Abstract
The Pharmacogenomics Center of the University of California, San Francisco (CA, USA) fosters research and educational activities focused on the genomic basis for variation in drug response. Investigators in the Center conduct multidisciplinary and multicenter research on a diverse array of clinically used drugs with the goal of understanding the genetic factors that contribute to variation in therapeutic and adverse drug response. The Center houses the large NIH-supported Pharmacogenomics of Membrane Transporters Project, which is a leader in understanding genetic variation in membrane transporters that are important in clinical drug response. Center investigators study racially and ethnically diverse populations, are pioneers in the education of PharmD, MD and PhD students in pharmacogenomics, and have led the establishment of unique graduate and postdoctoral training programs focused on pharmacogenomics. A key emphasis of the Center is on biological mechanisms with a goal of facilitating the development of safer and more effective medications.
Keywords: membrane transporters, personalized medicine, pharmacogenomics, polymorphisms
The completion of the Human Genome project and the commencement of multiple large-scale genome-wide studies focused on human genetic variation have stimulated a wide array of research activities in pharmacogenomics, the genetic basis for variation in drug response. The mission of the University of California San Francisco (UCSF; CA, USA) Pharmacogenomics Center is to advance discoveries in pharmacogenomics by fostering research and educational activities focused on human genetic variation that is of particular relevance to variation in drug response. The research goal in the Center is to identify genetic determinants of variation in therapeutic and adverse drug response with a long-term goal of supporting the development of personalized medicine, where genetic information is used to inform the selection and dosing of drugs. Investigators in the UCSF Pharmacogenomics Center conduct multidisciplinary research on a wide variety of therapeutic drugs, including anticancer, antiviral and antidiabetic drugs. A major emphasis of the Center is on understanding biological mechanisms; therefore, strong interactions with both computational and experimental scientists are critical in supporting the research and educational mission of the Center. The Center investigators lead and participate in large clinical and translational studies, which span ethnically diverse populations globally. This brief profile describes the history of the Center as well as the major research and educational programs within it. Interacting programs and departments at UCSF that enhance, support and facilitate the Center’s activities are also briefly described.
The Pharmacogenomics Center was established in 1999 by Ira Herskowitz and Kathy Giacomini, two UCSF scientists with complementary backgrounds in genetics and pharmacology, respectively. Together, Giacomini (then chair of the Department of Biopharmaceutical Sciences) and Herskowitz (co-director of the Program in Human Genetics) developed a visionary plan for a multidisciplinary center in pharmacogenomics. The plan was enthusiastically supported by the faculty in the department, the Dean of the School of Pharmacy, Mary Anne Koda Kimble, and the other co-director of the Program in Human Genetics, Charles Epstein. The department, school and program all contributed critical resources to establish the Center, including space at the planned Mission Bay campus and at Parnassus Heights. From its inception, Giacomini and Herskowitz partnered in writing grants, stimulating research and educational activities and in recruitment of faculty to the Center. Two major NIH-funded grants encouraged recruitment and activity in the Center – The Pharmacogenomics of Membrane Transporters (PMT) project and the Pharmacogenomics Research Facility, both of which are described below. Although many UCSF faculty members participated in Center-led teaching and research activities, seven full-time core faculty members are now a part of the Center:
Nadav Ahituv: mechanistic pharmacogenetic studies centered on noncoding regions of the genome;
Esteban Burchard: genetic studies of drug response in Latino populations;
Xin Chen: cancer genomics and pharmacogenomics;
Kathy Giacomini: basic and translational pharmacogenetic studies with a focus on membrane transporters;
Su Guo: vertebrate genetics in zebrafish;
Deanna Kroetz: translational and clinical pharmacogenetic studies in large populations including genome-wide association studies;
Andrej Sali: the bioinformatics of the effects of genetic variants on protein structure and function.
The Center is located on both the Mission Bay (Figure 1) and Parnassus Heights campuses at UCSF. On the Parnassus campus, the National Center for Research Resources (NCRR) funded a large grant to construct a large Pharmacogenomics Research Facility, which houses laboratory and computational space for the Center. This facility, located one floor below the Institute for Human Genetics, has become a focal point for activity at the Parnassus Heights campus. At Mission Bay, Center faculty members are located on the fourth and fifth floor of the new Rock Hall building and within the computational spaces of the California Institute of Quantitative Biosciences in Byers Hall.
Figure 1.

Rock Hall (lower right) on the Mission Bay Campus of the University of California San Francisco.
Four core faculties of the Pharmacogenomics Center (CA, USA) have laboratories in Rock Hall. The Mission Bay Campus, home for Rock Hall and Byers Hall (housing the Institute for Quantitative Biology) is shown in the upper picture.
Pharmacogenomics of Membrane Transporters Project
A large center grant, PMT, funded by the NIH from 2000, is a major focus for Center activities. Many of the Center’s faculty members participate in the NIH-funded project, which is led by Kathy Giacomini and Deanna Kroetz. This multidisciplinary research project is focused on membrane transporters that play a critical role in drug disposition and response. The primary goal of the PMT is to discover genetic variants in membrane transporters that contribute to variation in clinical drug response. Research involves integrated computational and mechanistic studies in cells, animal models and in ethnically diverse human populations. The PMT project has made a number of discoveries that have contributed to the field of pharmacogenomics and is particularly noted for its rich mechanistic and translational studies [1,2]. In particular, the project has resulted in the identification of many new coding- and noncoding-region variants in membrane transporter genes in ethnically diverse US populations. A large body of work focused on the functional genomics of membrane transporters has revealed that: approximately 15–20% of nonsynonymous variants in membrane transporters exhibit reduced function; rare nonsynonymous variants are more likely to exhibit functional changes than common variants; strong promoters have more genetic variation than weak promoters; and many common promoter-region variants with altered function are present in membrane transporter genes. In 2003, the PMT project established a unique cohort of healthy volunteers from populations of mixed ethnicity – studies of pharmacogenomics in ethnically diverse populations (SOPHIE) – who donated DNA samples and agreed to participate in further pharmacogenomic studies. SOPHIE has formed the basis for the large translational effort of PMT and is described in more detail later. Studies using SOPHIE have led to discoveries of variants that contribute to variations in drug disposition and response for the antidiabetic drug, metformin, and the antiepileptic drug, gabapentin, in human populations. PMT has also developed a large database of pharmacogenomic membrane transporters [101], which is a repository for genetic and functional information on genetic variation in membrane transporters.
Emphasis on biological mechanisms
The UCSF Center for Pharmacogenomics has a strong emphasis on biological mechanisms and exploits both computational and experimental methods. When conducting clinical pharmacogenomic studies (genotype – phenotype), such as those conducted by the PMT project, high-throughput phenotypic assays are needed to rapidly determine the pharmacological outcomes of different nucleotide variants. Moreover, pathway interrogations using model organisms can further lead to biologically relevant candidate genes for subsequent human association studies. These mechanistically oriented studies are becoming an even greater necessity with next-generation sequencing technologies being on the verge of providing whole-genome datasets. Our multidisciplinary center has been developing and implementing these mechanistic assays by taking advantage of advanced computational analyses coupled with high-throughput functional assays. For example, recently, investigators in the Center used computational modeling coupled with experimental assays to understand the functional impact of mutations in ABC transporters, where the majority of mutations had not yet been experimentally characterized [Kelly L et al.: Functional hotspots in human ABC transporter nucleotide binding domains. Manuscript submitted.]. A computational structure-based approach to discriminate between disease-associated and neutral point mutations tailored to this superfamily was developed. From this approach, the impact of 39 unannotated point mutations in seven ABC transporters identified was predicted. Three of the predictions were experimentally tested, using HEK293 cells stably transfected with the reference multidrug resistance transporter ABCC4 and its variants, to examine functional differences in the transport of the antiviral drug tenofovir. The results indicated that two of the three predictions agreed with the experimental results, demonstrating the utility of the computational model to predict functional consequences of point mutations.
Various animal models are used for functional assays; for example, zebrafish, a vertebrate model organism for molecular genetic studies, presents multiple salient features for pharmacogenomic studies [3,4]. Zebrafish have been used in the Center to understand the mechanisms of action of small molecules, including abused substances (alcohol and morphine) [5 – 8], antipsychotics [9] and neurotoxins [10]. In addition to zebrafish, mouse models are also widely used for pharmacogenomic studies. An organic cation transporter 1 (OCT1)-knockout mouse was used to explore the interaction between metformin, an antidiabetic drug, and OCT1 [11]. In addition, we are taking advantage of rapid-mouse techniques, such as the hydrodynamic tail-vein injection technique [12], to test for the functional effects of promoter and enhancer variants in liver membrane transporters, which is discussed later.
A less-explored pharmacogenomic terrain is the noncoding portion of the genome that is known to have important roles in gene regulation [13]. A small number of pharmacogenomic studies have focused on understanding the role of variants in regulatory elements, such as enhancers (sequences that regulate gene promoters), miRNAs and proximal promoters. However, these studies have largely focused on drug targets and drug metabolizing enzymes [14 – 20], but not on membrane transporters. To uncover regulatory variants in the membrane transporters, the Center investigators are using various computational tools to identify potential regulatory regions (e.g., enhancers, promoters, miRNAs and their binding sites). These regions are then assayed using both in vitro and in vivo analyses, such as the hydrodynamic tail-vein technique [14], for their function and also to determine how nucleotide variation within them can alter that function.
Translational studies in racially & ethnically diverse populations
The rich racial and ethnic diversity in the San Francisco Bay area (CA, USA) provides a unique resource for investigating population differences in drug response. As part of the PMT project, the UCSF Center for Pharmacogenomics developed the SOPHIE cohort, which is a local resource for studying pharmacogenetic traits. SOPHIE consists of over 1000 young, healthy individuals from Caucasian, African – American, Chinese – American and Mexican – American ancestry. In addition, SOPHIE includes a few individuals with ancestry in the Philippines. Inclusion criteria requires that each subject has four grandparents with the same ancestry. DNA from SOPHIE subjects serves as a resource for re-sequencing projects within the Center and facilitates the estimation of minor allele frequencies and haplotype distributions in these ethnic populations. Upon enrollment, all subjects also consent to be called back for potential participation in genotype – phenotype studies of interesting genetic variants. The SOPHIE cohort has proven invaluable in ‘proof-of-concept’ studies, where genetic variants that have been demonstrated, in laboratory investigations, to alter transporter function or expression and are tested for their effects on pharmacokinetic or pharmacodynamic properties of specific drug substrates. Recently published studies on metformin illustrate the importance of SOPHIE to our Center. A total of seven SLC22A1 (OCT1) variants were found to have a significant reduction of metformin transport in vitro, and Caucasians with at least one reduced-function allele had an attenuated response to the glucose lowering effect of metformin [11]. Another area of expertise within the Center is in genetic admixture and the use of individual ancestry estimates as a quantitative trait. The consideration of genetic estimates of ancestry as covariates is increasingly applied in pharmacogenetic analyses and is particularly relevant in admixed populations (i.e., Latinos, African – Americans, and Filipinos). Ongoing studies within the Center are applying these estimates to understand the differences in disease severity and response to asthma medications in various Hispanic ethnic groups (e.g., Puerto Rican vs Mexican) [21 – 24]. The importance of studying large and ethnically diverse populations for association of genetic polymorphisms with drug response typically necessitates a collaborative approach for both the collection of phenotypic data and genetic analysis. UCSF investigators have been instrumental in developing a formal collaboration with the Riken Center for Genomic Medicine in Yokohama, Japan and the Pharmacogenetic Research Network (PGRN) to conduct genome-wide association analyses of drug response phenotypes. The Global Alliance in Pharmacogenomics-Japan (GAP-J) capitalizes on the strength of the PGRN in collecting large, well-phenotyped patient populations and the powerful high-throughput genome-wide methods of the Riken Center for Genomic Medicine. Investigators in the UCSF Center for Pharmacogenomics lead three of the 14 current collaborative projects in GAP-J and are involved in three additional projects.
A major goal of the UCSF Center is to extend pharmacogenetic studies into understudied populations. In addition to the asthma pharmacogenetic studies in Hispanic populations, as described previously, efforts are ongoing to identify genetic predictors of response and toxicity to antiretroviral medications used in sub-Saharan Africa. In collaboration with clinical researchers at UCSF, detailed phenotypic data on viral and immune response and adverse events are being collected in a rural HIV/AIDS clinic in Uganda. The identification of genetic markers of response or toxicity can be used for the informed selection and proper use of the most appropriate antiretroviral combinations in these vulnerable populations.
Educational programs associated with the Center
Education is a core component of the UCSF Center for Pharmacogenomics, and the faculty is involved in didactic and research training in all aspects of the field. The Pharmaceutical Sciences and Pharmacogenomics (PSPG) graduate program was established in 1999 as the first graduate program in the USA dedicated to doctorate-level training in pharmacogenomics [102]. The program is supported, in part, by a NIH predoctoral training grant and is attractive to students with broad backgrounds ranging from human genetics to engineering. Strong didactic training in pharmacokinetics, drug metabolism and drug transport is coupled with courses in human genetics and pharmacogenetics. The latter is focused on the application of cutting-edge human genetic approaches (e.g., high-throughput SNP genotyping and expression arrays), the use of genetic estimates of admixture, an emphasis on biological mechanisms and the proper design of clinical studies for the investigation of genetic variability in drug response and toxicity. Graduate students can select from a wide range of research projects in over 20 different laboratories, including computational analysis of patterns of genetic variability in human populations, the development of novel methods for the analysis of pharmacogenetic association studies, functional genomic studies to understand the molecular basis for genetic associations with drug response genes and both phenotype – genotype and genotype – phenotype clinical studies that apply candidate gene and whole-genome analyses. Graduates of the PSPG program are leaders in the application of pharmacogenetic principles in drug development and discovery within academia, industry and regulatory agencies. PSPG graduates, with their strong background in pharmacokinetics, pharmacodynamics, drug metabolism, drug transport and pharmacogenomics, are highly sought after for positions in academia, industry and at the US FDA.
A postdoctoral clinical pharmacology training program focuses on the education of fellows [103]. The program has been supported for 35 years by a NIH training grant in clinical pharmacology, which traditionally focused on the education of MDs and PhDs. However, recently, the program expanded to include an emphasis on the training of PharmD and pediatric postdoctoral fellows. Didactic courses in all aspects of clinical pharmacology are required, including a strong emphasis on pharmacogenetic principles and applications. A large proportion of the fellows carry out pharmacogenetic-based clinical research across all therapeutic disciplines. Clinical pharmacology fellows are also involved in campus-wide efforts to incorporate pharmacogenetic testing into clinical practice. Since its inception in 1965, this program has graduated 120 fellows, most of whom are actively engaged in academic research and training, and are highly qualified to lead efforts for the application of pharmacogenomic discoveries in clinical medicine.
Units at UCSF that support the Center
At UCSF, several units support the activities of the Center for Pharmacogenomics: the Department of Bioengineering and Therapeutic Sciences, the Institute for Human Genetics, the California Institute of Quantitative Biosciences (QB3), and the Clinical and Translational Sciences Institute (CTSI) (Figures 1 & 2). The Center is housed in the Department of Bioengineering and Therapeutic Sciences, which consists of five research foci including the Pharmacogenomics Center – drug development sciences, therapeutic bioengineering, computational and systems biology as well as cellular and molecular engineering. The department is a joint department between the School of Pharmacy and the School of Medicine, and provides state-funded faculty salary lines, financial and administrative support, and facilitates cross-disciplinary scientific interactions relevant to therapeutic sciences. The Institute for Human Genetics in the School of Medicine is also a critical unit for the Center. The Institute, which grew out of the Program in Human Genetics, is headed by Neil Risch, a visionary leader who strongly endorses and supports the Center and has provided many resources to it. The Genomics Core Facility, which is managed by the Institute for Human Genetics, provides DNA banking, high- and medium-throughput genotyping and sequencing, and a host of other technologies and support that is necessary for modern genome-wide pharmacogenomic studies. Strong computational support is critical in modern pharmacogenomic studies and to the research conducted in the Pharmacogenomics Center. Computational support provided by the QB3 faculty members has fostered the development of the database of pharmacogenomic membrane transporters, a premier database in the field of pharmacogenomics. Furthermore, the QB3 faculty provide an array of expertise in systems and computational biology, protein structure and in the development of algorithms for functional predictions of genetic variants in membrane transporters. These computational activities are increasingly important with the emergence of multiple human genetic variation projects and genome-wide association studies. To understand the contribution of functionally important variants to clinical drug disposition and response, translational and clinical studies are essential. Support for these studies is provided by the UCSF NIH-funded Clinical and Translational Sciences Institute (CTSI), which includes a Clinical Research Center (CRC), a key unit in translating Pharmacogenomics Center discoveries to clinical studies. The CRC unit has assisted the Center in study design and, most importantly, in the implementation of clinical studies. This unit is critical for the success of the UCSF Pharmacogenomics Center.
Figure 2.
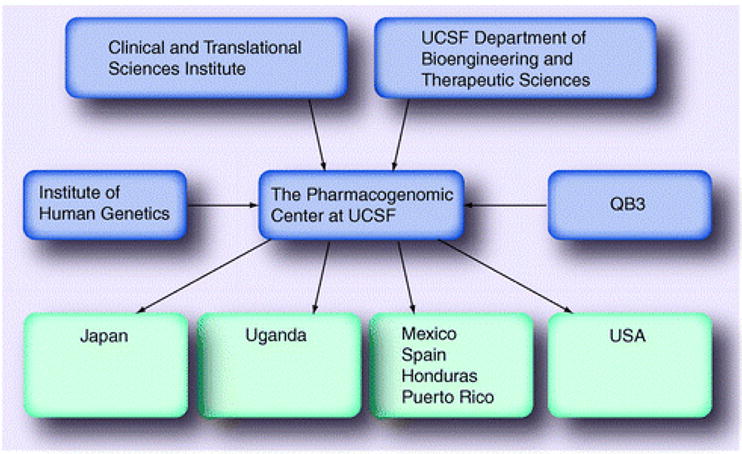
Relationship between the various UCSF institutes and departments that support the Pharmacogenomics Center.
QB3: California Institute of Quantitative Biosciences; UCSF: University of California San Francisco.
Conclusions and future directions
The UCSF Pharmacogenomics Center continues to play a pivotal role in the field of pharmacogenomics. The Center is a world leader in functional and computational genomics as well as in the translation of new discoveries in pharmacogenomics to clinical studies of therapeutic and adverse drug response. Center faculty members are key contributors in educating the next generation of pharmaceutical scientists, clinical pharmacologists and pharmacists, and scientists in computational and systems biology, who serve in academia, industry and regulatory agencies, to advance the field of pharmacogenomics and foster its translation to personalized medicines.
Footnotes
Financial & competing interests disclosure
The authors would like to acknowledge the NIH Pharmacogenomics of Membrane Transporters grant, GM61390, for partial support of this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Urban TJ, Sebro R, Hurowitz EH, et al. Functional genomics of membrane transporters in human populations. Genome Res. 2006;16(2):223–230. doi: 10.1101/gr.4356206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leabman MK, Huang CC, DeYoung J, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA. 2003;100(10):5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grunwald DJ, Eisen JS. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet. 2002;3(9):717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 4.Guo S. Linking genes to brain, behavior and neurological diseases: what can we learn from zebrafish? Genes Brain Behav. 2004;3(2):63–74. doi: 10.1046/j.1601-183x.2003.00053.x. [DOI] [PubMed] [Google Scholar]
- 5.Lockwood B, Bjerke S, Kobayashi K, Guo S. Acute effects of alcohol on larval zebrafish: a genetic system for large-scale screening. Pharmacol Biochem Behav. 2004;77(3):647–654. doi: 10.1016/j.pbb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Lau B, Bretaud S, Huang Y, Lin E, Guo S. Dissociation of food and opiate preference by a genetic mutation in zebrafish. Genes Brain Behav. 2006;5(7):497–505. doi: 10.1111/j.1601-183X.2005.00185.x. [DOI] [PubMed] [Google Scholar]
- 7.Bretaud S, Li Q, Lockwood BL, Kobayashi K, Lin E, Guo S. A choice behavior for morphine reveals experience-dependent drug preference and underlying neural substrates in developing larval zebrafish. Neuroscience. 2007;146(3):1109–1116. doi: 10.1016/j.neuroscience.2006.12.073. [DOI] [PubMed] [Google Scholar]
- 8.Peng J, Wagle M, Mueller T, et al. Ethanol-modulated camouflage response screen in zebrafish uncovers a novel role for cAMP and extracellular signal-regulated kinase signaling in behavioral sensitivity to ethanol. J Neurosci. 2009;29(26):8408–8418. doi: 10.1523/JNEUROSCI.0714-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacomini NJ, Rose B, Kobayashi K, Guo S. Antipsychotics produce locomotor impairment in larval zebrafish.Neurotoxicol. Teratol. 2006;28(2):245–250. doi: 10.1016/j.ntt.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Bretaud S, Lee S, Guo S. Sensitivity of zebrafish to environmental toxins implicated in Parkinson’s disease.Neurotoxicol. Teratol. 2004;26(6):857–864. doi: 10.1016/j.ntt.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Shu Y, Sheardown SA, Brown C, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117(5):1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10(10):1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 13.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 14.Yee SW, Shima JE, Hesselson S, et al. Identification and characterization of proximal promoter polymorphisms in the human concentrative nucleoside transporter 2 (SLC28A2) J Pharmacol Exp Ther. 2009;328(3):699–707. doi: 10.1124/jpet.108.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drazen JM, Yandava CN, Dube L, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22(2):168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 16.Tahara H, Yee SW, Urban TJ, et al. Functional genetic variation in the basal promoter of the organic cation/carnitine transporters OCTN1 (SLC22A4) and OCTN2 (SLC22A5) J Pharmacol Exp Ther. 2009;329(1):262–271. doi: 10.1124/jpet.108.146449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. A promoter polymorphism in cholesterol 7α-hydroxylase interacts with apolipoprotein E genotype in the LDL-lowering response to atorvastatin. Atherosclerosis. 2005;180(2):407–415. doi: 10.1016/j.atherosclerosis.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Kim TW, Innocenti F. Insights, challenges, and future directions in irinogenetics. Ther Drug Monit. 2007;29(3):265–270. doi: 10.1097/FTD.0b013e318068623b. [DOI] [PubMed] [Google Scholar]
- 19.Lencz T, Robinson DG, Xu K, et al. DRD2 promoter region variation as a predictor of sustained response to antipsychotic medication in first-episode schizophrenia patients. Am J Psychiatry. 2006;163(3):529–531. doi: 10.1176/appi.ajp.163.3.529. [DOI] [PubMed] [Google Scholar]
- 20.Whale R, Quested DJ, Laver D, Harrison PJ, Cowen PJ. Serotonin transporter (5-HTT) promoter genotype may influence the prolactin response to clomipramine. Psychopharmacology (Berl) 2000;150(1):120–122. doi: 10.1007/s002130000432. [DOI] [PubMed] [Google Scholar]
- 21.Corvol H, De Giacomo A, Eng C, et al. Genetic ancestry modifies pharmacogenetic gene-gene interaction for asthma.Pharmacogenet. Genomics. 2009;19(7):489–496. doi: 10.1097/FPC.0b013e32832c440e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naqvi M, Tcheurekdjian H, DeBoard JA, et al. Inhaled corticosteroids and augmented bronchodilator responsiveness in Latino and African American asthmatic patients. Ann Allergy Asthma Immunol. 2008;100(6):551–557. doi: 10.1016/S1081-1206(10)60055-5. [DOI] [PubMed] [Google Scholar]
- 23.Naqvi M, Thyne S, Choudhry S, et al. Ethnic-specific differences in bronchodilator responsiveness among African Americans, Puerto Ricans, and Mexicans with asthma. J Asthma. 2007;44(8):639–648. doi: 10.1080/02770900701554441. [DOI] [PubMed] [Google Scholar]
- 24.Tcheurekdjian H, Thyne SM, Williams LK, et al. Augmentation of bronchodilator responsiveness by leukotriene modifiers in Puerto Rican and Mexican children. Ann Allergy Asthma Immunol. 2009;102(6):510–517. doi: 10.1016/S1081-1206(10)60126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Pharmacogenetics of Membrane Transporters Database. http://pharmacogenetics.ucsf.edu.
- 102.University of California; San Francisco: http://bts.ucsf.edu/pspg/ [Google Scholar]
- 103.Division of clinical pharmacology. University of California; San Francisco: http://medicine.ucsf.edu/sfgh/divisions/clinpharm/index.html. [Google Scholar]


