Abstract
Background
Previous studies examining genetic associations with MRI-defined brain infarct have yielded inconsistent findings. We investigated genetic variation underlying covert MRI-infarct, in persons without histories of transient ischemic attack or stroke. We performed meta-analysis of genome-wide association studies of white participants in 6 studies comprising the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium.
Methods
Using 2.2 million genotyped and imputed SNPs, each study performed cross-sectional genome-wide association analysis of MRI-infarct using age and sex-adjusted logistic regression models. Study-specific findings were combined in an inverse-variance weighted meta-analysis, including 9401 participants with mean age 69.7, 19.4% of whom had ≥1 MRI-infarct.
Results
The most significant association was found with rs2208454 (minor allele frequency: 20%), located in intron 3 of MACRO Domain Containing 2 gene and in the downstream region of Fibronectin Leucine Rich Transmembrane Protein 3 gene. Each copy of the minor allele was associated with lower risk of MRI-infarcts: odds ratio=0.76, 95% confidence interval=0.68–0.84, p=4.64×10−7. Highly suggestive associations (p<1.0×10−5) were also found for 22 other SNPs in linkage disequilibrium (r2>0.64) with rs2208454. The association with rs2208454 did not replicate in independent samples of 1822 white and 644 African-American participants, although 4 SNPs within 200kb from rs2208454 were associated with MRI-infarcts in African-American sample.
Conclusions
This first community-based, genome-wide association study on covert MRI-infarcts uncovered novel associations. Although replication of the association with top SNP failed, possibly due to insufficient power, results in the African American sample are encouraging, and further efforts at replication are needed.
Keywords: genome-wide association study, brain infarction, MRI, cohort study, meta-analysis
Vascular disease of the brain is a leading cause of long-term disability and death. The burden of brain vascular disease is far greater than suggested by occurrence of acute neurological events such as stroke.1 Brain imaging techniques, especially magnetic resonance imaging (MRI), have revealed that brain infarcts are common in the elderly, especially small subcortical infarcts.2 While the majority of these MRI-infarcts do not produce acute clinical symptoms leading to a diagnosis of stroke, they cannot be considered benign, silent, or asymptomatic, as they are associated with an increased risk for cognitive deficits, motor impairments, and future stroke.2 The pathogenesis of these covert brain infarcts remains poorly understood.
Whereas several monogenic disorders are known to cause brain infarcts, the genes underlying brain infarcts in the general population remain undetermined.3 A genetic component is suggested by increased risk of covert MRI-infarcts among individuals whose parents or siblings have experienced clinically overt infarcts.4,5 As we will detail, previous candidate gene studies of covert MRI-infarcts have yielded inconsistent findings. Genome-wide association studies (GWAS) of MRI-infarcts are lacking and would permit an unbiased search for genetic variants associated with this phenotype, without relying on a priori hypotheses about underlying pathophysiology.6
To study genetics of these infarcts, we adapted an analytic approach used in a prior study of stroke from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium7 and combined GWAS from 6 prospective population-based cohort studies: the Aging Gene-Environment Susceptibility-Reykjavik Study (AGES-Reykjavik); the Atherosclerosis Risk in Communities (ARIC) Study; the Austrian Stroke Prevention Study (ASPS); the Cardiovascular Health Study (CHS); the Framingham Heart Study (FHS); and the Rotterdam Study. We present results from this meta-analysis that included 9401 stroke-free white participants.
METHODS
Consortium
The CHARGE consortium includes large prospective community-based cohort studies having genome-wide variation data coupled with extensive data on multiple phenotypes.8 All participating studies agreed on phenotype harmonization, covariate selection, pre-specified analytic plans for within-study analyses, and meta-analysis of results. Each study secured approval from institutional review boards, and all participants provided written informed consent for study participation, MRI scanning, and use of DNA for genetic research.
Setting
Details of cohort selection, risk factor assessment, and outcome determination in the 6 studies have been reported previously (Appendix, Section 1). Briefly, the AGES-Reykjavik Study is a single center prospective continuation of the Reykjavik Study, which included persons born 1907–1935 and living in Reykjavik, Iceland in 1967, when the study was started. In 2002–2006, 5764 participants from the cohort were reexamined as a part of the AGES-Reykjavik Study.9 The ARIC study enrolled adults aged 45 and 64 years, from 4 U.S. communities (N=15,792, including 11,478 whites). The baseline examination was in 1987–1989.10 The ASPS enrolled 2007 inhabitants of Graz, Austria who lacked neuropsychiatric disease. Between 1991–1994 and 1999–2003, an extended diagnostic work-up including neuroimaging was done in a subset of participants aged 45 to 85 years.11,12 The CHS enrolled adults who were 65 years or older and from 4 U.S. communities (N=5,888 including 4,925 whites). The baseline examination was either in 1989–90 or 1992–93.13 The FHS is a U.S.-based single-site study that comprises 3 generations of participants. Members of the Original cohort followed since 1948 (N=5,209)14,15 and the Offspring cohort followed since 1971 (N=5,124),14 were invited to undergo an initial brain MRI in 1999–2005. The Rotterdam Study enrolled inhabitants from a district of Rotterdam (Ommoord), The Netherlands, aged 55 years or older (N=7,983) at the baseline examination in 1990–93 (Rotterdam Study I).16 In 2001, the Rotterdam Study cohort was expanded by 3,011 newly eligible persons (Rotterdam Study II).
MRI scans
In each study, eligible participants were invited to undergo MRI scans, which were performed and interpreted in a standardized fashion without knowledge of demographic or clinical information (Appendix, Section 2). Infarct on MRI scan was defined as an area of abnormal signal intensity in a vascular distribution that lacked mass effect. Infarcts had to be 3–4 millimeters in size or greater. Efforts were made in all studies to distinguish infarcts from dilated perivascular spaces. All participants were categorized as having or not at least 1 MRI-infarct.
Genotyping
The consortium was formed after individual studies had finalized their GWAS platforms, which differed across studies. All studies used their genotype data to impute to the 2.5 million non-monomorphic, autosomal SNPs described in HapMap's CEU panel. Extensive quality control (QC) analyses were performed in each cohort. Because the top SNP was imputed in all of the cohorts, we directly genotyped it in studies where the quality of imputation was judged to be poor. Details on the genotyping, imputation, and QC efforts can be found in the Appendix, Section 3.
Study population
Participants were eligible for these analyses if they had genotyping, an MRI, and lacked a history of transient ischemic attack or stroke prior to their MRI (covert MRI-infarcts). Participants were entirely or almost entirely European whites in the AGES-Reykjavik Study, ASPS, FHS and Rotterdam Study, so African American participants from ARIC and CHS were not included in these analyses. In addition, CHS did not genotype participants with any form of clinical cardiovascular disease at baseline. By design, ASPS did not perform MRI scans in patients with transient ischemic attack or stroke. Also ASPS and Rotterdam Study did not perform MRI scans in participants with dementia. The number and characteristics of participants from each cohort are shown in Table 1.
Table 1.
Characteristics of Study Participants in Analysis of covert MRI-defined brain infarcts.
| Characteristics | AGES- Reykjavik |
ARIC | ASPS | CHS | FHS | Rotterdam Study I |
Rotterdam Study II |
|---|---|---|---|---|---|---|---|
| Number with MRI and genotyping | 2866 | 751 | 787 | 2122 | 2291 | 481 | 591 |
| Excluded for TIA or stroke | 310 | 21 | 0† | 0† | 87 | 46 | 24 |
| Number in these analyses | 2556 | 730 | 787 | 2122 | 2204 | 435 | 567 |
| Number (%) with MRI-infarcts | 714 (27.9%) | 65 (8.9%) | 88 (11.2%) | 555 (26.2%) | 252 (11.4%) | 89 (20.4%) | 59 (10.4%) |
| Number (%) with lacunar infarcts | NA | 58 (8.0%) | 73 (9.3%) | 482 (22.7%) | 211 (9.6%) | 82 (18.9%) | 48 (8.5%) |
| Mean Age (±SD) at MRI | 76.2 (5.4) | 63.2 (±4.4) | 65.3 (±8.0) | 71.7 (±4.8) | 63.9 (±11.3) | 72.9 (7.9) | 67.2 (5.3) |
| Women, % | 1510 (59.1%) | 433 (59.3%) | 449 (57.1%) | 1304 (61.5%) | 1195 (54.2%) | 224 (51.5%) | 284 (50.1%) |
| Dementia at MRI, % | 107 (4.2%) | 0 | 0‡ | 77 (3.6%) | 6 (0.3%) | 0‡ | 0‡ |
| Cardiovascular risk factor at MRI* | |||||||
| Systolic BP (mean ±SD) | 142 (20) | 119 (±17) | 143 (±22.6) | 133.7 (±20.5) | 127 (±19) | 146 (20) | 145 (18) |
| Hypertension (mean ± SD) | 2025 (79.2%) | 193 (26.7%) | 524 (66.6%) | 1043 (49.2%) | 943 (43.6%) | 310 (71.3%) | 391 (69%) |
| Diabetes Mellitus, % | 271 (10.6%) | 74 (10.2%) | 75 (9.5) | 203 (9.7%) | 261 (12.2%) | 18 (4.1%) | 51 (9%) |
| Current smoker, % | 320 (12.5%) | 136 (18.7%) | 92 (11.7) | 218 (10.3%) | 252 (11.7%) | 81 (18.6%) | 167 (30%) |
| Prevalent CVD at MRI, % | 397 (15.5%) | 41 (5.8%) | 269 (34.2%) | 0† | 242 (11.1%) | 31 (7.2%) | 38 (6.7%) |
AGES-Reykjavik, Aging Gene-Environment Susceptibility-Reykjavik Study; ARIC, Atherosclerosis Risk in Communities Study; ASPS, Austrian Stroke Prevention Study; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; MRI, magnetic resonance imaging; TIA, transient ischemic attack; SD, standard deviation; BP, blood pressure; CVD, cardiovascular disease other than transient ischemic attack or stroke, including clinically evident coronary artery disease, congestive heart failure and peripheral vascular disease; NA, not available.
Definition of baseline characteristics was uniform across all studies: Hypertension was defined using standard criteria17 as systolic blood pressure ≥140 mm Hg, diastolic blood pressure ≥90 mm Hg or being on antihypertensive treatment; Diabetes mellitus was defined as a casual or 2 hour post-prandial blood glucose ≥200mg/dl (11 mmol/L), a fasting blood glucose ≥126mg/dl (7 mmol/L), or use of insulin or oral hypoglycemic agents; CVD (cardiovascular disease) was defined as presence of congestive heart failure, coronary heart disease or intermittent claudication.
In ASPS and CHS, participants with prevalent TIA or stroke were not included. In CHS, participants with other prevalent CVD were also not included.
In ASPS and Rotterdam studies, participants with dementia did not undergo cranial MRI.
Statistical analyses within studies
Each study fit an additive genetic model with a 1-degree of freedom trend test relating genotype dosage, 0 to 2 copies of the minor allele, to having or not at least 1 MRI-infarct. We used logistic regression models to calculate odds ratios (OR) with corresponding 95% confidence intervals (CI). Initial analyses were adjusted only for age and sex to avoid adjusting for covariates that might lie along a causal pathway. In addition, ARIC and CHS also adjusted for study site, and FHS adjusted for familial structure. To explore potential mechanisms, we additionally adjusted our most significant association in one model for systolic blood pressure and in another model for the presence or absence of hypertension, defined as systolic blood pressure ≥140 mm Hg, diastolic blood pressure ≥90 mm Hg or use of antihypertensive medications.17 All studies screened for latent population substructure, which was negligible (Appendix, Section 4).
Meta-analysis
We conducted a fixed-effects meta-analysis of results from the 6 studies, with 7 cohorts counting the Rotterdam Study II, using inverse-variance weighting. After QC, filtering, and imputation within each study, we restricted our meta-analysis to 2,217,889 autosomal SNPs that were common to all studies and had an average minor allele frequency (MAF) greater than 2%. Details on the meta-analysis strategy and functional annotation of SNPs are available in the Appendix, Section 5. As suggested by others,6 we decided a priori on a genome-wide significance threshold of 5×10−8. SNPs with 5×10−8<p<1×10−5 were considered highly suggestive associations. As available in meta-analysis, we also examined associations with candidate SNPs, or their proxies, previously reported to be significantly associated with covert MRI-infarcts.
Replication
We attempted to replicate findings for our top SNP by genotyping it in 1822 elderly white participants from the 3C-Dijon study18,19 and 644 African American participants from the ARIC study.10 We also 59 explored SNPs within 300kb of our top SNP using in-silico replication in the African American sample (Appendix, Section 7).10 We set the threshold for replication at a one-sided p-value of 0.05.
RESULTS
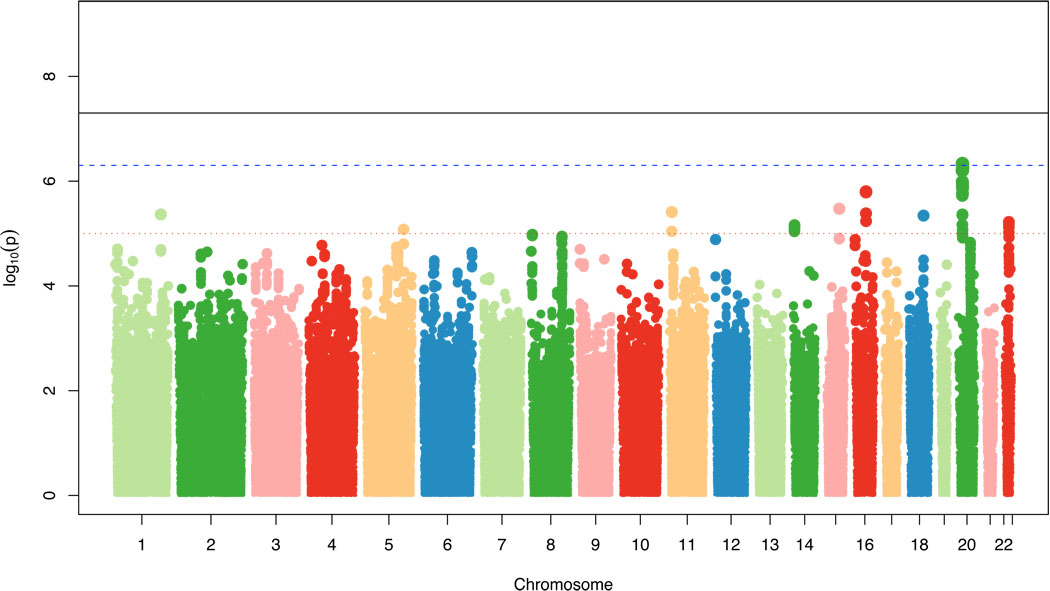
Among 9401 participants whose mean age was 69.7 years and who were 53.4% women, 1822 (19.4%) had at least 1 MRI-infarct (Table 1). After meta-analysis the genomic inflation factor lambda was 0.996, indicating no significant inflation of p-values. Figure 1 shows the genome-wide plot of p-values for individual SNPs against their genomic position. None of the peaks cleared the threshold for genome-wide significance, but 51 SNPs had highly suggestive associations with p<1×10–5 (Table 2 and Appendix, Section 6 and Table A). There was no significant heterogeneity across studies for the association with these SNPs (Appendix, Section 6).
Figure 1.
Genome-wide signal intensity (Manhattan) plot showing individual p-values against their genomic position for covert MRI-defined brain infarcts. Within each chromosome (x-axis), results are plotted left to right from p-terminal end. Solid black line indicates preset threshold for genome-wide significance, p=5.0×10−8; dashed blue line, more liberal threshold for genome-wide significance also used in the literature, p=5.0×10−7; dotted red line, threshold for highly suggestive associations, p=1.0×10−5.
Table 2.
Strongest single nucleotide polymorphism (SNP)-phenotype associations in meta-analysis for covert MRI-defined brain infarcts.
| SNP rs# | SNP function | Chr position |
Minor allele |
MAF | OR (95%CI) |
p-value | PAR | Closest Gene | 2nd Closest Gene | Additional SNPs at p<10−5 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Distance | Name | Distance | |||||||||
| rs2208454 | intronic | 20:14213415 | T | 0.20 | 0.76 (0.68–0.84) |
4.64E-07 | 0.12 | MACROD2 | in gene | FLRT3 | 39.2 | 22 intronic |
| rs1834018 | intronic | 16:56864743 | G | 0.12 | 1.32 (1.18–1.48) |
1.59E-06 | 0.07 | CCDC113 | in gene | KLKBL4 | 6.7 | 1 intronic, 2 upstream |
| rs2869036 | intergenic | 15:76454627 | G | 0.22 | 0.76 (0.68–0.85) |
3.36E-06 | 0.13 | CRABP1 | 27.0 | IREB2 | 62.9 | 0 |
| rs1471895 | intronic | 11:11297762 | A | 0.06 | 1.44 (1.23–1.68) |
3.90E-06 | 0.05 | GALNTL4 | in gene | EIF4G2 | 510.6 | 1 intronic |
| rs4335430 | intronic | 1:215166854 | T | 0.16 | 1.27 (1.15–1.41) |
4.31E-06 | 0.08 | ESRRG | in gene | USH2A | 503.5 | 0 |
| rs17695069 | intronic | 18:54801916 | G | 0.11 | 1.34 (1.18–1.52) |
4.54E-06 | 0.07 | ZNF532 | in gene | SEC11C | 156.2 | 0 |
| rs2284038 | intronic | 22:35965001 | G | 0.36 | 1.20 (1.11–1.30) |
5.98E-06 | 0.13 | RAC2 | in gene | SSTR3 | 26.7 | 3 intronic |
| rs12885474 | intronic | 14:24451217 | A | 0.12 | 0.75 (0.66–0.85) |
6.90E-06 | 0.08 | STXBP6 | in gene | GZMB | 277.9 | 13 intronic |
| rs11746929 | intronic | 5:149114067 | A | 0.27 | 0.81 (0.73–0.89) |
8.35E-06 | 0.12 | PPARGC1B | in gene | PDE6A | 103.6 | 0 |
The reference single nucleotide polymorphism (SNP) number (rs#), function, and chromosome (chr) position are listed. Odds ratios (OR), 95% confidence intervals (CI), and p-values as powers of 10 (E) are based on the meta-analysis. Each row lists only the SNP-phenotype association with the lowest p-value for that locus. The last column shows the number of additional SNPs at the same locus, within 250 kb of the specified SNP, that were also associated with the phenotype with a p-value <10−5. Complete details for these additional SNPs are provided online in the Appendix, Table A. Alleles were identified based on the plus strand of the NCBI build #36. The minor allele was also the coded allele, and minor allele frequency (MAF) is based on allele frequency in meta-analysis sample. The Appendix, Section 8, has details on calculating the population attributable risk (PAR). For SNPs whose minor allele had an inverse association with MRI-infarcts (OR<1.0), the PAR has been calculated using the major allele as the risk allele. The Human Gene Organization (HUGO) Gene Nomenclature System symbols is used for the 2 genes located closest to each SNP, and the distance of the associated SNP from the 5′ end (start) or 3’ end (stop) of the gene (the closest of the 2). Standardized gene annotations for all SNP results were derived programmatically from the UCSC Genome Browser RefSeq gene track (hg18). Distances to genes are given in kilo-base (kb) pairs, based on NCBI build #36.
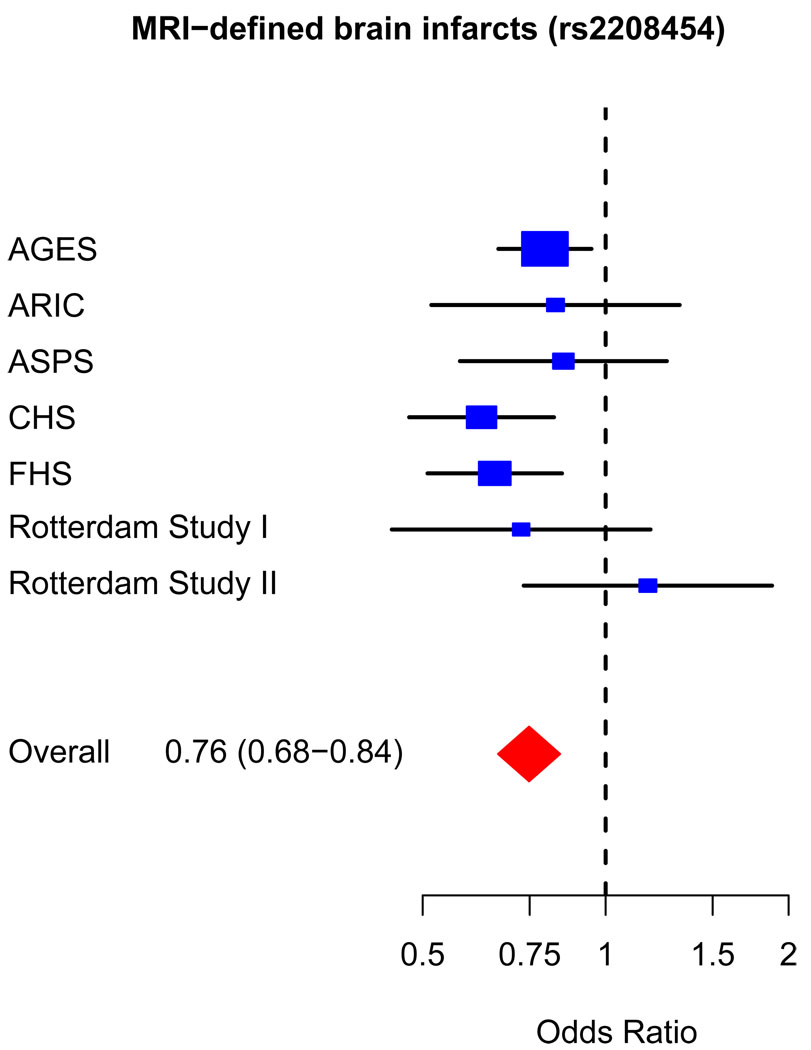
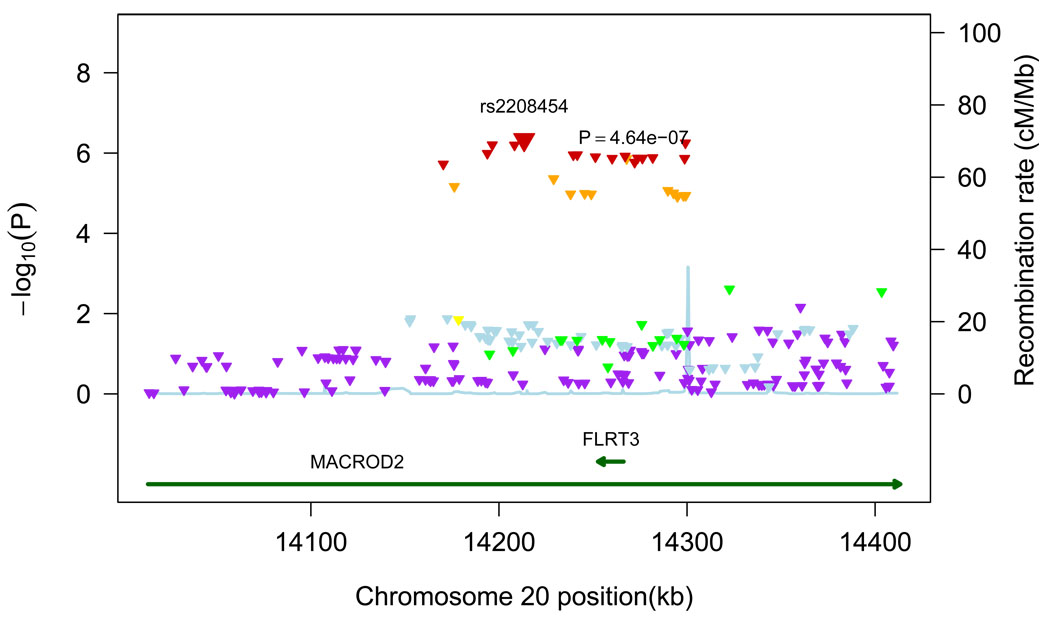
The most significant association was found on chromosome 20p12 with SNP rs2208454, located in intron 3 of MACRO Domain Containing 2 (MACROD2) gene and in the downstream region of Fibronectin Leucine Rich Transmembrane Protein 3 (FLRT3) gene. The OR for MRI-infarcts was 0.76 (95% CI=0.68–0.84, p=4.64×10−7). Additional adjustment for systolic blood pressure (OR=0.76, 95% CI=0.68–0.85) or hypertension (OR=0.76, 95% CI=0.68–0.84) did not change results. Figure 2 shows a forest plot of risk estimates for rs2208454 across the 7 cohorts. Twenty-two other SNPs in intron 3 of MACROD2 were also associated with MRI-infarcts with a p-value of less than 1.0×10−5 (Table 2 and Appendix, Table A). All were in linkage disequilibrium with rs2208454: r2>0.64 for all and r2>0.8 for 17 of the SNPs. Of these 22 SNPs, 1 was intronic within FLRT3 (rs6110247), and 3 were potential transcription factor binding sites (rs6110247, rs743216, and rs3789335). Figure 3 shows all SNPs within a 200kb region on either side of the top hit, together with p-values, recombination rates, and known genes in that region.
Figure 2.
Forest plot for top hit (rs2208454). Individual studies are plotted against individual effect sizes (odds ratios). Size of blue boxes is inversely proportional to variance. Horizontal lines are 95% confidence intervals.
Figure 3.
Regional plot for associations in region centered on top hit. All SNPs (triangles) are plotted with their meta-analysis p-values against their genomic position. The color of the triangles represents the linkage disequilibrium between SNPs: purple: r2≤0.05, light blue: 0.05<r2≤0.10, green: 0.10<r2≤0.30, yellow: 0.30<r2≤0.60, orange: 0.60<r2≤0.80, red: r2>0.80. Light blue line represents estimated recombination rates. Genes are shown as dark green arrows.
Although estimated quality of imputation for rs2208454 was excellent in most studies (O/E ratio > 0.95), it was poor in CHS (O/E ratio = 0.48). Therefore, rs2208454 was genotyped in CHS. When incorporating this result in the meta-analysis instead of the imputed data, the OR was the same at 0.76 but the p-value smaller at 1.44×10−7.
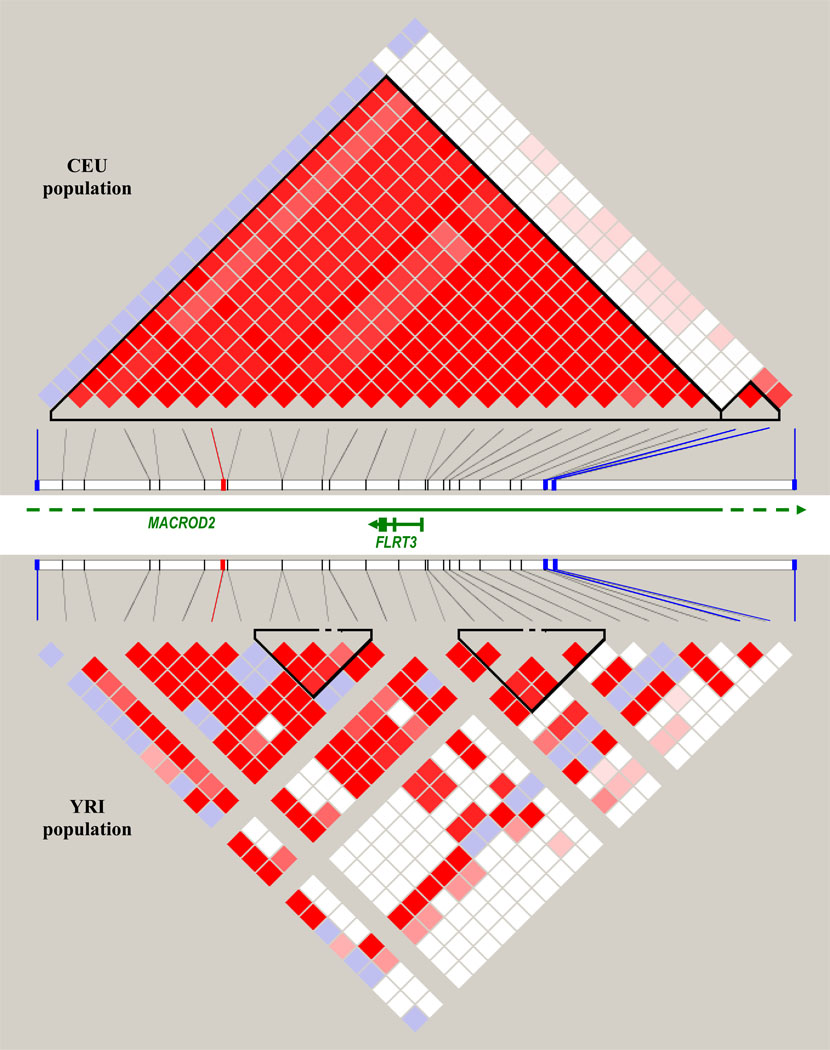
We failed to replicate findings for the top SNP either in 1822 white participants from 3C-Dijon for whom mean age was 72.5 years and 9.4% had at least 1 MRI-infarct (MAF=22%, OR=0.99, 95% CI=0.76–1.29, p=0.92) or in 644 African-American participants from the ARIC study for whom mean age was 61.5 years and 15.5% had at least 1 MRI-infarct (MAF=6.5%, OR=1.26, 95% CI=0.74–2.15, p=0.40). However, four other SNPs in intron 3 of MACROD2 within 50–151kb from rs2208454 were significantly associated with MRI-infarcts in the African-American sample (rs7268327, p=0.045; rs1998237, p=0.006; rs4464346, p=0.0006; rs8116105, p=0.01). In the discovery sample, rs8116105 was also associated with MRI-infarcts, although the direction of the association was opposite (p=0.02) (Appendix, Table B). The linkage disequilibrium pattern in this region differs substantially between Caucasian and African populations (Figure 4). Details on the replication effort are contained in the Appendix, Section 7.
Figure 4.
Linkage disequilibrium (LD) plot of region in MACROD2 including top hit (rs2208454), 22 other SNPs with p<10−5 in meta-analysis, and 4 SNPs with p<0.05 in African-American sample, using HapMap release 22. Plot on top depicts LD in European population (CEU) while plot on bottom depicts LD in African population (YRI). The color scheme is white for D’<1 and LOD<2, blue for D’=1 and LOD<2, shades of pink and red for D’<1 and LOD≥2, and bright red for D’=1 and LOD≥2. The SNP marked in red is rs2208454, the 4 SNPs marked in blue are SNPs with p<0.05 in African-American sample, from left to right: rs7268327, rs1998237, rs4464346, rs8116105. LD could not be measured for 3 SNPs in the YRI population because of a minor allele frequency <0.001 (rs6135125, rs6110247, rs12624446). Green lines represent genes in region.
We repeated these analyses including, rather than excluding, participants with a history of a transient ischemic attack or stroke (Table 1). The results were similar although the associations were slightly weaker, in general (data not shown). We also examined associations with previously reported candidate SNPs, or their proxies, none of which was significant after correction for multiple testing (Appendix, Section 9). In addition, we examined associations with top SNPs in the Ninjurin-2 gene associated with ischemic stroke in a recent CHARGE GWAS meta-analysis7 and found none significantly associated with MRI-infarcts. Finally, we explored relations between ischemic stroke and the 23 SNPs in MACROD2 (Table 2) in the recent CHARGE GWAS meta-analysis on ischemic stroke.7 No significant association of these SNPs with ischemic stroke was identified.
DISCUSSION
This meta-analysis of GWAS data on covert MRI-defined brain infarcts included 9401 participants without a history of transient ischemic attack or stroke from 6 community-based studies. The most significant association (p=4.64×10−7) was found for SNP rs2208454 on chromosome 20p12, located in intron 3 of MACROD2 and in the downstream region of FLRT3. The less common allele was associated with a lower risk. Twenty-two SNPs in linkage disequilibrium with rs2208454 were also associated with MRI-infarcts with p-values less than 1.0×10−5, as were 28 other SNPs in 8 different loci. No association reached our preset threshold for genome-wide significance of 5.0×10−8. In 2 replication samples of 1822 white participants from 3C-Dijon and 644 African-American participants from ARIC, we did not observe an association with rs2208454, although 4 SNPs within 200kb from rs2208454 were associated with MRI-infarcts in the African-American sample. Finally, we failed to observe an association with SNPs previously reported to be associated significantly with covert MRI-infarcts in candidate genes studies.
The function of the protein encoded by MACROD2 is poorly understood. It contains a macro domain that is evolutionarily conserved and expressed in fetal and adult human brain.20,21 Macro domains bind ADP-ribose, suggesting a role in ADP-ribosylation, a post-translational modification involved in many processes including DNA repair, transcriptional activation and repression, and telomere and chromatin biology.22 Nested in intron 3 of MACROD2, FLRT3 encodes fibronectin leucine rich transmembrane protein 3.23 The gene is expressed in various tissues, including brain, and is well conserved across species.24 The protein it encodes modulates homotypic cell adhesion and promotes fibroblast growth factor signaling,24 which is potentially involved in angiogenesis and neurogenesis.25 In animal experiments, FLRT3 was shown to promote neurite outgrowth after axonal injury.26,27
Intriguingly, even though both FLTR3 and Ninjurin-2 appear to modulate response to neuronal injury, the Ninjurin-2 SNPs identified in the recently published ischemic stroke GWAS within the CHARGE consortium were not significantly associated with covert MRI-infarcts and the MACROD2 SNPs identified through the present analysis were not associated with overt ischemic stroke. A possible explanation for this discrepancy could be that MRI-infarcts comprise mainly small subcortical infarcts, while ischemic stroke represents a much more heterogeneous entity. Further investigations are needed, beginning with replication of the associations in external cohorts of MRI-defined infarcts, ischemic stroke, and subtypes of ischemic stroke.
None of the 23 SNPs in MACROD2 with p<10−5 was significantly associated with gene expression in publicly available genome-wide expression quantitative trait loci (eQTL) datasets (Appendix, Section 6), but this finding should be interpreted cautiously as less than half of these SNPs were present on any of the genotyping arrays used in these studies and eQTL may be tissue and insult specific.
This meta-analysis has strengths. It included 6 large cohort studies with similar MRI protocols. The analyses were restricted to white participants to minimize the risk of population stratification. Genotyping was subjected to rigorous quality control. Our study also has limitations. Despite having close to 10,000 participants of whom almost 2,000 had covert MRI-infarcts, we had limited power to detect associations with small effect sizes and associations with rare variants. An important caution is that the association with the top SNP was not directly replicated in 2 independent samples. Hence we cannot exclude the possibility that this association was a chance finding. However, the power to detect an association with rs2208454 in the 2 replication samples was relatively low: 61% for the white participants from 3C-Dijon and 18% for the African-American participants from ARIC, assuming the same effect size as in the discovery sample, which is likely an overestimation.28 The definition of infarcts may also have differed across discovery and replication cohorts, especially concerning the discrimination of infarcts from enlarged perivascular spaces. Furthermore, identified SNPs may not be the causal variants but merely markers in linkage disequilibrium with causal variants. Substantial differences in linkage patterns between whites and African-Americans could result in different markers being in linkage disequilibrium with the causal variant in the 2 populations. Interestingly, 4 SNPs located in the same locus as rs2208454 were associated with MRI-infarcts in the African-American sample. Although these exploratory results do not provide direct replication, as these SNPs were in weak LD with rs2208454, they suggest that the locus may be worthy of further exploration. If replication can be obtained in the future, including in African Americans samples, the latter could perhaps help refine the signal and prove useful for the identification of a causal variant. Finally, even though the studies included in the meta-analysis are population-based, the samples are not perfectly representative of the total cohort, as they include subsets of individuals who were able to undergo brain MRI and agreed to do so.
This meta-analysis of GWAS shows highly suggestive association of covert MRI-infarcts with rs2208454 on chromosome 20p12 (p=4.64×10−7). Attempted replication of the top SNP in an independent white sample failed, and additional attempts in larger samples to replicate this finding, as well as associations with other suggestive loci, are needed. Extending replication efforts not only in white but also African-American populations may prove useful for fine mapping purposes. The molecular, clinical, and epidemiological correlates of confirmed associations may permit new insights into the pathophysiology and prevention of covert brain infarcts.
Supplementary Material
Acknowledgements
Aging Gene-Environment Susceptibility-Reykjavik Study: The research has been funded by NIA contract N01-AG-12100 with contributions from NEI, NIDCD and NHLBI, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).
The Atherosclerosis Risk in Communities Study: The research is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022 and grants R01HL087641 and R01HL093029; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
The Austrian Stroke Prevention Study: The research reported in this article was funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180. The Medical University of Graz supports the databank of the ASPS. The authors thank the staff and the participants of the ASPS for their valuable contributions. We thank Birgit Reinhart for her long-term administrative commitment and Ing Johann Semmler for the technical assistance at creating the DNA-bank.
Cardiovascular Health Study: The research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01-HC-15103, N01-HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Framingham Heart Study: From the Boston University School of Medicine and the Framingham Heart Study. This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project and the authors are particularly grateful for the contribution of Dr. Qiong Yang and Dr. Ming-Huei Chen. This study was also supported by grants from the National Institute of Neurological Disorders and Stroke (NS17950) and the National Institute of Aging (AG08122, AG16495, and AG033193).
Rotterdam Study: The generation and management of genome-wide association study genotype data for the Rotterdam Study is supported by the Netherlands Organization of Scientific Research Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (project nr. 050-060-810), and NWO grants (918-46-615, 948-00-010). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands, Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The authors are grateful to the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists.
3C-Dijon Study: The Three-City (3C) Study is conducted under a partnership agreement between the Institut National de la Santé et de la Recherche Médicale (INSERM), the Victor Segalen–Bordeaux II University, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Generale de l’Education Nationale (MGEN), Institut de la Longévité, Conseils Régionaux of Aquitaine and Bourgogne, Fondation de France, and Ministry of Research–INSERM Programme "Cohortes et collections de données biologiques."
Affiliations
For authors representing the Aging Gene-Environment Susceptibility-Reykjavik Study
Icelandic Heart Association (SS, AS, GE, TA, VG), Kopavogur, Iceland; Department of Radiology (MAvB), Leiden University Medical Center, Leiden, The Netherlands; University of Iceland (TA, VG), Reykjavik, Iceland; Intramural Research Program, Laboratory of Epidemiology, Demography and Biometry (LJL, TBH), National Institute of Aging, Bethesda, Md.
For authors representing the Atherosclerosis Risk in Communities Study
Brown Foundation Institute of Molecular Medicine and Human Genetics Center (MF, EB), University of Texas Health Science Center at Houston, TX; Department of Epidemiology (GH), University of North Carolina, Chapel Hill, NC; Department of Neurology (DSK), Mayo Clinic, Rochester, MN; Department of Biostatistics (DJC), University of North Carolina, Chapel Hill, NC; Department of Radiology (DKS), University of Washington Medical Center, Seattle, WA; Department of Neurology (RFG), Johns Hopkins University School of Medicine, Baltimore, MD Department of Medicine (Geriatrics) (THM), University of Mississippi Medical Center, Jackson, MS.
For authors representing the Austrian Stroke Prevention Study
Department of Neurology, (HS, CE, FF), Department of Neurogeriatrics (RS), Department of Neuroradiology (RS, CE), Institute of Molecular Biology and Biochemistry (HS, MT), Medical University Graz, Austria; Department of Epidemiology & Biostatistics, Department of Forensic Molecular Biology (MS), the Erasmus MC University Medical Center, Rotterdam, The Netherlands.
For authors representing the Cardiovascular Health Study
Departments of Medicine (JCB, BMP, WTL), Biostatistics (TL, KR), Epidemiology (SRH, BMP, WTL), Health Services (BMP), and Neurology (WTL), University of Washington, Seattle, WA; the Center for Health Studies, Group Health (SRH, BMP), Seattle, WA; Brown Foundation Institute of Molecular Medicine and Human Genetics Center (MF), University of Texas Health Science Center at Houston, TX; the Department of Medicine and Pathology (MC), University of Vermont, Burlington, VT; the Medical Genetics Institute, Cedars-Sinai Medical Center (JIR), Los Angeles, CA; the Departments of Neurology and Psychiatry (OL), University of Pittsburgh School of Medicine, Pittsburgh, PA.
For authors representing the Framingham Heart Study
Department of Neurology (SD, AB, ALD, LDA, JRR, MK-H, RA, PAW, SS), Boston University School of Medicine, and Department of Biostatistics (AB, ALD), Boston University School of Public Health, Boston, MA; Department of Neurology and Center for Neuroscience (CD), University of California at Davis, CA.
For authors representing the Rotterdam Study
Departments of Epidemiology (MAI, YSA, AH, CMvD, MMBB), Neurology (PJK), Internal Medicine (AGU, FR), Radiology (MAI, AvdL), and Clinical Chemistry (AGU), from the Erasmus MC University Medical Center, Rotterdam, The Netherlands; The Netherlands Consortium of Healthy Aging (MAI, YSA, FR, AGU, AH, CMvD, MMBB), The Netherlands.
For authors representing the 3C-Dijon Study
Inserm, Unit 708 (CD, CT, YZ), Paris, France; Université Pierre et Marie Curie-Paris (CD, CT, YZ), Paris, France; Service de Neurologie (YZ), Hopital Lariboisière, Paris, France; Department of Neurology (YZ), Peking Union Medical College Hospital, China.
Corresponding Authors
Aging Gene-Environment Susceptibility-Reykjavik Study
Lenore J. Launer, PhD, Laboratory of Epidemiology, Demography and Biometry, National Institute on Aging, 7201 Wisconsin Avenue, 3C-309, Bethesda, MD 20892, launerl@nia.nih.gov.
Atherosclerosis Risk in Communities Study
Thomas H. Mosley, PhD, Department of Medicine (Geriatrics), University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216-4505, tmosley@medicine.umsmed.edu.
Austrian Stroke Prevention Study
Reinhold Schmidt, MD, Department of Neurology, Medical University of Graz, Auenbruggerplatz 22, A-8036 Graz, Austria, reinhold.schmidt@medunigraz.at
Cardiovascular Health Study
WT Longstreth, Jr, MD, Department of Neurology, Box 359775, Harborview Medical Center, 325 Ninth Avenue, Seattle, WA 98104-2420, wl@u.washington.edu
Framingham Heart Study
Sudha Seshadri, MD, Department of Neurology, Boston University School of Medicine, 72 East Concord Street, Boston, MA 02118, suseshad@bu.edu
The Rotterdam Study
Monique M.B. Breteler, MD, PhD, Professor of Neuroepidemiology, Department of Epidemiology, Erasmus MC, University Medical Center Rotterdam, PO Box 2040, 3000 CA Rotterdam, The Netherlands, m.breteler@erasmusmc.nl
Footnotes
Conflicts of Interest and Disclosures
None.
REFERENCES
- 1.Longstreth WT., Jr Brain vascular disease overt and covert. Stroke. 2005;36:2062–2063. doi: 10.1161/01.str.0000179040.36574.99. [DOI] [PubMed] [Google Scholar]
- 2.Vermeer SE, Longstreth WT, Jr, Koudstaal PJ. Silent brain infarcts: a systematic review. Lancet Neurol. 2007;6:611–619. doi: 10.1016/S1474-4422(07)70170-9. [DOI] [PubMed] [Google Scholar]
- 3.Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–161. doi: 10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- 4.Morrison AC, Fornage M, Liao D, Boerwinkle E. Parental history of stroke predicts subclinical but not clinical stroke: the Atherosclerosis Risk in Communities Study. Stroke. 2000;31:2098–2102. doi: 10.1161/01.str.31.9.2098. [DOI] [PubMed] [Google Scholar]
- 5.Leistner S, Huebner N, Faulstich A, Ludwig D, Rees M, Marx P, Langer B, Nikolova A, Hartmann A, Koennecke HC. Increased prevalence of microangiopathic brain lesions among siblings of patients with lacunar stroke. A prospective multicenter study. Eur Neurol. 2008;59:143–147. doi: 10.1159/000111877. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 7.Ikram MA, Seshadri S, Bis JC, Fornage M, Destefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, Decarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunians T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinden AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT, Jr, Wolf PA. Genomewide Association Studies of Stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JCM, Boerwinkle E on Behalf of the CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:273–280. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, Thorgeirsson G, Aspelund T, Garcia ME, Cotch MF, Hoffman HJ, Gudnason V. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 11.Schmidt R, Lechner H, Fazekas F, Niederkorn K, Reinhart B, Grieshofer P, Horner S, Offenbacher H, Koch M, Eber B, et al. Assessment of cerebrovascular risk profiles in healthy persons: definition of research goals and the Austrian Stroke Prevention Study (ASPS) Neuroepidemiology. 1994;13:308–313. doi: 10.1159/000110396. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt R, Schmidt H, Pichler M, Enzinger C, Petrovic K, Niederkorn K, Horner S, Ropele S, Watzinger N, Schumacher M, Berghold A, Kostner GM, Fazekas F. C-reactive protein, carotid atherosclerosis, and cerebral small-vessel disease: results of the Austrian Stroke Prevention Study. Stroke. 2006;37:2910–2916. doi: 10.1161/01.STR.0000248768.40043.f9. [DOI] [PubMed] [Google Scholar]
- 13.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 14.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 15.Dawber TR, Kannel WB. The Framingham study. An epidemiological approach to coronary heart disease. Circulation. 1966;34:553–555. doi: 10.1161/01.cir.34.4.553. [DOI] [PubMed] [Google Scholar]
- 16.Hofman A, Breteler MM, van Duijn CM, Janssen HL, Krestin GP, Kuipers EJ, Stricker BH, Tiemeier H, Uitterlinden AG, Vingerling JR, Witteman JC. The Rotterdam Study: 2010 objectives and design update. Eur J Epidemiol. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, Roccella EJ. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 18.The 3C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003;22:316–325. doi: 10.1159/000072920. [DOI] [PubMed] [Google Scholar]
- 19.Godin O, Dufouil C, Maillard P, Delcroix N, Mazoyer B, Crivello F, Alperovitch A, Tzourio C. White matter lesions as a predictor of depression in the elderly: the 3C-Dijon study. Biol Psychiatry. 2008;63:663–669. doi: 10.1016/j.biopsych.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Front Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 21.Maas NM, Van de Putte T, Melotte C, Francis A, Schrander-Stumpel CT, Sanlaville D, Genevieve D, Lyonnet S, Dimitrov B, Devriendt K, Fryns JP, Vermeesch JR. The C20orf133 gene is disrupted in a patient with Kabuki syndrome. J Med Genet. 2007;44:562–569. doi: 10.1136/jmg.2007.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- 23.Lacy SE, Bonnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2, and FLRT3: a novel family of transmembrane leucine-rich repeat proteins. Genomics. 1999;62:417–426. doi: 10.1006/geno.1999.6033. [DOI] [PubMed] [Google Scholar]
- 24.Karaulanov EE, Bottcher RT, Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006;7:283–290. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZL, Cheng SM, Ma MM, Ma YP, Yang JP, Xu GL, Liu XF. Intranasally delivered bFGF enhances neurogenesis in adult rats following cerebral ischemia. Neurosci Lett. 2008;446:30–35. doi: 10.1016/j.neulet.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Tsuji L, Yamashita T, Kubo T, Madura T, Tanaka H, Hosokawa K, Tohyama M. FLRT3, a cell surface molecule containing LRR repeats and a FNIII domain, promotes neurite outgrowth. Biochem Biophys Res Commun. 2004;313:1086–1091. doi: 10.1016/j.bbrc.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 27.Robinson M, Parsons Perez MC, Tebar L, Palmer J, Patel A, Marks D, Sheasby A, De Felipe C, Coffin R, Livesey FJ, Hunt SP. FLRT3 is expressed in sensory neurons after peripheral nerve injury and regulates neurite outgrowth. Mol Cell Neurosci. 2004;27:202–214. doi: 10.1016/j.mcn.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Xiao R, Boehnke M. Quantifying and correcting for the winner's curse in genetic association studies. Genet Epidemiol. 2009;33:453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.