Abstract
Commercial HIV-1 genotypic resistance assays are very expensive, particularly for use in resource-constrained settings like India. Hence a cost effective in-house assay for drug resistance was validated against the standard ViroSeq™ HIV-1 Genotyping System 2.0 (Celera Diagnostics, CA, USA). A total of 50 samples were used for this evaluation (21 proficiency panels and 29 clinical isolates). Known resistance positions within HIV-1 protease (PR) region (1–99 codons) and HIV-1 reverse-transcriptase (RT) region (1–240 codons) were included. The results were analysed for each codon as follows: (i) concordant; (ii) partially concordant; (iii) indeterminate and (iv) discordant. A total of 2750 codons (55 codons per patient sample × 50 samples) associated with drug resistance (1050 PR and 1700 RT) were analysed. For PR, 99% of the codon results were concordant and 1% were partially concordant. For RT, 99% of the codon results were concordant, 0.9% were partially concordant and 0.1% were discordant. No indeterminate results were observed and the results were reproducible. Overall, the in-house assay provided comparable results to those of US FDA approved ViroSeq™, which costs about a half of the commercial assay ($ 100 vs. $ 230), making it suitable for resource-limited settings.
Keywords: ViroSeq™ HIV-1 genotyping, In-house HIV-1 drug resistance assay, Concordance, Mixtures, Indeterminate rate, HIV-1 genotyping evaluation
1. Introduction
Antiviral resistance is one of the primary reasons for highly active antiretroviral therapy (HAART) failure over time (Shafer et al., 1998; Vella, 2002). Mutations at drug resistance positions result from continued use of antiretroviral (ARV) agents with incomplete viral suppression favouring selection of drug-resistant viruses. The management of virologic failure, particularly when viral load testing is relatively infrequent, represents a serious challenge in current clinical practice, especially in resource-limited countries. Resistance testing is an important component of management of antiretroviral-experienced patients, assisting in selection of appropriate regimens in patients in whom treatment is failing due to resistant virus (Kuritzkes, 2004; Gallant, 2006). Genotyping has also become an increasingly important part of determining initial antiretroviral therapy given the high (and rising) rates of resistant virus transmission in developed countries (Little et al., 2002; Bennett et al., 2006; Bowles et al., 2006). However, cost limits the accessibility of resistance testing in resource-limited settings like India. Two types of resistance assays, genotypic and phenotypic, are used to determine HIV sensitivity or resistance to individual antiretroviral drugs. The most frequent genotypic resistance assays are based on reverse transcription of viral RNA and cDNA sequencing (TRUGENE®, ViroSeq™, and in-house) or reverse hybridization (LiPA).
HIV-1 genotypic resistance testing detects resistance mutations in the reverse-transcriptase (RT) and protease (PR) genes by comparing the gene sequences of a resistant virus with those of a wild-type reference subtype B strain. The clinical trials, responsible for demonstrating the utility of drug resistance genotyping for the selection of antiretroviral therapy, were supported by centralized reference laboratories with extensive experience in viral genotypic testing (Tural et al., 2002; Baxter et al., 2000; Durant et al., 1999). In clinical practice, drug resistance genotypic testing is performed using a wide variety of methods whose performance characteristics have not been established. Completely significant variations exist between laboratories in the extent of technical training, the reagents used, the amplification and sequencing protocols, and datum review and reporting procedures. This variation of methods may contribute to the variable quality of resistance assay results that have been observed in international evaluations (Schuurman et al., 1999).
Many in-house genotyping assays with low running cost have been designed (Lindstrom and Albert, 2003; Steegen et al., 2006) and they were found to have a high success rate (>85.3%) of amplifying and sequencing subtype B and non-B HIV-1 samples (Steegen et al., 2006). However, their performance was not validated against internationally approved reference systems. The present study aimed to evaluate a cost effective in-house genotyping assay for routine drug resistance-related mutation detection on plasma samples with genetically diverse HIV-1 and also to study the accuracy and reproducibility of results obtained in both systems. The present study evaluating the cost effective in-house assay may be useful for considering this genotyping system in India where majority of the HIV infected patients cannot afford the commercial genotypic resistance testing.
2. Materials and methods
2.1. Clinical specimens
Fifty stored plasma samples (30 subtype C, 18 subtype B and 2 subtype D) including both clinical (ART treated and failing patients (n = 29)) and proficiency testing (PT) panels (VQA, Rush University, USA (n = 10); Teragenix, Abbott, USA (n = 6) and TAQAS, NRL Australia (n = 5)) with a viral loads ranging from >1500 to 500,000 copies/mL were recovered from storage (−75 ± 5 °C) for the purpose of assay validation.
2.2. 2.2: RNA extraction, RT-PCR amplification, sequencing and HIV-1 drug resistance mutation analysis
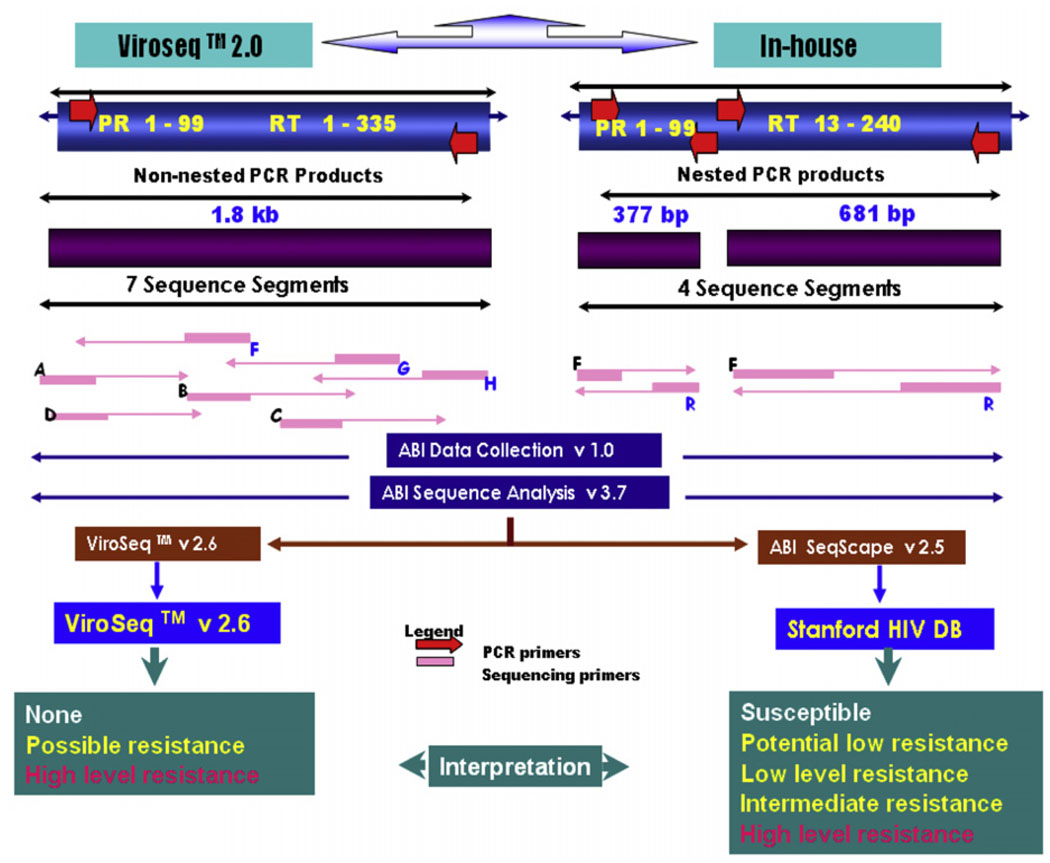
One millilitre of plasma was centrifuged at 4 °C for 1 h at 25,000 × g. Sequencing was performed using a method as described previously (Boden et al., 1999; Larder et al., 1991). Briefly, viral RNA was extracted from plasma (QIAamp viral RNA extraction kit, Qiagen Inc., USA) and reverse transcribed to complementary DNA (cDNA) with random hexamer primers (Fig. 1). PR and RT regions were amplified using nested PCR along with known low copy HIV-1 RNA plasma (1500 cps/mL) as a positive control. For PR region outer primers (forward) 5′-ACCAGAGCCAACAGCCCCACCA-3′ and (reverse) 5′-CTTTTGGGCCATCCATTCCTGGC-3′ and inner primers (forward) 5′-GAAGCAGGAGCCGATAGACAAGG-3′ and (reverse) 5′-CTTTTGGGCCATCCATTCCTGGC-3′ were used. Cycling condition for the first and nested PR PCR were 95 °C 1 min, [95 °C 15 s/65 °C 30 s/72 °C 45 s] × 35 and 95 °C 1 min [95 °C 15 s/55 °C 30 s/72 °C 45s] × 25 respectively. Amplification of RT region included outer primers (forward) 5′-CCTATTGAAACTGTACCAGT-3′ and (reverse) 5′-ACTGTCCATTTATCAGGATG-3′ and inner primers (forward) 5′-AAGCCAGGAATGGATGGCCCA-3′ and (reverse) 5′-CCATTTATCAGGATGGAGTTC-3′ with cycling profile of 95 °C 3 s, [95 °C 20 s/55 °C 30 s/72 °C 30 s] × 40 and 95 °C 3 s, [95 °C 20 s/55 °C 30 s/72 °C 30 s] × 35 for first and nested PCR of the RT region respectively (Boden et al., 1999; Larder et al., 1991). The column (Intron, Korea) purified nested amplicons (PR and RT) were bidirectionally sequenced with 1/4 dilution of the commercially available BDT version 3.1 (Applied Biosystems, CA, USA) on an automated ABI 3100-Avant Genetic analyser (Applied Biosystems, CA, USA). The inner amplification primers were used as sequencing primers for both PR and RT sequencing PCR. The pol sequences were then multiple aligned by SeqScape v2.5 (Applied Biosystems, CA, USA) with HXB-2 (K03455) sequence as a reference, assembled, edited and then the exported consensus fasta sequences were analysed through the Stanford University HIV Drug Resistance Database HIVdb program(version 4.2.6 [http://hivdb.stanford.edu]) for genotypic resistance interpretation. The HIV-1 subtypes of the pol sequences were identified and phylogenetically analysed using Mega software version 3.1 (Kumar et al., 2004). Parallel extraction, amplification, sequencing and interpretation for each sample were performed using ViroSeq™ HIV-1 genotyping v2.0 (Celera diagnostics, CA, USA) following the manufacturers instruction (Fig. 1). The ViroSeq™ HIV-1 genotyping system software v2.6 (Celera Diagnostics, CA, USA) that comes with the kit, assembles, edits, and identifies mutations within this 1.8 kb sequence. The software compares the consensus sequence with a known reference, HXB-2 (K03455), to determine mutations present in the sample. Finally, the ViroSeq™ software uses a proprietary algorithm to analyse the mutations and generate a drug resistance report. For in-house assay the same HXB-2 reference sequence was used for editing to minimize biases towards the wild-type during manual editing.
Fig. 1.
Schematic representation showing protease (PR) and reverse-transcriptase (RT) codons covered by ViroSeq™ v2.0 and in-house assay.
2.3. Data analysis
For analysis, codons 1–99 in PR region and 1–240 in RT region were considered for both the assays. Validation was done by comparing the base called codons/amino acids (synonymous/non-synonymous substitutions) at known drug resistance positions in PR and RT regions as per ViroSeq™ version 2.6 by both assays. A total of 2750 codons (55 codons per patient sample × 50 samples) of HIV drug resistance positions [1050 PR (21 × 50) and 1700 RT (34 × 50)] were analysed under the following categories:
Concordant (if both assays identified the same result).
Partially concordant (mixture by one assay but not by other).
Indeterminate (no result by in-house).
Discordant (the two assays detected different amino acids).
In order to assess reproducibility, few samples were selected at random and tested by both systems independently.
3. Results
All the 50 samples of different subtypes included for the validation were successfully amplified by both the ViroSeq™ and the in-house assays. A total of 55 codons which are drug resistance-related mutations in the PR and RT regions were identified by the ViroSeq™ and the in-house assay and were reproducible when repeated. The in-house assay was able to amplify samples with a plasma viral load of >1500 copies/mL similar to ViroSeq™.
Out of the 1050 PR codons, 1042 (99%) were concordant, while in RT out of 1700 codon positions analysed, 1683 (99%) were concordant (Table 1). Both the assays showed identical base substitutions and amino acids in these positions. Partial concordance was observed in 8 (1%) and 15 (0.9%) codon positions in the PR and RT regions, respectively. In the RT region, of 15 amino acid mixtures detected, ViroSeq™ detected more (10/15; 67%) mixtures than in-house (5/15; 33%). The details of mixtures detected by both the assays are shown in (Table 2).
Table 1.
Comparison of the results of HIV-1 drug resistance testing by homebrew and ViroSeq™ assay (n = 50 specimens).
| pol Gene | Codons analysed | Concordant | Partially concordant | Discordant | |
|---|---|---|---|---|---|
| Mixture by vsa only | Mixture by ih onlyb | ||||
| PR regionc | 1050 | 1042 (99%) | 8 (1%) | Nil | 0 |
| RT regiond | 1700 | 1683 (99%) | 10 (0.6%) | 5(0.3%) | 2(0.1%) |
vs, ViroSeq™ HIV-1 genotyping assay.
ih, in-house HIV-1 genotyping assay.
PR, protease region.
RT, reverse-transcriptase region.
Table 2.
Details of synonymous and non-synonymous substitutions at drug resistance positions among the partially concordant and discordant mutations.
| Category | Gene | Codon | Amino acid/bases detected by both assays | |||
|---|---|---|---|---|---|---|
| ViroSeq™ | In-house | |||||
| Amino acid | Base | Amino acid | Base | |||
| Partial concordance | PR | 10 | LE | YTT | L | CTT |
| 33 | F | TTY | F | TTT | ||
| 48 | G | GGR | G | GGG | ||
| 63 | PL | CYT | L | CTT | ||
| 63 | PA | SCT | A | GCT | ||
| 63 | P/L/A/S/ | SYC | A | GYC | ||
| 71 | A | GCW | A | GCA | ||
| 71 | IV | RTT | V | GTT | ||
| RT | 44 | ML | WTG | L | TTG | |
| 69 | IT | AYT | T | ACT | ||
| 74 | IL | WTA | I | ATA | ||
| 75 | TA | RCA | T | ACA | ||
| 98 | AG | GSA | G | GGA | ||
| 98 | AG | GSA | G | GGA | ||
| 101 | KE | RAA | K | AAA | ||
| 101 | E | GAA | EQ | SAA | ||
| 103 | N | AAC | KN | AAM | ||
| 106 | V | GTR | V | GTA | ||
| 118 | IV | RTT | V | GTT | ||
| 190 | AG | GSA | G | GGA | ||
| 190 | A | GCA | A | GCR | ||
| 190 | G | GGA | AG | GSA | ||
| 210 | L | TTA | LV | STG | ||
| Discordance | RT | 74 | VL | KTA | IL | RTA |
| 75 | V | GTA | I | ATA | ||
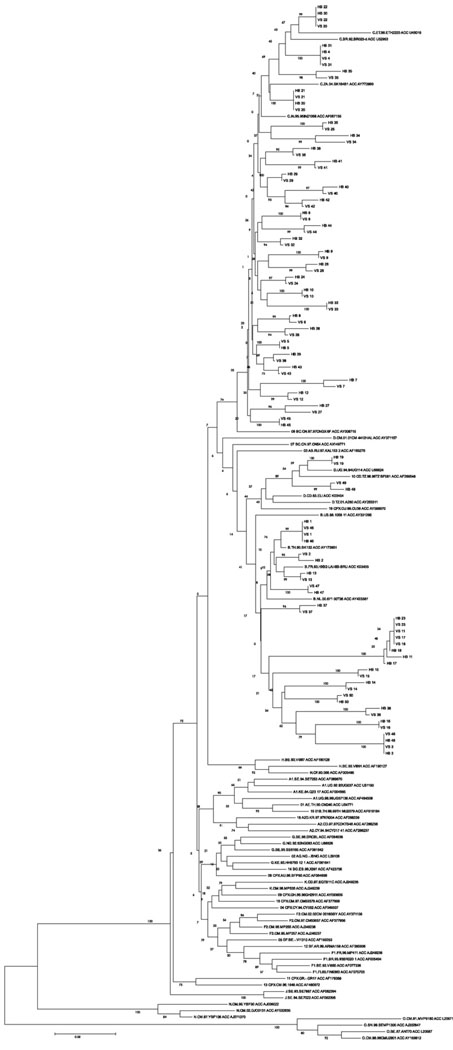
The rate of discordance between the two assays was negligible, none in PR and 0.1% (2/1700) in RT regions respectively. The discordance in RT was observed at codons 74 and 75. The ViroSeq™ detected L74VL and in-house detected L74IL at codon 74 while at codon 75, ViroSeq™ detected wild type while in-house detected V75I. No indeterminate results were observed. Phylogenetic analysis showed that sequences of each sample from both the assays monophyletically clustered together (Fig. 2).
Fig. 2.
Phylogenetic analysis of sequences from both the assays.
4. Discussion
With the improved availability of generic HAART in India (Kumarasamy et al., 2005), the demand for accurate but affordable monitoring of patients under treatment is becoming a priority (Kent et al., 2003). Indeed, increasing access to antiretroviral drugs without adequate monitoring may well result in transmission of drug resistant virus which has been observed in North America and Europe (Bennett et al., 2006; Geretti, 2007). The transmission of drug-resistant HIV strains in developing countries may be mitigated by measures that provide guidance for the prescription of highly effective drug combinations, promote adherence and, support the uninterrupted supply of the drugs. The availability of laboratory methods to monitor side effects and the success of treatment and clear guidelines for actions in case of therapeutic failure are also critical. WHO guidelines (WHO, 2008) suggest that monitoring the success or failure of therapy should include CD4 count at minimum and viral load determinations where possible. When viral load monitoring is performed, genotyping of virologic failures can guide further treatment decisions and provide a direct measure of drug resistance. However, access to commercial assays for viral load monitoring and drug resistance in India is limited primarily by cost.
The present study demonstrated that there was a high concordance between ViroSeq™ and in-house genotyping systems. Mutations associated with the patient’s current failing therapy were more consistently identified by each assay and could be reproduced when repeated independently. Discordant mutations were identified to be less than 0.5% and mainly corresponded to secondary mutations and polymorphisms. ViroSeq™ has detected more mixtures (78%) of WT and mutant virus than by the in-house genotyping (22%), suggesting possibly increased sensitivity of commercial assay in terms of detecting minority variants. In addition to the design of the amplification and sequencing primers there are also differences in the labour and analytic time between the genotyping systems. One advantage of the ViroSeq™ is that it comes with the software that is designed specifically for editing and resitance interpretation, while the in-house assay requires a separate software for assembling sequenced fragments, manual editing and exporting consensus sequence, which needs to be considered. However, this software (SeqScape v2.5), is relatively user friendly and allows simple compilation of all sequence fragments from one sample into one contig, inspection of the sequence traces, and translation and check for frame shifts. The high concordance of mutations (99%) detected by the in-house assay (using SeqScape v2.5 package and Stanford HIV DB) and the ViroSeq™ system showed that the two systems had similar performance on interpreting all the clinically important resistance mutations. There was no significant difference in the time scales required for the assay process to be completed by both the systems. The turn around time for the in-house assay versus the standard assay was 16 and 15 h, respectively.
On the other hand, when genotyping assays are performed, it is important to minimize contamination of reaction mixtures with previously amplified products. Sample cross-contamination is minimized in the ViroSeq™ system by using a single non-nested PCR for amplification, and by using a dUTP/UNG contamination control system. However, the in-house assay, which uses nested PCR amplification and which does not have UNG based contamination control system, requires more vigilance to avoid cross-contamination and strict adherence to good clinical laboratory practices (GCLP) is crucial.
To conclude, the in-house assay performed well in comparison to a gold-standard, US-FDA approved ViroSeq™ system. All the clinically relevant mutations were concordant by both assays and reproducible. Despite very minimal discrepancies, the in-house assay, which costs about 50% ($ 100) of the ViroSeq™ ($ 230), demonstrated a similar capacity to identify clinically relevant mutations compared to the ViroSeq™. Studies spanning a wider spectrum of viral loads and known quantified resistant mutants are required to compare the analytical sensitivity for detection of HIV-1 drug resistance mutations. In addition to this, comparison between the two assays using a wider panel of the different HIV-1 subtypes and CRFs would be useful. Additional issues include the development of new and emerging assays for minority variants (Church et al., 2008; Johnson et al., 2007; Villahermosa et al., 2001; Beck et al., 2002; Vega et al., 2005) and assays for genotyping in integrase and envelope, particularly as new drugs against these targets become available (Grant and Zolopa, 2008; Paar et al., 2008; He et al., 2008). In-house approaches to implementing molecular diagnostics can build capacity and provide training in resource-limited settings at a cost substantially lower than commercial assays ($ 100 vs. $ 230). The successful validation and implementation of the in-house assay will also serve to lower the prices and increase competition among commercial suppliers. Wide applications of low-cost genotyping will facilitate surveillance studies and improve clinical decision making for HAART, and thus reduce the risk of further drug resistance-related mutations.
Acknowledgement
This work was supported by research grant from YRG CARE, Chennai, India.
Footnotes
Conflict of interest
None declared.
References
- Baxter JD, Mayers DL, Wentworth DN, Neaton JD, Hoover ML, Winters MA, Mannheimer SB, Thompson MA, Abrams DI, Brizz BJ, Ioannidis JP, Merigan TC. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–F93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- Beck IA, Mahalanabis M, Pepper G, Wright A, Hamilton S, Langston E, Frenkel LM. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J. Clin. Microbiol. 2002;40:1413–1419. doi: 10.1128/JCM.40.4.1413-1419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DE, Smith AJ, McCormick L, Prachand N, Sey E. Categorization of HIV drug resistance using the WHO/CDC HIV drug resistance threshold survey method. Antivir. Ther. 2006;11:S116. [Google Scholar]
- Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Jones E, Tsay J, Ip J, Farthing C, Limoli K, Parkin N, Markowitz M. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- Bowles E, Wensing AMJ, van de Vijver DAMC. WATCH: a worldwide database for collecting and analysing data on transmission of drug resistant HIV using standardized methods. Program and abstracts of the 16th International AIDS Conference; August 13–18; Toronto, Canada. 2006. (Abstract MOPE0388). [Google Scholar]
- Church JD, Towler WI, Hoover DR, Hudelson SE, Kumwenda N, Taha TE, Eshleman JR, Eshleman SH. Comparison of LigAmp and an ASPCR assay for detection and quantification of K103N-containing HIV variants. AIDS Res. Hum. Retroviruses. 2008;24:595–605. doi: 10.1089/aid.2007.0224. [DOI] [PubMed] [Google Scholar]
- Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher CA, Schapiro JM, Dellamonica P. Drug resistance genotyping in HIV-1 therapy: the VIRADAPT randomized controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- Gallant JE. Antiretroviral drug resistance and resistance testing. Top. HIV Med. 2006;13:138–142. [PubMed] [Google Scholar]
- Geretti AM. Epidemiology of antiretroviral drug resistance in drug-naïve persons. Curr. Opin. Infect. Dis. 2007;20:22–32. doi: 10.1097/QCO.0b013e328013caff. [DOI] [PubMed] [Google Scholar]
- Grant P, Zolopa A. Integrase inhibitors: a clinical review of raltegravir and elvitegravir. J. HIV Ther. 2008;13:36–39. [PubMed] [Google Scholar]
- He Y, Cheng J, Lu H, Li J, Hu J, Qi Z, Liu Z, Jiang S, Dai Q. Potent HIV fusion inhibitors against Enfuvirtide-resistant HIV-1 strains. Proc.Natl. Acad. Sci. U.S.A. 2008;105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Li JF, Wei X, Lipscomb J, Bennett D, Brant A, Cong ME, Spira T, Shafer RW, Heneine W. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS ONE. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent DM, McGrath D, Ioannidis JP, Bennish ML. Suitable monitoring approaches to antiretroviral therapy in resource-poor settings: setting the research agenda. Clin. Infect. Dis. 2003;37:S13–S24. doi: 10.1086/375368. [DOI] [PubMed] [Google Scholar]
- Kumarasamy N, Solomon S, Chaguturu SK, Cecelia AJ, Vallabhaneni S, Flanigan TP, Mayer KH. The changing natural history of HIV disease: before and after the introduction of generic antiretroviral therapy in southern India. Clin. Infect. Dis. 2005;15:1525–1528. doi: 10.1086/497267. [DOI] [PubMed] [Google Scholar]
- Kuritzkes DR. Preventing and managing antiretroviral drug resistance. AIDS Patient Care STDS. 2004;18:259–273. doi: 10.1089/108729104323076007. [DOI] [PubMed] [Google Scholar]
- Larder BA, Kellam P, Kemp SD. Zidovudine resistance predicted by direct detection of mutation in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- Lindstrom A, Albert J. A simple and sensitive ‘in-house’ method for determining genotypic drug resistance of HIV-1. J. Virol. Methods. 2003;107:45–51. doi: 10.1016/s0166-0934(02)00188-x. [DOI] [PubMed] [Google Scholar]
- Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, Conway B, Kilby M, Wang L, Whitcomb JM, Hellmann NS, Richman DD. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- Paar C, Palmetshofer C, Flieger K, Geit M, Kaiser R, Stekel H, Berg J. Genotypic antiretroviral resistance testing for human immunodeficiency virus Type 1 integrase inhibitors on the TRUGENE® sequencing system. Clin. Microbiol. 2008;46:4087–4090. doi: 10.1128/JCM.01246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer RW, Winters MA, Palmer S, Merigan TC. Multiple concurrent reverse transcriptase and protease mutations multidrug resistance of HIV-1 isolates from heavily treated patients. Ann. Intern. Med. 1998;128:906–911. doi: 10.7326/0003-4819-128-11-199806010-00008. [DOI] [PubMed] [Google Scholar]
- Steegen K, Demecheleer E, De Cabooter N, Nges D, Temmerman M, Ndumbe P, Mandaliya K, Plum J, Verhofstede C. A sensitive in-house RT-PCR genotyping system for combined detection of plasma HIV-1 and assessment of drug resistance. J. Virol. Methods. 2006;133:137–145. doi: 10.1016/j.jviromet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Tural C, Ruiz L, Holtzer C, Schapiro J, Viciana P, González J, Domingo P, Boucher C, Rey-Joly C, Clotet B Havana Study Group. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS. 2002;16:209–218. doi: 10.1097/00002030-200201250-00010. [DOI] [PubMed] [Google Scholar]
- Vega Y, Pérez-Alvárez L, Delgado E, Muñoz M, Casado G, Carmona R, Sierra M, Vázquez de Parga E, Pinilla M, García V, Medrano L, Contreras G, Thomson M, Nájera R. Oligonucleotide ligation assay for detection of mutations associated with reverse transcriptase and protease inhibitor resistance in non-B subtypes and recombinant forms of human immunodeficiency virus type 1. J. Clin. Microbiol. 2005;43:5301–5304. doi: 10.1128/JCM.43.10.5301-5304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella S. Antiretroviral drug resistance and HIV/AIDS in the developing world. J. HIV Ther. 2002;7:53–55. [PubMed] [Google Scholar]
- Villahermosa ML, Beck I, Pérez-Alvarez L, Contreras G, Frenkel LM, Osmanov S, de Parga EV, Delgado E, Manjon N, Cuevas MT, Thomson MM, Medrano L, Najera R. Detection and quantification of multiple drug resistance mutations in HIV-1 reverse transcriptase by an oligonucleotide ligation assay. J. Hum. Virol. 2001;4:238–248. [PubMed] [Google Scholar]
- WHO. Panel on antiretroviral guidelines for adults and adolescents. Department of Health and Human Services; Guidelines for the use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents, November 3. 2008:1–139.