated into nuclei through Golgi apparatus during apoptosis induced by 6-hydroxydopamine in human dopaminergic SH-SY5Y cells. Neurosci Lett. 2002;321:29–32. doi: 10.1016/s0304-3940(01)02490-9. [DOI] [PubMed] [Google Scholar]
- 169.Smith J. Human Sir2 and the ‘silencing’ of p53 activity. Trends Cell Biol. 2002;12:404–406. doi: 10.1016/s0962-8924(02)02342-5. [DOI] [PubMed] [Google Scholar]
- 170.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McCann SM. The nitric oxide hypothesis of brain aging. Exp Gerontol. 1997;32:431–440. doi: 10.1016/s0531-5565(96)00154-4. [DOI] [PubMed] [Google Scholar]
- 172.Guix FX, Uribesalgo I, Coma M, Munoz FJ. The physiology and pathophysiology of nitric oxide in the brain. Prog Neurobiol. 2005;76:126–152. doi: 10.1016/j.pneurobio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 173.Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 174.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Calabrese V, Cornelius C, Rizzarelli E, Owen JB, Dinkova-Kostova AT, Butterfield DA. Nitric Oxide in Cell Survival: A Janus Molecule. Antioxid Redox Signal. 2009 doi: 10.1089/ars.2009.2721. [DOI] [PubMed] [Google Scholar]
- 176.Molina y Vedia L, McDonald B, Reep B, Brune B, Di Silvio M, Billiar TR, Lapetina EG. Nitric oxide-induced S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase inhibits enzymatic activity and increases endogenous ADP-ribosylation. J Biol Chem. 1992;267:24929–24932. [PubMed] [Google Scholar]
- 177.McDonald LJ, Moss J. Stimulation by nitric oxide of an NAD linkage to glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1993;90:6238–6241. doi: 10.1073/pnas.90.13.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vaidyanathan VV, Sastry PS, Ramasarma T. Inverse relationship of the dehydrogenase and ADP-ribosylation activities in sodium-nitroprusside-treated glyceraldehyde-3-phosphate dehydrogenase is coincidental. Biochim Biophys Acta. 1993;1203:36–44. doi: 10.1016/0167-4838(93)90033-n. [DOI] [PubMed] [Google Scholar]
- 179.Dimmeler S, Brune B. Characterization of a nitric-oxide-catalysed ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1992;210:305–310. doi: 10.1111/j.1432-1033.1992.tb17422.x. [DOI] [PubMed] [Google Scholar]
- 180.Zhang J, Snyder SH. Nitric oxide stimulates auto-ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1992;89:9382–9385. doi: 10.1073/pnas.89.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dimmeler S, Lottspeich F, Brune B. Nitric oxide causes ADP-ribosylation and inhibition of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1992;267:16771–16774. [PubMed] [Google Scholar]
- 182.Moss J, Vaughan M. ADP-ribosylation of guanyl nucleotide-binding regulatory proteins by bacterial toxins. Adv Enzymol Relat Areas Mol Biol. 1988;61:303–379. doi: 10.1002/9780470123072.ch6. [DOI] [PubMed] [Google Scholar]
- 183.Brune B, Lapetina EG. Properties of a novel nitric oxide-stimulated ADP-ribosyltransferase. Arch Biochem Biophys. 1990;279:286–290. doi: 10.1016/0003-9861(90)90493-i. [DOI] [PubMed] [Google Scholar]
- 184.Melino G, Bernassola F, Knight RA, Corasaniti MT, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 185.Liu L, Stamler JS. NO: an inhibitor of cell death. Cell Death Differ. 1999;6:937–942. doi: 10.1038/sj.cdd.4400578. [DOI] [PubMed] [Google Scholar]
- 186.Nagano Y, Yamashita H, Takahashi T, Kishida S, Nakamura T, Iseki E, Hattori N, Mizuno Y, Kikuchi A, Matsumoto M. Siah-1 facilitates ubiquitination and degradation of synphilin-1. J Biol Chem. 2003;278:51504–51514. doi: 10.1074/jbc.M306347200. [DOI] [PubMed] [Google Scholar]
- 187.Moriyoshi K, Iijima K, Fujii H, Ito H, Cho Y, Nakanishi S. Seven in absentia homolog 1A mediates ubiquitination and degradation of group 1 metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2004;101:8614–8619. doi: 10.1073/pnas.0403042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah-1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 189.Fiucci G, Beaucourt S, Duflaut D, Lespagnol A, Stumptner-Cuvelette P, Geant A, Buchwalter G, Tuynder M, Susini L, Lassalle JM, Wasylyk C, Wasylyk B, Oren M, Amson R, Telerman A. Siah-1b is a direct transcriptional target of p53: identification of the functional p53 responsive element in the siah-1b promoter. Proc Natl Acad Sci U S A. 2004;101:3510–3515. doi: 10.1073/pnas.0400177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, Aggrawal N, Hester L, Dore S, Snyder SH, Sawa A. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63:81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mielke R, Schroder R, Fink GR, Kessler J, Herholz K, Heiss WD. Regional cerebral glucose metabolism and postmortem pathology in Alzheimer’s disease. Acta Neuropathol. 1996;91:174–179. doi: 10.1007/s004010050410. [DOI] [PubMed] [Google Scholar]
- 192.Mercer RW, Dunham PB. Membrane-bound ATP fuels the Na+/K+ pump. Studies on membrane-bound glycolytic enzymes on inside-out vesicles from human red cell membranes. J Gen Physiol. 1981;78:547–568. doi: 10.1085/jgp.78.5.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hardin CD, Zhang C, Kranias EG, Steenaart NA, Raeymaekers L, Paul RJ. Regulation of glycolytically fueled Ca2+ uptake in smooth muscle plasmalemmal vesicles by phosphorylation. Am J Physiol. 1993;265:H1326–1333. doi: 10.1152/ajpheart.1993.265.4.H1326. [DOI] [PubMed] [Google Scholar]
- 194.Galli F, Rovidati S, Ghibelli L, Canestrari F. S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase decreases the enzyme affinity to the erythrocyte membrane. Nitric Oxide. 1998;2:17–27. doi: 10.1006/niox.1997.0148. [DOI] [PubMed] [Google Scholar]
- 195.Xu KY, Zweier JL, Becker LC. Functional coupling between glycolysis and sarcoplasmic reticulum Ca2+ transport. Circ Res. 1995;77:88–97. doi: 10.1161/01.res.77.1.88. [DOI] [PubMed] [Google Scholar]
- 196.Kant JA, Steck TL. Specificity in the association of glyceraldehyde-3-phosphate dehydrogenase with isolated human erythrocyte membranes. J Biol Chem. 1973;248:8457–8464. [PubMed] [Google Scholar]
- 197.Rogalski AA, Steck TL, Waseem A. Association of glyceraldehyde-3-phosphate dehydrogenase with the plasma membrane of the intact human red blood cell. J Biol Chem. 1989;264:6438–6446. [PubMed] [Google Scholar]
- 198.Tsai IH, Murthy SN, Steck TL. Effect of red cell membrane binding on the catalytic activity of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1982;257:1438–1442. [PubMed] [Google Scholar]
- 199.Brorson JR, Schumacker PT, Zhang H. Nitric oxide acutely inhibits neuronal energy production. The Committees on Neurobiology and Cell Physiology. J Neurosci. 1999;19:147–158. doi: 10.1523/JNEUROSCI.19-01-00147.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Selak I, Skaper SD, Varon S. Pyruvate participation in the low molecular weight trophic activity for central nervous system neurons in glia-conditioned media. J Neurosci. 1985;5:23–28. doi: 10.1523/JNEUROSCI.05-01-00023.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kauppinen RA, Enkvist K, Holopainen I, Akerman KE. Glucose deprivation depolarizes plasma membrane of cultured astrocytes and collapses transmembrane potassium and glutamate gradients. Neuroscience. 1988;26:283–289. doi: 10.1016/0306-4522(88)90145-5. [DOI] [PubMed] [Google Scholar]
- 202.Kahlert S, Reiser G. Requirement of glycolytic and mitochondrial energy supply for loading of Ca(2+) stores and InsP(3)-mediated Ca(2+) signaling in rat hippocampus astrocytes. J Neurosci Res. 2000;61:409–420. doi: 10.1002/1097-4547(20000815)61:4<409::AID-JNR7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 203.Weiss JN, Lamp ST. Cardiac ATP-sensitive K+ channels. Evidence for preferential regulation by glycolysis. J Gen Physiol. 1989;94:911–935. doi: 10.1085/jgp.94.5.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Canzoniero LM, Snider BJ. Calcium in Alzheimer’s disease pathogenesis: too much, too little or in the wrong place? J Alzheimers Dis. 2005;8:147–154. doi: 10.3233/jad-2005-8207. discussion 209–115. [DOI] [PubMed] [Google Scholar]
- 205.Mattson MP, Chan SL. Neuronal and glial calcium signaling in Alzheimer’s disease. Cell Calcium. 2003;34:385–397. doi: 10.1016/s0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 206.Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008;31:454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 208.Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 209.Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Gogic G, Chan J, Cravchik A, Ross D, Lau K, Kwok S, Chang SY, Catanese J, Sninsky J, White TJ, Hardy J, Powell J, Lovestone S, Morris JC, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101:15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lin PI, Martin ER, Bronson PG, Browning-Large C, Small GW, Schmechel DE, Welsh-Bohmer KA, Haines JL, Gilbert JR, Pericak-Vance MA. Exploring the association of glyceraldehyde-3-phosphate dehydrogenase gene and Alzheimer disease. Neurology. 2006;67:64–68. doi: 10.1212/01.wnl.0000223438.90113.4e. [DOI] [PubMed] [Google Scholar]
- 211.Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM. Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem. 1996;66:928–935. doi: 10.1046/j.1471-4159.1996.66030928.x. [DOI] [PubMed] [Google Scholar]
- 212.Sunaga K, Takahashi H, Chuang DM, Ishitani R. Glyceraldehyde-3-phosphate dehydrogenase is over-expressed during apoptotic death of neuronal cultures and is recognized by a monoclonal antibody against amyloid plaques from Alzheimer’s brain. Neurosci Lett. 1995;200:133–136. doi: 10.1016/0304-3940(95)12098-o. [DOI] [PubMed] [Google Scholar]
- 213.Tamaoka A, Endoh R, Shoji S, Takahashi H, Hirokawa K, Teplow DB, Selkoe DJ, Mori H. Antibodies to amyloid β protein (Aβ) crossreact with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Neurobiol Aging. 1996;17:405–414. doi: 10.1016/0197-4580(96)00031-0. [DOI] [PubMed] [Google Scholar]
- 214.Ponte P, Gonzalez-DeWhitt P, Schilling J, Miller J, Hsu D, Greenberg B, Davis K, Wallace W, Lieberburg I, Fuller F. A new A4 amyloid mRNA contains a domain homologous to serine proteinase inhibitors. Nature. 1988;331:525–527. doi: 10.1038/331525a0. [DOI] [PubMed] [Google Scholar]
- 215.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 216.Gandy S, Czernik AJ, Greengard P. Phosphorylation of Alzheimer disease amyloid precursor peptide by protein kinase C and Ca2+/calmodulin-dependent protein kinase II. Proc Natl Acad Sci U S A. 1988;85:6218–6221. doi: 10.1073/pnas.85.16.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kitaguchi N, Takahashi Y, Tokushima Y, Shiojiri S, Ito H. Novel precursor of Alzheimer’s disease amyloid protein shows protease inhibitory activity. Nature. 1988;331:530–532. doi: 10.1038/331530a0. [DOI] [PubMed] [Google Scholar]
- 218.Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL. Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer’s disease. Nature. 1988;331:528–530. doi: 10.1038/331528a0. [DOI] [PubMed] [Google Scholar]
- 219.Naletova I, Schmalhausen E, Kharitonov A, Katrukha A, Saso L, Caprioli A, Muronetz V. Non-native glyceraldehyde-3-phosphate dehydrogenase can be an intrinsic component of amyloid structures. Biochim Biophys Acta. 2008;1784:2052–2058. doi: 10.1016/j.bbapap.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 220.Cumming RC, Schubert D. Amyloid-β induces disulfide bonding and aggregation of GAPDH in Alzheimer’s disease. FASEB J. 2005;19:2060–2062. doi: 10.1096/fj.05-4195fje. [DOI] [PubMed] [Google Scholar]
- 221.Trojanowski JQ, Lee VM. Phosphorylation of paired helical filament tau in Alzheimer’s disease neurofibrillary lesions: focusing on phosphatases. FASEB J. 1995;9:1570–1576. doi: 10.1096/fasebj.9.15.8529836. [DOI] [PubMed] [Google Scholar]
- 222.Yang G, Wang L, Zhu M, Xu D. Identification of non-Alzheimer’s disease tauopathies-related proteins by proteomic analysis. Neurol Res. 2008;30:613–622. doi: 10.1179/174313208X284124. [DOI] [PubMed] [Google Scholar]
- 223.Chen YH, He RQ, Liu Y, Xue ZG. Effect of human neuronal tau on denaturation and reactivation of rabbit muscle D-glyceraldehyde-3-phosphate dehydrogenase. Biochem J. 2000;351:233–240. doi: 10.1042/0264-6021:3510233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Karp G. Cell and Molecular Biology. Concepts and Experiments. John Wiley & Sons, Inc; New York, NY, U. S. A: 2003. [Google Scholar]
- 225.Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer’s disease: an initial assessment. J Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- 226.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer’s disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 227.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Merchant M, Markesbery WR, Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer’s disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 228.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II: dihydropyrimidinase-related protein 2, α-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 229.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 230.Reed TT, Pierce WM, Jr, Turner DM, Markesbery WR, Butterfield DA. Proteomic identification of nitrated brain proteins in early Alzheimer’s disease inferior parietal lobule. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer’s disease. Neurobiol Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]