Abstract
Background
The effectiveness of rotavirus vaccines will be dependent upon the immunity conferred against prevalent and emergent variants causing severe diarrhoeal disease. Longitudinal surveillance of disease causing strains is a pre-requisite to intervention.
Methods
Molecular characterization was conducted on rotavirus positive stool samples from children admitted with diarrhea to a rural district hospital 2002-04. Extracted viral RNA was separated by polyacrylamide gel electrophoresis (PAGE) and specific rotavirus VP4 (P-types) and VP7 (G-types) determined.
Results
Of 558 cases investigated the predominant genotype was P[8]G1 (42%), followed by P[8]G9 (15%), P[4]G8 (7%), P[6]G8 (6%), and P[8]G8 (4%), with 10% mixed strains. In total there were 6 different P types and 7 G types. No association was identified between genotype and child age, sex or severity of diarrhea. The P and G genotypes and PAGE electropherotypes showed significant temporal variation in frequency: P[8]G1 declined from 51% (95%CI 43-58) in 2002 to 30% (24-37) in 2004, and P[4]G8 increased from 2% (0-5) in 2002 to 13% (9-19). Quarterly data revealed seasonally endemic and emergence/decay patterns.
Conclusions
Our study of rotaviruses causing severe diarrhea in rural Kenyan children shows a predominance of P[8]G1, and confirms the importance of G8 and G9 strains in sub-Saharan Africa. Considerable genetic diversity of rotaviruses was observed, including substantial mixed and unusual types, coupled to significant temporal strain variation and emergence. These results warn of variable vaccine efficacy and the need for long-term surveillance of circulating rotavirus genotypes.
Keywords: rotavirus genotypes, diversity, vaccination, Kenya children
Introduction
Rotavirus is the leading cause of acute severe diarrhea in children under the age of 5 years, and estimated to be a major cause of childhood death in the developing world, in particular the Indian sub-continent and sub-Saharan Africa[1, 2]. Group A rotaviruses (GARV), which account for the vast majority of human disease, are classified into different P and G-types based on the two immunodominant outer capsid proteins, VP4 and VP7, respectively. A total of 28 P-types and 20 G-types[3-6] have been identified worldwide, with the P[8]G1, P[8]G3, P[8]G4, P[4]G2, P[8]G9 and P[6]G9 commonly detected in annual epidemics[7].
Studies have shown wide geographical variation in G and P-type prevalence across continents, and local and global temporal changes in the frequency of dominant strains and emergence of unusual P and G types and combinations[7]. Globally, 85% of rotavirus strains studied carry P[8]G1, P[4]G2, P[8]G3 or P[8]G4 specificity. However, in South America the proportion of the common genotypes is 68% while in Africa only 50% of strains belong to one of these genotypes. Furthermore, studies in Africa show only 23% of strains with P[8]G1 (compared to 65% worldwide) and significant proportions of P[6], G8, unusual and mixed infections [7-15].
Within the last 15 years, G9 strains have emerged as an important disease causing variant and recently genotype G12 strains have also emerged globally in a manner similar to the emergence of G9 strains in early nineties[7]. Within Africa, both of these genotypes have been detected and G9 strains have been shown to predominate in certain settings, replacing the more traditional genotypes for a season or two or circulating at low background levels over a number of seasons. In addition, the G9 genotype has been found in combination with P[6] or P[8] VP4 specificity, subgroup I or subgroup II VP6 specificity and either long or short electropherotypes, suggesting a “promiscuous” nature and a predilection for reassortment[16, 17].
Assessing the impact of a rotavirus vaccine should take into account the natural temporal variability in P and G types. The compositions of the two live oral rotavirus vaccines, Rotarix® and Rotateq™, present very different immunization strategies, with the former based primarily on heterotypic immunity and the latter based more on homotypic immunity[18, 19]. While both have demonstrated safety and proved to be efficacious in various developed and developing country settings, vaccine effectiveness following implementation will have to be closely monitored. In this way, any influence on vaccine efficacy by the emergence and temporal increase in the frequency of new variants, both naturally occurring and induced through the process of genotype replacement, can be determined. Longitudinal genetic surveillance of disease causing rotaviruses is a clear priority, and particularly so in Africa prior to and after wide scale vaccine introduction.
Materials and Methods
Study samples
The present study makes use of rotavirus positive stool samples arising from surveillance of pediatric (0-12 years) admissions to Kilifi District Hospital (KDH), coastal Kenya between January 2002 and December 2004[20]. Among 2,039 diarrhea cases and 620 non-diarrhea controls tested, 588 (29%) and 19 (3%), respectively, were group A rotavirus (GARV) antigen positive as determined by enzyme-immunoassay (DAKO Rotavirus IDEIA™, Oxoid, Ely, United Kingdom). Contemporaneous controls were selected from pediatric admissions without a history of diarrhea frequency matched by age to cases with a ratio of 1:3. Samples were stored unprocessed at -80°C prior to shipping on dry ice to Medical Research Council - Diarrhoeal Pathogens Research Unit, South Africa for further analysis. The present study undertook molecular characterization on 558 (95%) and 12 (63%) GARV isolates obtained from patients with or without severe diarrhea, respectively.
Life-threatening diarrhea was assigned to a case with one or more of the following clinical features: hypoxia (<90% saturation), prostration or coma, hyponatremia, hypoglycemia, severe malnourishment, or bacteremia. Full details of the sampling and testing procedures, clinical definitions and the socio-demographic, and clinical and laboratory characteristics of the children are reported elsewhere [20].
Polyacrylamide gel electrophoresis (PAGE)
The dsRNA genome was extracted from 10% fecal suspensions using the standard phenol-chloroform method, followed by ethanol precipitation in the presence of 0.3M sodium acetate. The extracted RNA was resolved on 10% polyacrylamide gels with 3% stacking gels, using a discontinuous buffer system at 100 V for 18 hours at ambient temperature[21]. RNA segments were then stained by silver nitrate according to published method[22].
Viral RNA extraction for RT-PCR
Viral dsRNA was extracted from 250μl of rotavirus-positive faecal suspensions using Tri-Reagents-LS (Molecular Research Centre Ohio, USA) according to the manufacturers’ instructions. The extracted dsRNA was further precipitated in ice-cold isopropyl alcohol and the air dried pellet resuspended in 40 μl of deionised water.
RT-PCR
The extracted dsRNA was subjected to reverse transcription and amplification by polymerase chain reaction (RT-PCR) using several primer pairs taken from highly conserved regions of the RNA genome. The primer pair, sBeg/End9, were used to generate full-length copies of the VP7 gene (1062bp), while 9con1/EndA were utilized to generate 903bp VP7 gene fragments in difficult to type specimens[23-25]. Con2/Con3 and VP4F/VP4R primer pairs were used to amplify VP4 gene fragments of the 876bp and 663bp, respectively [26, 27]
Rotavirus typing
Genotyping of the cDNA was carried out by nested multiplex PCR using type-specific VP7 and VP4 primers described previously[10, 23, 25, 26, 28-30]. The non-typeable G and P genotypes were further analysed using animal G and P primers described by Gouvea and colleagues [31, 32]. Combinations of P and G genotypes have been defined as usual or unusual based on delineations described by Santos and Hoshino[7].
Statistical analysis
Data were analyzed using STATA 10.1 (Statacorp TX, USA). Exact 95% confidence intervals (95%CI) were defined for genotype frequencies. Chi-square was used to test for lack of independence in cross-tabulations (i.e. for possible association between two variables), pooling marginal totals with frequency less than 15, and the Wilcoxon rank-sum test for equality of distributions.
Results
Among 558 group A rotavirus (GARV) positive diarrhea cases (median age 10 months IQR 7-15; 60% male), 82 were PAGE negative, 26 were G non-typeable (GNT), 30 were P non-typeable (P[NT]) (including 6 both GNT and P[NT]), and 508 were positive for G and P types. Within 12 GARV positive non-diarrhea controls (median age 25 months IQR 2-39; 42% male), 2 were negative for PAGE and P type.
PAGE distribution in diarrhea cases
A total of 86/476 (18%) and 390/476 (82%) rotavirus strains displayed short and long electropherotypes, respectively (Table S1). Three long electropherotypes (L1, L2 and L3) were detected in 63%, 11% and 4.4% of cases, respectively, while short electropherotypes (S3, S2 and S1) were identified in 8%, 4.6% and 3.6% of cases. The remainder of the electropherotypes detected (L4-6, mixed and S4) revealed prevalences of less than 2%. There is evidence of temporal variation in the PAGE composition between the three years, in particular L1 and L2 and S3 types (χ2(10) 123.681 P<0.001, Table S1). The proportion PAGE negative did not change by year (χ2(2) 3.493 P=0.174).
Genotype distribution in diarrhea cases
For the 558 cases analyzed for both VP4 (P) and VP7 (G) genes, 6 different P types and 7 G types were identified in both single and mixed infections (Tables 1-2). Among the P types, P[8] was predominant (68% of all samples) followed by P[6] and P[4] (both 12%), with the other types (P[9], P[11], P[14]) with low prevalence and only in mixed infections (Table 2). Among the G types, G1 predominated (48%), followed by G9 (19%), G8 (18%), with the remainder (G2, G3, G4, G10) at low prevalence and G3, G4 and G10 only as mixed infections (Table 2).
Table 1.
P and G combinations for 558 group A rotavirus positive pediatric diarrhea cases admitted to Kilifii District Hospital, Kenya 2002-2004
| Year | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | 2002 | % | 2003 | % | 2004 | % | Total | % | |
| Usual* | |||||||||
| P[4]G2x | 1 | 1 | 4 | 2 | 0 | 0 | 5 | 1 | |
| P[6]G1 | 3 | 2 | 0 | 0 | 6 | 3 | 9 | 2 | |
| P[6]G9xq | 3 | 2 | 4 | 2 | 5 | 2 | 12 | 2 | |
| P[8]G1 | 90 | 51 | 79 | 47 | 63 | 30 | 232 | 42 | |
| P[8]G9 | 23 | 13 | 36 | 21 | 25 | 12 | 84 | 15 | |
| sub-total | 120 | 67 | 123 | 73 | 99 | 47 | 342 | 61 | |
| Unusual* | |||||||||
| P[4]G1 | 0 | 0 | 0 | 0 | 10 | 5 | 10 | 2 | |
| P[4]G8xq | 3 | 2 | 7 | 4 | 28 | 13 | 38 | 7 | |
| P[4]G9xq | 0 | 0 | 2 | 1 | 1 | 0 | 3 | 1 | |
| P[6]G8xq | 17 | 10 | 1 | 1 | 15 | 7 | 33 | 6 | |
| P[8]G2 | 4 | 2 | 1 | 1 | 1 | 0 | 6 | 1 | |
| P[8]G8 | 8 | 4 | 1 | 1 | 13 | 6 | 22 | 4 | |
| sub-total | 32 | 18 | 12 | 7 | 68 | 32 | 112 | 20 | |
| Non-typeable & mixed% | |||||||||
| P[4]GNTxq | 0 | 0 | 0 | 0 | 6 | 3 | 6 | 1 | |
| P[6]GNTxq | 2 | 1 | 0 | 0 | 3 | 1 | 5 | 1 | |
| P[8]GNT | 8 | 4 | 0 | 0 | 1 | 0 | 9 | 2 | |
| P[NT]G1 | 5 | 3 | 5 | 3 | 2 | 1 | 12 | 2 | |
| P[NT]G8xq | 1 | 1 | 1 | 1 | 1 | 0 | 3 | 1 | |
| P[NT]G9xq | 2 | 1 | 2 | 1 | 2 | 1 | 6 | 1 | |
| P[NT]GNT | 1 | 1 | 2 | 1 | 3 | 1 | 6 | 1 | |
| mixed | 7 | 4 | 24 | 14 | 26 | 12 | 57 | 10 | |
| sub-total | 26 | 15 | 34 | 20 | 44 | 21 | 104 | 19 | |
| Total | 178 | 100 | 169 | 100 | 211 | 100 | 558 | 100 | |
Notes:
Usual and unusual genotypes worldwide as defined by Santos and Hoshino[7]
P[NT], GNT, P[NT]GNT - non-typeable for VP4(P), VP7(G) or both, respectively.
types not included in Rotarix® (x) or Rotateq™ (q) vaccines
Table 2.
P-G type combinations in 57 mixed infections from pediatric diarrhea cases admitted to Kilifii District Hospital, Kenya 2002-2004
| P-type | G-type | 2002 | 2003 | 2004 |
|---|---|---|---|---|
| P[4] | G1,8 | 0 | 1 | 2 |
| P[4] | G1,9 | 0 | 1 | 1 |
| P[4] | G1,10 | 0 | 1 | 0 |
| P[4] | G8,9 | 0 | 0 | 1 |
| P[4,6] | G8 | 0 | 0 | 2 |
| P[4,8] | G1 | 1 | 0 | 0 |
| P[4,11] | G4 | 0 | 0 | 1 |
| P[4,11] | G8 | 0 | 1 | 0 |
| P[4,14] | G2 | 1 | 0 | 0 |
| P[6] | G1,2 | 0 | 0 | 1 |
| P[6] | G1,8 | 0 | 1 | 1 |
| P[6] | G1,9 | 0 | 1 | 1 |
| P[6] | G8,9 | 0 | 0 | 1 |
| P[6,8] | G1 | 0 | 1 | 0 |
| P[6,8] | G9 | 0 | 0 | 2 |
| P[6,8] | G1,8 | 0 | 1 | 0 |
| P[6,8] | G2,8,3 | 0 | 1 | 0 |
| P[6,8,9] | G9 | 0 | 0 | 1 |
| P[6,9] | G1 | 0 | 0 | 1 |
| P[8] | G1,2 | 1 | 1 | 0 |
| P[8] | G1,8 | 0 | 9 | 9 |
| P[8] | G1,9 | 2 | 2 | 1 |
| P[8] | G8,9 | 0 | 2 | 0 |
| P[8,9] | G1 | 1 | 0 | 0 |
| NT | G1,8 | 0 | 1 | 1 |
| NT | G1,9 | 1 | 0 | 0 |
| Total | 7 | 24 | 26 |
The predominant P-G genotype was P[8]G1 detected in 42% of cases, followed by P[8]G9 at 15%, P[4]G8 at 7%, P[6]G8 at 7% and P[8]G8 at 4%. Usual and unusual genotype combinations were identified in 61% and 20% of isolates, respectively, or if excluding mixed isolates in 68% and 25%, respectively (Table 1). The remaining single genotypes occurred at low frequency. Mixed infections occurred in 10% (n=57) of cases, with 56% of these involving P[8] and 32% including P[8] in combination with G1 (Table 2). Specimens untypeable for at least one of VP4 or VP7 genes occurred with a frequency of 9% (Table 1).
Genotype association with age, sex, or disease severity
Data on genotypes with a case frequency of <15 were pooled for analysis. Comparing 350 infants (<1 year) and 208 older children (1-5 years), there was no significant difference in the distribution of genotypes (χ2(6) 3.745 P=0.711). Similarly, the distribution of genotypes in 334 males compared to 224 females did not significantly differ (χ2(6) 5.992 P=0.424). There was no association between genotype and cases with (n=119) or without (n=439) life-threatening diarrhea (χ2(6) 4.453 P=0.616).
Temporal variation in genotype distribution
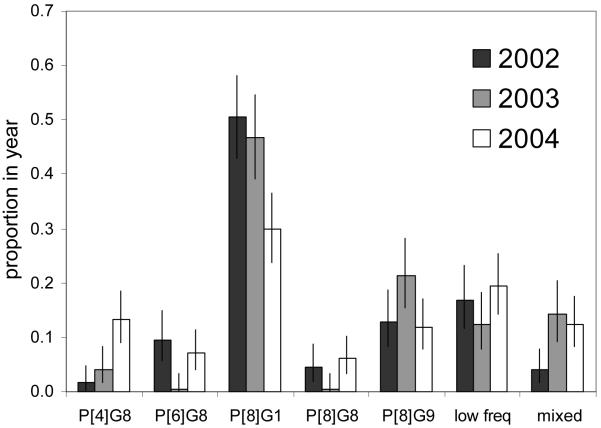
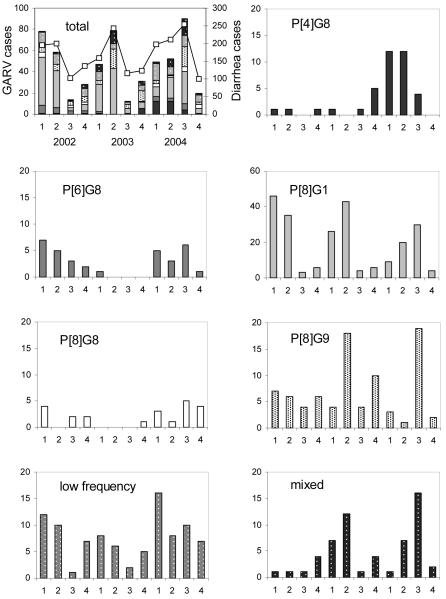
The distribution of rotavirus genotypes, by year, is shown in Tables 1 & 2. Pooling low frequency observations (<15 cases over the 3 years) there is evidence for significant temporal variation in P-G genotype composition (χ2(12) = 72.841, P<0.001), in particular an increase in P[4]G8 from 2% in 2002 to 13% in 2004, and a decrease in P[8]G1 from 51% in 2002 to 30% in 2004 (Fig.1). Other genotypes, e.g. P[6]G8, showed significant fluctuation. Genotype observations stratified by year and quarter (Fig.2) provide further detail of the temporal variation, including seasonal patterns (P[8]G1), emergence (P[4]G8), and decay and re-emergence (P[6]G8). The diversity of single genotype infections was roughly constant at 9 different genotypes in 2002-3 and 10 genotypes in 2004 (Table 1). However, as seen in Fig.2 and Fig S1a-c, year 2004 showed a decrease in the dominance of one (primarily P[8]G1) or a few genotypes coupled to altered seasonality in total diarrhea admissions and rotavirus cases, compared to 2002-3.
Fig. 1.
Variation by year (2002-04) in P-G genotype frequencies (with exact 95% CI) determined for 558 rotavirus positive samples from Kilifi District Hospital, Kenya 2002-04. Low frequency types include all single strain genotypes with a frequency over the 3 years of less than 15 cases.
Fig. 2.
Seasonal variation in P-G genotypes determined for 558 group A rotavirus (GARV) positive samples from Kilifi District Hospital, Kenya 2002-04. The top left panel details totals for all variants by year and quarter (bars) and total cases of diarrhea (lines) from which GARV positives identified.
During 2004, an increase in G8 strains in combination with P[4], P[6] and P[8] were noted (Table 1). Investigation of mixed infections during 2003 provides possible evidence for human-human reassortment events as G1G8P[6]P[8], G2G8G3P[6]P[8] and G8P[4]P[6] infections were detected. In addition, the increase in G1P[4] infections in 2004 may have arisen from mixed infections with G8, G9 and G10 in 2003. Additional analysis of the full genomes of unusual human-human reassortants and investigation of the population of rotaviruses in mixed infections will be required.
Controls
Among the 12 positive controls with genetic results, 7 were L1 and 3 L2, and 4 were P[8]G1, 2 P[8]G9, 2 P[NT]G1 and 4 mixed (3 P[8]G1,9; 1 P[8]G8,9), approximately corresponding to the distribution in cases. The asymptomatic controls were noticeably older than the symptomatic cases (25 months vs. 10 months).
Vaccine type prevalence
The total number of cases with types in common with those in the Rotarix® (P[8]G1) and Rotateq™ (P[5]G1-4, P[8]G6) vaccines was 77% (432/558) and 79% (439/558), respectively. The proportion not represented in Rotarix® was 16% (29/178), 12% (20/169) and 33% (70/211) in 2002-4, respectively (χ2(2) 24.291 P=<0.001), and, correspondingly, the proportion not represented in Rotateq™ was 17% (31/178), 14% (24/169), and 34% (71/211) (χ2(2) 29.420, P=<0.001).
Discussion
We have investigated the genetic characteristics of group A rotavirus in clinically well defined severe pediatric diarrhea cases in a rural Kenyan setting over a three year period. The study reveals no evidence of an association between genotype and age, sex or disease severity. However, we identify considerable genotypic diversity with patterns of prevalence both consistent and in contrast to elsewhere in Africa. Similar to Africa in general and specifically to Kenya we find a predominance of genotype combinations G1, G8, G9 with P[4], P[6] and P[8][7, 11]. The G-P combinations found in over 90% of isolates in temperate countries of Europe, North America and parts of Australia/Oceania were identified at a proportion of only 68%; consistent with the African context[7]. Yet the prevalence of P[8]G1 (42%) and P[8]G9 (15%) were unusually high, there was an absence of two usually frequent genotypes, P[8]G3 and P[8]G4, and no case of the emergent G12 type was identified. Those genotypes recognized as unusual worldwide[7], in particular G8 variants (P[4]G8, P[8]G8 and P[6]G8), comprised 25% of all single strains (Table 1), and the prevalence of mixed G-P combinations was 10%; both proportions similar to that summarized for Africa[7].
Furthermore, we observed significant temporal variation in the prevalence of some genotypes. While P[8]G1 was the predominant variant throughout, its prevalence declined markedly from ~50% in 2002-3 to 30% in 2004. By contrast P[4]G8 increased from 2-4% in 2002-3 to 13% in 2004, and both P[6]G8 and P[8]G8 had moderate prevalence in 2002 and 2004 and were practically absent in 2003. Of further interest is that mixed genotypes showed a significant increase from 2002 to 2003-4. Temporal variation of this nature has been observed previously[33]. Kenya, for example, has seen the emergence of G8 and G9, and a shift in predominance of P[8] to P[6], accompanied by considerable year on year variation[34]. However, our data show local patterns inconsistent with the national picture: for example P[8] remains the predominant P-type in Kilifi for each of the years. All these studies demonstrate the limitation of short-term surveillance in discerning patterns of prevalence, but in addition, our results show the importance of widespread monitoring to capture local variation, which may be of importance in assessing vaccine impact.
Rotavirus hospitalizations are often observed to have marked seasonality, particularly in developed countries. In Kilifi, we observed peak group A rotavirus admissions between March and May in 2002-3, with higher prevalence throughout 2004 particular in the 3rd quarter prevalence, all of which mirrored total diarrhea admissions over this period (Fig. 2)[20]. Year 2004 was also associated with greater proportional representation of a wide group of variants (Fig.2), with the emergence of P[4]G8 early in 2004 associated with a delay in the peak in P[8]G1 to the 3rd quarter. Clearly, longer-term surveillance has the capacity to identify genotype emergence, decay and altering dominance patterns, which have a bearing on potential vaccine effectiveness and the value of before–after studies in assessing vaccine impact.
The diversity of GARV types identified in Kilifi is supplemented by a high proportion of unusual G-P combinations, among which are likely reassortant strains. These include rare occurrences of P[4]G9 and P[8]G2 and emergence of P[4]G1, all probable human-human reassortants. The presence, among isolates of mixed G-P type, of strains inclusive of these unusual and emergent combinations (eg P[4]G1,8; P[4]G8,9; P[6,8]G1,8; P[6,8]G2,3,8; P[8]G1,8; and P[8]G8,9), provides strong circumstantial evidence for natural reassortant events arising from co-circulating local strains. The prevalence of 9% of strains untypeable for at least one of VP4 or VP7 genes, presumably a result of antigenic drift, provides further evidence for extensive natural variation in rotavirus strains in this location.
Esona and colleagues[35] recently investigated the characteristics of all 11 genes of G8 rotavirus strains detected in Cote d’ Ivoire, Cameroon, Ethiopia and Tunisia. The study found that genes for VP7, NSP2 and NSP5 were closely related to cognate genes of animal strains and suggested that African G8 strains may have arisen through reassortment of VP7 and VP4 genes. In this study, large numbers of P[4]G8 (emerging in 2004) and P[6]G8 and P[8]G8 (showing marked year to year variation) were detected. Furthermore, G10, P[11] and P[14] genotypes, more commonly associated with infections in animals, were also detected in mixed infections Although additional analysis on the full genomes of these strains will be required, the possibility exists that these strains are human-animal reassortants and may provide evidence of animal rotaviruses acting as reservoirs for one or several genes of human rotavirus strains.
Severe rotavirus disease represents a major burden in the developing world and is an identified target for vaccine intervention[36]. Antigenic diversity of strains in co-circulation, antigenic drift and emergence of new variants through reassortment (human with human, and human with animal) and animal introductions[33], represent considerable potential for impaired vaccine efficacy. Our study provides support for the presence of each of these elements of variation in the Kilifi setting, which together with evidence of geographical and temporal variation in prevalent genotype composition, have a bearing on potential vaccine effectiveness and on the potential to measure vaccine impact through surveillance.
Supplementary Material
Acknowledgements
We would like to thank the participants involved in the study and their caregivers, and the rotavirus study staff (in the ward and the laboratory), pediatric wards, hospital administration. The study is published with permission of the Director of KEMRI.
Funding: Financial support was provided by the Wellcome Trust (grant# 076278 (DJN) and 076934 (TNW) and the Rotavirus Vaccine Program, PATH (GAV.1142-01-07211-SPS). The funding agency had no role in study design, data collection or preparation of this manuscript. The authors declare there to be no conflict of interest in relation to the publication of this work.
Footnotes
Ethical approval: The Kenyan National Research Ethical Committee and the Coventry Research Ethics Committee, UK, granted ethical approval for the study.
Referenecs
- 1.Molbak K, Fischer TK, Mikkelsen CS. The estimation of mortality due to rotavirus infections in sub-Saharan Africa. Vaccine. 2000;19:393–5. doi: 10.1016/s0264-410x(00)00199-7. [DOI] [PubMed] [Google Scholar]
- 2.Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304–6. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estes MK, Kapikian AZ. Chapter 53: Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MM, editors. Fields Virology. 5th ed. Vol. 2. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 1917–1958. [Google Scholar]
- 4.Khamrin P, Maneekarn N, Peerakome S, et al. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology. 2007;361:243–52. doi: 10.1016/j.virol.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Ciarlet M, Rahman M, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153:1621–9. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solberg OD, Hasing ME, Trueba G, Eisenberg JN. Characterization of novel VP7, VP4, and VP6 genotypes of a previously untypeable group A rotavirus. Virology. 2009;385:58–67. doi: 10.1016/j.virol.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 8.Adah MI, Rohwedder A, Olaleye OD, Durojaiye OA, Werchau H. Further characterization of field strains of rotavirus from Nigeria VP4 genotype P6 most frequently identified among symptomatically infected children. J Trop Pediatr. 1997;43:267–74. doi: 10.1093/tropej/43.5.267. [DOI] [PubMed] [Google Scholar]
- 9.Armah GE, Pager CT, Asmah RH, et al. Prevalence of unusual human rotavirus strains in Ghanaian children. J Med Virol. 2001;63:67–71. [PubMed] [Google Scholar]
- 10.Cunliffe NA, Gondwe JS, Broadhead RL, et al. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57:308–12. [PubMed] [Google Scholar]
- 11.Kiulia NM, Kamenwa R, Irimu G, et al. The epidemiology of human rotavirus associated with diarrhoea in Kenyan children: a review. J Trop Pediatr. 2008;54:401–5. doi: 10.1093/tropej/fmn052. [DOI] [PubMed] [Google Scholar]
- 12.Nakata S, Gatheru Z, Ukae S, et al. Epidemiological study of the G serotype distribution of group A rotaviruses in Kenya from 1991 to 1994. J Med Virol. 1999;58:296–303. doi: 10.1002/(sici)1096-9071(199907)58:3<296::aid-jmv17>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Page NA, Steele AD. Antigenic and genetic characterization of serotype G2 human rotavirus strains from South Africa from 1984 to 1998. J Med Virol. 2004;72:320–7. doi: 10.1002/jmv.10571. [DOI] [PubMed] [Google Scholar]
- 14.Steele AD, Ivanoff B. Rotavirus strains circulating in Africa during 1996-1999: emergence of G9 strains and P[6] strains. Vaccine. 2003;21:361–7. doi: 10.1016/s0264-410x(02)00616-3. [DOI] [PubMed] [Google Scholar]
- 15.Desselberger U, Iturriza-Gomara M, Gray JJ. Rotavirus epidemiology and surveillance. Novartis Found Symp. 2001;238:125–47. doi: 10.1002/0470846534.ch9. discussion 147-52. [DOI] [PubMed] [Google Scholar]
- 16.Oka T, Nakagomi T, Nakagomi O. Apparent re-emergence of serotype G9 in 1995 among rotaviruses recovered from Japanese children hospitalized with acute gastroenteritis. Microbiol Immunol. 2000;44:957–61. doi: 10.1111/j.1348-0421.2000.tb02590.x. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara M, Cubitt D, Steele D, et al. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J Med Virol. 2000;61:510–7. doi: 10.1002/1096-9071(200008)61:4<510::aid-jmv15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- 19.Vesikari T, Matson DO, Dennehy P, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 20.Nokes DJ, Abwao J, Pamba A, et al. Incidence and clinical characteristics of group A rotavirus infections among children admitted to hospital in Kilifi, Kenya. PLoS Med. 2008;5:e153. doi: 10.1371/journal.pmed.0050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steele AD, Alexander JJ. Molecular epidemiology of rotavirus in black infants in South Africa. J Clin Microbiol. 1987;25:2384–7. doi: 10.1128/jcm.25.12.2384-2387.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–7. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das BK, Gentsch JR, Cicirello HG, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–2. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gault E, Chikhi-Brachet R, Delon S, et al. Distribution of human rotavirus G types circulating in Paris, France, during the 1997-1998 epidemic: high prevalence of type G4. J Clin Microbiol. 1999;37:2373–5. doi: 10.1128/jcm.37.7.2373-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouvea V, Glass RI, Woods P, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–82. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentsch JR, Glass RI, Woods P, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–73. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmonds MK, Armah G, Asmah R, et al. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol. 2008;42:368–73. doi: 10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee I, Ramani S, Primrose B, et al. Modification of rotavirus multiplex RT-PCR for the detection of G12 strains based on characterization of emerging G12 rotavirus strains from South India. J Med Virol. 2007;79:1413–21. doi: 10.1002/jmv.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iturriza-Gomara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31:259–65. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Mphahlele MJ, Peenze I, Steele AD. Rotavirus strains bearing the VP4P[14] genotype recovered from South African children with diarrhoea. Arch Virol. 1999;144:1027–34. doi: 10.1007/s007050050565. [DOI] [PubMed] [Google Scholar]
- 31.Gouvea V, Santos N, Timenetsky Mdo C. VP4 typing of bovine and porcine group A rotaviruses by PCR. J Clin Microbiol. 1994;32:1333–7. doi: 10.1128/jcm.32.5.1333-1337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 1994;32:1338–40. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Ryan M. The ever-changing landscape of rotavirus serotypes. Pediatr Infect Dis J. 2009;28:S60–2. doi: 10.1097/INF.0b013e3181967c29. [DOI] [PubMed] [Google Scholar]
- 34.Kiulia NM, Peenze I, Dewar J, et al. Molecular characterisation of the rotavirus strains prevalent in Maua, Meru North, Kenya. East Afr Med J. 2006;83:360–5. doi: 10.4314/eamj.v83i7.9447. [DOI] [PubMed] [Google Scholar]
- 35.Esona MD, Geyer A, Page N, et al. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: relationship to animal rotaviruses. J Med Virol. 2009;81:937–51. doi: 10.1002/jmv.21468. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organisation Rotavirus vaccines. Wkly Epidemiol Rec. 2007;82:285–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.