Abstract
The cytoplasmic tail domain (CTD) of retroviral envelope (Env) proteins has been implicated in modulating Env incorporation into viral particles. We generated a panel of murine leukemia virus (MLV) Env mutants and analyzed their ability to be recruited to human immunodeficiency virus-1 (HIV-1) assembly sites. Surprisingly, the entire CTD was dispensable for recruitment to assembly sites, but a mutation that disrupted the furin cleavage site in Env abolished recruitment. To determine if MLV Env can show selectivity for homologous assembly sites, cells were co-transfected with both HIV-1 and MLV assembly components along with each MLV Env construct and assayed for infectious particle production. MLV Env selectively formed infectious particles with the MLV components at the expense of infectious HIV-1 infectious particle production, but truncation of the CTD progressively reduced this selectivity. Collectively these data suggest that there are two separable mechanisms that govern MLV Env recruitment to viral assembly sites.
Keywords: Pseudotype, Assembly, RD114, Env, MLV, HIV-1, Recruitment, Retrovirus, cytoplasmic domain
Introduction
Fundamentally, the formation of an infectious retrovirus is the result of the coordinated assembly of at least three viral components: the genome, the core structural proteins (Gag and GagPol) and an envelope glycoprotein (Env). Although Env is expendable for nascent particle assembly and budding, it is required for selectively binding to specific receptors and mediating entry into the host cell. MLV Env (gp85) forms a heterotrimeric, type-1 transmembrane protein and is produced as a single protein that is cleaved into a transmembrane (TM or p15E) subunit and an extracellular surface (SU or gp70) subunit. The SU subunit of ecotropic MLV Env mediates binding to the host cell surface receptor, murine cationic amino acid transporter-1 (mCAT-1), and the TM subunit of Env contributes to membrane fusion following receptor binding. The MLV Env TM subunit contains an N-terminal ectodomain followed by a membrane-spanning domain and a 35 amino acid C-terminal cytoplasmic tail domain (CTD).
In a process known as pseudotyping, some enveloped viruses are capable of incorporating heterologous viral glycoproteins during assembly. The mechanism by which particular viral glycoproteins are sorted to assembly sites of unrelated viruses remains unknown. Using a scanning electron microscopy technique (SEM), we recently demonstrated that MLV Env is specifically recruited to HIV-1 budding sites during assembly (Jorgenson et al., 2009). It has been shown for several Env proteins, including those of HIV-1, RD114, GaLV and others, that the CTD of Env dictates pseudotyping restrictions (Christodoulopoulos and Cannon, 2001; Sandrin et al., 2006; Sandrin and Cosset, 2006; Sandrin et al., 2004; Schnierle et al., 1997; Stitz et al., 2000). These studies suggest that amino acid sequences in the CTD mediate Env inclusion into viral particles, possibly through cytoplasmic interactions with Gag, GagPol or cellular cytoplasmic proteins.
Within the MLV Env CTD, several elements regulate Env incorporation and fusogenicity. The C-terminus of the MLV Env CTD is known as the R peptide, a 16 amino acid sequence that is cleaved off by the viral protease during virus maturation (Bobkova et al., 2002; Green et al., 1981; Henderson et al., 1984). Among gammaretroviruses, the R peptide includes a conserved leucine-valine dipeptide cleavage site and a tyrosine (YXXL) motif that has been implicated in promoting endocytosis of the Env protein (Aguilar et al., 2003; Blot et al., 2006; Bobkova et al., 2002; Kubo et al., 2007; Lodge et al., 1997; Loving et al., 2008). Mutation of the tyrosine to an alanine does not disrupt fusogenicity, but does increase Env expression on the cell surface presumably through disruption of the R peptide endocytosis motif (Aguilar et al., 2003). Additionally, some mutations at the tyrosine position have been shown to reduce Env incorporation into viral particles, a possible result of a defect in Env trafficking (Taylor and Sanders, 2003). Although R-minus Env is highly fusogenic, large defects in the context of MLV particles have been reported. MLV produced with R-minus Env resulted in a decrease in cellular transduction relative to wildtype MLV Env particles (Aguilar et al., 2003; Januszeski et al., 1997; Kubo and Amanuma, 2003; Rein et al. 1994; Rozenberg-Adler et al., 2007). In cell culture, the R peptide is required as demonstrated by the rapid reversion of MLV R-minus Env mutants to an Env with a functional R peptide (Thomas et al., 1997). Fractionation studies have shown R peptide association with immature particles, suggesting a role for R peptide-mediated assembly between Env and budding particles (Andersen et al., 2006).
Prior to its cleavage, the R peptide functionally suppresses premature Env fusogenicity. MLV Env proteins become highly fusogenic in mutants that lack the R peptide (Kubo et al., 2007; Loving et al., 2008; Ragheb and Anderson, 1994; Rein et al., 1994; Taylor and Sanders, 2003; Yang and Compans, 1997). When R peptide cleavage is inhibited, MLV Env proteins are still incorporated into viral particles but have greatly diminished fusogenic activity (Loving et al., 2008; Rein et al., 1994). A conserved leucine, located in the second position of the R peptide N-terminus, is critical in the suppression of premature Env activation. Mutation of this amino acid results in enhance Env fusogenicity in the absence of R peptide cleavage (Kubo and Amanuma, 2003; Kubo et al., 2007; Taylor and Sanders, 2003; Yang and Compans, 1997). Some mutations in the R peptide can also alter the conformation of the Env ectodomain (Aguilar et al., 2003). The mechanistic action of R peptide removal on SU subunit activation has recently been elucidated further. Cleavage of the R peptide facilitates the isomerization of the disulfide bond bridging the SU and TM subunits by reducing the ability of the R peptide to maintain stability within the CTD alpha-helical coil and interfere with the activity of CXXC-thiol in the SU subunit (Loving et al., 2008).
Although the R peptide is a prominent factor in the activation of Env, other portions of the CTD contribute to incorporation and fusion. Truncations of the MLV CTD demonstrate a correlation between reduced tail length and a reduction in infectivity and incorporation, despite similar levels of Env expression at the cell surface (Januszeski et al., 1997; Rozenberg-Adler et al., 2008). Truncations in the CTD greater than the last 31 C-terminal amino acids result in a dramatic decrease in incorporation and infectivity (Rozenberg-Adler et al., 2008).
Like all retroviral Env proteins, the precursor MLV Env is assembled into a trimeric structure in the endoplasmic reticulum and is subsequently cleaved into SU and TM subunits by a cellular furin or furin-like protease. The cleavage of pr85 into subunits is required and considered to be the first step towards MLV Env activation (Freed and Risser, 1987; McCune et al., 1988; Sjoberg et al., 2006; Zavorotinskaya and Albritton, 1999). The SU and TM subunits remain associated with each other through a disulfide bond bridge by a CXXC-thiol located in the SU subunit. Interestingly, reports vary on whether the cleavage of retroviral Env proteins is required for trafficking to the plasma membrane or incorporation into viral particles (Dong et al., 1992; Dubay et al., 1995; Freed and Risser, 1987; Goodman et al., 1993; Guo et al., 1990; Machida and Kabat, 1982; McCune et al., 1988; Moulard et al., 1999; Zavorotinskaya and Albritton, 1999).
We investigated a series of MLV Env mutants with truncations of the CTD, a furin-cleavage defective construct, as well as an MLV Env chimera containing the CTD of the gammaretrovirus RD114. We demonstrate that all MLV Env truncations are robustly recruited to viral budding sites of HIV-1. Surprisingly, we found that the CTD is not required for recruitment of MLV Env to viral budding sites, but that the CTD does mediate Env discrimination between MLV and HIV-1 sites of assembly. Of the Env mutants investigated, only the furin-cleavage defective mutant failed to associate with viral budding sites or produce infectious particles. Our findings suggest that the minimal factors facilitating generic recruitment and incorporation of Env lie outside of the CTD while elements regulating viral specificity are located within the CTD.
Results
Properties of MLV Env mutants
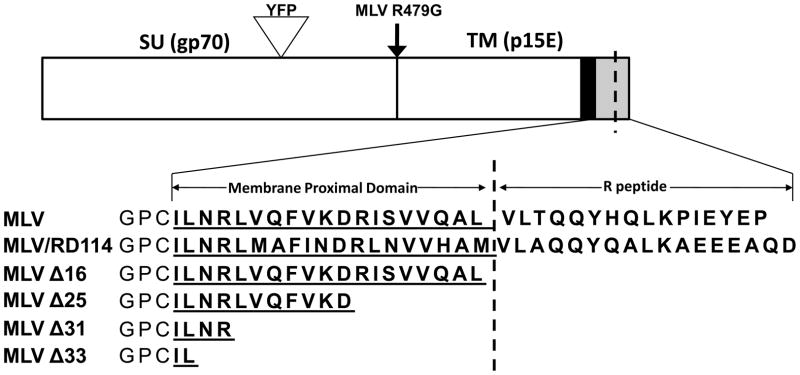
Previous studies have demonstrated that the Env CTD has a critical role in promoting Env protein incorporation into viral particles and modulating fusogenicity. We created a library of expression constructs containing sequential C-terminal truncations of the CTD as well as a processing defective MLV Env (R479G) that lacks the pr85 cleavage site required to produce the SU and TM subunits (Fig. 1). In addition, we engineered an MLV chimeric construct that contains the CTD of the feline gammaretrovirus RD114 (Fig. 1). An equivalent glycoprotein construct has been shown to be incompatible with the lentivirus simian immunodeficiency virus (SIVmac) (Bouard et al., 2007; Sandrin et al., 2006; Sandrin and Cosset, 2006; Sandrin et al., 2004).
FIG. 1.
Schematic of MLV Env protein constructs. Sequences are the C-terminal cytoplasmic tails of ecotropic MLV Env, an MLV/RD114 chimera and MLV Env mutants. Specific mutations are shown below MLV wildtype Env. The cytoplasmic tail domain is shown in bold, the membrane proximal domain is underlined, an arrow indicates the point mutation made at the furin cleavage site. The YFP insertion site is shown by the triangle for mutants used in fluorescent cell surface expression, syncytial and SEM assays. SU=surface domain, TM=transmembrane domain, black column= transmembrane spanning helix.
The six constructs were evaluated to characterize their relative infectivity, fusogenicity and surface expression (Table 1). These assays were performed primarily to confirm that our MLV Env constructs behave similarly to previously described MLV Env mutants. First, we determined whether each MLV Env construct was capable of forming infectious particles in the context of either lentiviral (HIV-1) core particles or homologous gammaretrovirus (MLV) core particles. Both gammaretroviral or lentiviral particles were able to pseudotype with all Env chimeras up to deletion of the C-terminal 31 amino acids. The furin-cleavage mutant, R479G, was unable to generate a significant viral titer. These findings are generally consistent with previous reports (Christodoulopoulos and Cannon, 2001; Freed and Risser, 1987; Januszeski et al., 1997; Rozenberg-Adler et al., 2008). Surprisingly, we did not observe a decrease in infectivity with HIV-1 upon substitution of the MLV CTD with the RD114 Env CTD. Although a similar MLV Env construct has been reported to be incompatible with SIVmac (Sandrin et al., 2004), its compatibility with HIV-1 has not been reported.
Table 1.
Properties of MLV Envelope mutations
| Env construct | Infectivity a |
Syncytia b | Cell surface expression (FACS) c | |
|---|---|---|---|---|
| HIV-1 Core | MLV Core | |||
| MLV Env | 100 | 100 | 0±0 | 100 |
| MLV/RD114 | 128±14 | 98±2 | 0.01±0.01 | 123 |
| MLV Δ16 | 117±37 | 71±28 | 0.53±0.15 | 108 |
| MLV Δ25 | 198±50 | 64±17 | 0.36±0.09 | 110 |
| MLV Δ31 | 70±28 | 4±2 | 0.23±0.17 | 89 |
| MLV Δ33 | 8±4 | 1±1 | 0±0 | 87 |
| MLV R479G | 0±0 | 0±0 | 0±0 | 51 |
I nfectivity values for each construct are shown as infectious units (IU) per ml relative to wildtype MLV Env (±SD, n=3).
The relative cell fusion index (CFI) was calculated per field by first determining the CFI relative to the CFI of each treatment group as described in materials and methods.
Truncations in the Env cytoplasmic tail fail to alter cell surface expression. Calculated as mean channel FL-2 fluorescence to mean channel GFP fluorescence for each construct relative to wildtype expression levels. Values shown at the average of two independent experimental replicates.
Syncytial assays were performed to determine the fusogenic properties of the MLV Env constructs. As expected, the removal of the R peptide (Δ16) promoted the formation of syncytia. Some syncytia were also observed with the other truncation mutants that lacked the R peptide, but essentially no syncytia were observed with wildtype MLV Env, R479G, or the MLV/RD114 chimera. These findings are consistent with previous reports (Januszeski et al., 1997; Kubo and Amanuma, 2003; Rein et al., 1994; Rozenberg-Adler et al., 2008; Taylor and Sanders, 2003; Yang and Compans, 1997).
To exclude the possibility that reductions in infectivity could be explained by a decrease in MLV Env present at the cellular plasma membrane, the surface abundance of the MLV Env proteins was quantified via flow cytometry. Each of the six proteins displayed significant surface expression, although there was some reduction with select truncations and with the R479G mutant. These data are consistent with reports by others using similar MLV Env modifications demonstrating that Env truncations are capable of plasma membrane expression (Januszeski et al., 1997; Rozenberg-Adler et al., 2008).
The MLV Env cytoplasmic tail is dispensable for recruitment to HIV-1 assembly sites
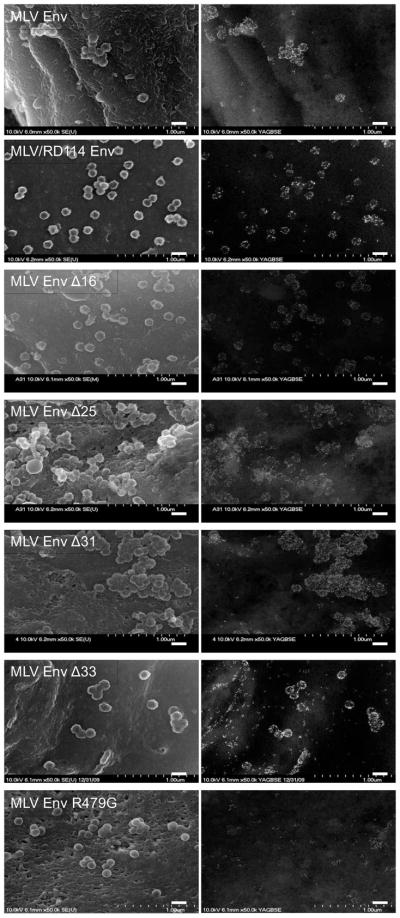
Because all MLV Env truncations, with the exception of Δ33, readily produced infectious particles with HIV-1, we next sought to determine if infectivity of pseudotyped particles was the result of specific or non-specific recruitment of each Env to HIV-1 assembly sites. It has been estimated that a single infectious retroviral particle requires inclusion of less than ten Env trimers (Klasse, 2007; Magnus et al., 2009; Yang et al., 2005) and it is plausible that sufficient levels of Env could be passively incorporated into particles as they bud from the cellular plasma membrane. However, we have previously demonstrated, using an SEM imaging technique, that MLV Env incorporation into HIV-1 particles is not random, but MLV Env is in fact robustly recruited to HIV-1 assembly sites (Jorgenson et al., 2009). To determine if the CTD of MLV Env modulates this recruitment, we imaged the distribution of each of the MLV Env constructs relative to HIV-1 assembly sites by SEM. Surprisingly, we found that all truncation mutants, including the mutant lacking its entire CTD, were recruited to budding HIV-1 particles (Fig. 2). The MLV/RD114 chimera was also robustly recruited to HIV-1 assembly sites. The only Env mutant that was not recruited to HIV-1 assembly sites was R479G. This protein appeared somewhat clustered in distribution on the surface of cells, but these clusters did not coincide with HIV-1 assembly sites. Although this result is consistent with some previous studies that have suggested that furin-cleavage defective Env proteins are excluded from viral particles despite cell surface expression (Dubay et al., 1995; Moulard et al., 1999), it was nonetheless remarkable to see the dramatic shift of protein away from HIV-1 assembly sites due to an alteration in the ectodomain of the protein. These images demonstrate that the CTD of Env is not required for recruitment of MLV Env to HIV-1 budding sites.
FIG. 2.
Cell surface distribution of MLV Env mutants relative to HIV-1 assembly sites. Left, secondary electron images of HIV-1 assembly sites. Right, backscatter electron images of gold-labeled Env. Scale bars = 200-nm.
MLV Env displays selectivity for MLV particles
Because MLV Env is known to be a promiscuous viral glycoprotein that is robustly recruited to HIV-1 assembly sites (Fig. 2) and was able to efficiently form infectious particles when combined with HIV-1 (Table 1), we wished to determine if MLV Env demonstrates intrinsic selectivity for MLV cores. Because the SEM assay relies on identifying assembly sites based on their surface topology, it was not feasible to compare recruitment of Env to two different types of viral assembly that would be morphologically similar. Instead, we employed an infectivity-based competition assay to determine whether MLV Env displays viral selectivity. To produce MLV and HIV-1 particles, we utilized an MLV reporter construct that expresses the red fluorescent protein TdTomato in infected cells and an HIV-1 reporter that expresses green fluorescent proteins (GFP) in infected cells. Along with their respective fluorescent reporters, plasmids encoding the MLV and HIV-1 structural components were transfected into cells and allowed to pseudotype with a single viral glycoprotein. Viral medium was transferred to 293T mCAT-1 cells and the relative viral infectivity output was determined by flow cytometry. As a control, the promiscuous rhabdoviral glycoprotein VSV-G was utilized instead of MLV Env. Infected cells were gated as either green or red fluorescent populations and cells co-infected with HIV-1 and MLV were excluded from infectivity analyses for consistency.
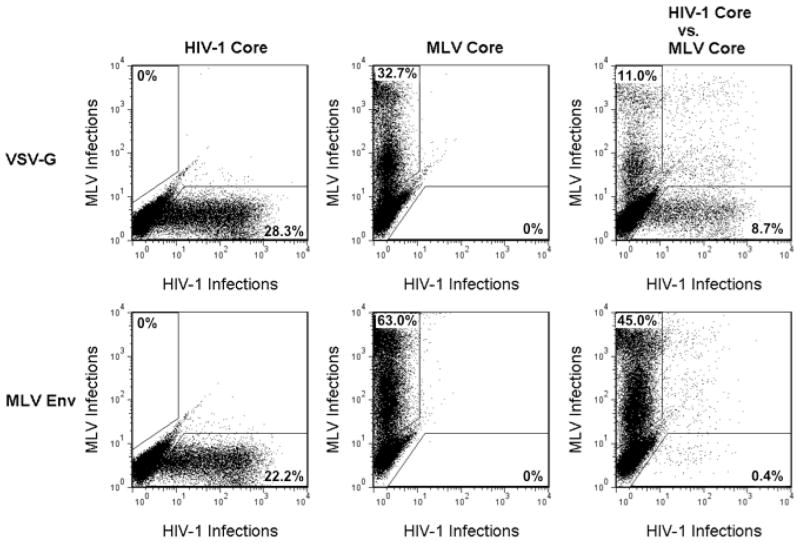
Individually, both HIV-1 and MLV cores were able to efficiently form infectious particles with VSV-G, and when the two viral cores were simultaneously introduced, the output of infectious particles was equivalent for the two types of virus (Fig. 3, top). When these transfections were conducted with MLV Env, both individually expressed MLV and HIV-1 particles were able to form infectious particles. However, when viral particles were produced in the presence of both cores there was a dramatic shift towards the production of infectious MLV particles at the expense of infectious HIV-1 particle production (Fig. 3, bottom). The ratio of infectious MLV to HIV-1 was approximately 100-fold higher with MLV Env than with VSV-G. These data demonstrate that, although MLV Env is attracted to HIV-1 assembly sites, it displays a distinct preference for MLV assembly sites.
FIG. 3.
MLV cores outcompete HIV-1 cores for MLV Env proteins. Infectious particle output was determined by flow cytometry. The input viral cores, HIV-1 or MLV, are shown top and the input viral glycoprotein, VSV-G or MLV Env mutant, is shown left. Numbers represent the percentage of the total cells in the defined gates. 293FT cells were transfected with either MLV Env or VSV-G along with MLV proviral components expressing TdTomato and/or HIV proviral components expressing GFP. Viral media from transfected cells were transferred onto 293T mCAT-1 cells.
The CTD of MLV Env dictates viral selectivity
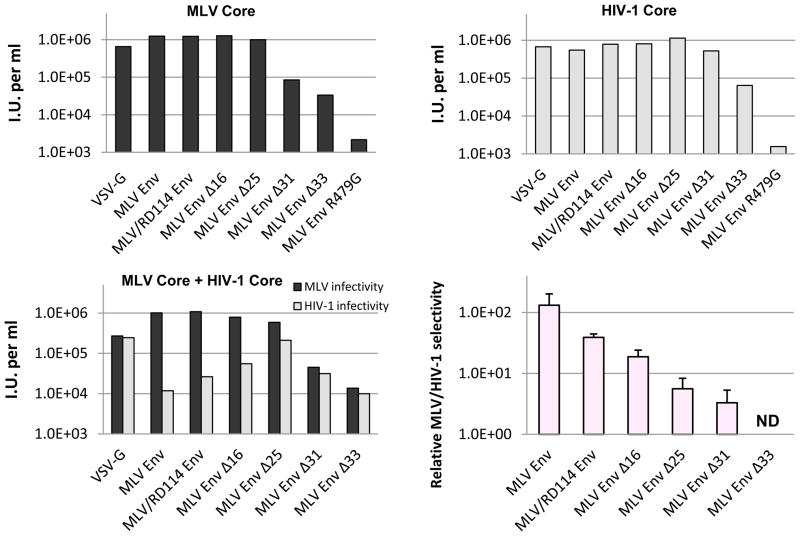
Although the MLV Env CTD clearly is not required for recruitment to HIV-1 budding sites, we questioned if the CTD contributes to the protein’s selectivity for MLV viral particles. To address this query, each of the MLV Env mutants was subjected to the competition assay described in the previous section. The results confirmed that MLV and HIV-1 cores formed infectious particles with each of the MLV Env constructs, except R479G (Fig. 4). In general, progressive truncation reduced the infectivity of MLV Env with both particle types (Fig. 4A and B). When the two types of particles were produced simultaneously in cells, MLV Env again showed a strong preference for forming infectious MLV particles. However, truncation of the MLV Env CTD progressively reduced this selectivity (Fig. 4C and D). In the complete absence of the CTD, MLV Env displayed little preference for MLV over HIV-1 cores. Collectively, these data suggest that determinants that contribute to MLV Env selectivity reside within the CTD.
FIG. 4.
Env cytoplasmic tail length affects MLV specificity and infectivity in pseudotyped particles. Infectivity assays were performed by transfecting 293FT cells and 48 h later transferring viral media onto 293T mCAT-1 cells. (A) MLV GagPol with a fluorescent MLV genome reporter or (B) an HIV-1 provirus with a GFP reporter was co-transfected with each Env construct into 293FT cells. (C) Competition assays were performed as described above, but both proviral components were co-transfected with each Env construct into each well. (D) Relative infectivity was calculated for each Env treatment by first calculating the ratio MLV IU (infectious units) per ml to HIV-1 IU per ml and then each value was normalized by the ratio of VSV Env with MLV core to HIV-1 core. A representative experimental result from one of three replicates is shown for parts A and B. Means and ±SD from all three experiments are reported in Table 1.
Discussion
Through the use of two novel assays, we have analyzed several MLV Env mutants that are similar or identical to constructs previously described. We utilized an SEM assay to determine whether the Env proteins were actively recruited to HIV-1 budding sites and an infectivity-based competition assay to determine whether selectivity between HIV-1 and MLV occurred. Using the SEM assay we observed that MLV Env is recruited to HIV-1 budding sites in a manner that is independent of the CTD. Using the competition assay we observed that MLV Env preferentially forms infectious particles with MLV cores and this selectivity is dependent on the CTD. Collectively these data suggest that at least two distinct and separable mechanisms modulate recruitment of MLV Env to retroviral budding sites. We will refer to these two mechanisms as generic recruitment (CTD independent) and specific recruitment (CTD dependent).
Generic recruitment
MLV Env is a promiscuous glycoprotein that is recruited to sites of retroviral assembly of viruses as divergent as HIV-1 and Rous sarcoma virus (RSV) (Jorgenson et al., 2009). Because the CTD of Env is not required for recruitment, the mechanism is likely not based on a direct protein-protein interaction between Env and GagPol. Although unlikely, we cannot exclude the possibility that Gag binds to a very short portion of the CTD that remains partially exposed in the cytoplasm. Instead, we favor the hypothesis that recruitment occurs indirectly. Several possible mechanisms could support indirect interaction between viral proteins, including the following: 1) a direct protein intermediate links viral proteins, 2) cellular trafficking factors independently target Gag and Env to the same cellular microenvironment, or 3) viral proteins independently co-associate with a non-protein intermediate.
The feasibility of a cellular protein intermediate that directly links Gag and Env is easy to propose and has some precedent. One such direct protein intermediate is TIP47, which has been proposed to function as a protein connector between HIV-1 Gag and the HIV-1 Env cytoplasmic tail (Lopez-Verges et al., 2006). However, because the CTD of MLV Env is not required for recruitment to budding HIV-1 particles, the relevant interaction domain of MLV Env would be restricted to the membrane spanning domain and/or the ectodomain. A direct protein intermediate would likely be complex in structure and/or function in order to mediate signaling between at least two distinct cellular domains in which viral Env and Gags reside. If this proposed intermediate were required to maintain the interaction between Gag and Env, it would be expected to be abundant in budded particles. However, extensive HIV-1 proteomic studies (Chertova et al., 2006; Saphire et al., 2006; Zhao et al., 2005) have identified no obvious cellular protein intermediate.
In an alternative model, a cellular factor separately targets MLV Env and HIV-1 Gag to the same cellular region, such as a specific site on the plasma membrane or a specific endocytic compartment, where Gag and Env co-assemble into particles. According to this model the cellular trafficking machinery would function transiently as an intermediate facilitator and thus might not be present during the budding process. This mechanism fails to readily explain why some viral glycoproteins can form pseudotypes with a wide variety of viruses, but others can only form pseudotypes with a subset of viruses.
Finally, in a third model, a non-protein intermediate fosters an environment favorable for pseudotyping. Multiple studies have suggested that retroviral assembly occurs at lipid rafts on the plasma membrane and that Env proteins are independently attracted to these sites (Briggs et al., 2003; Leung et al., 2008; Pickl et al., 2001; Waheed and Freed, 2009). Based on this hypothesis, a lipid microenvironment essentially serves as an intermediate during viral assembly. However, our previous and current data do not support a simple raft model. The raft hypothesis proposes that during assembly Gag targets Env proteins that are stably present in pre-existing clusters at the plasma membrane. However, we have previously shown that wildtype MLV Env protein is essentially random in its plasma membrane distribution in the absence of Gag (Jorgenson et al., 2009). In addition, the viral glycoprotein VSV-G is considered to be a non-raft protein (Benting et al., 1999; Scheiffele et al., 1999) and yet it efficiently pseudotypes with most viruses and is recruited to HIV-1 budding sites (Jorgenson et al., 2009).
Our data are consistent with a model where Gag assembly creates a unique lipid environment at the assembly site and that this environment is favored by certain viral glycoproteins. This unique lipid region is perhaps similar to what is defined as a lipid raft or a liquid ordered domain. If some glycoproteins, such as VSV-G or MLV Env, have a higher affinity for a specific microenvironment than other viral glycoproteins, it could explain why promiscuous glycoproteins pseudotype efficiently with many different types of virus.
This model does not exclude the possibility that additional factors either enforce (see next section) or inhibit Env incorporation. For instance, non-lentiviral retroviruses can be pseudotyped only with HIV-1 Env that lacks the cytoplasmic tail (Freed and Martin, 1996; Mammano et al., 1995; Wilk et al., 1992). This restriction could be explained if the HIV-1 Env is recruited to budding sites in a manner similar to that for MLV Env, but is excluded from incorporation because of steric hindrance from its long CTD. In addition, some cellular proteins may have an affinity for retroviral assembly sites but fail to be incorporated into particles due to interactions with other cellular proteins. For instance, this may explain why the protein human CD4 is efficiently incorporated into RSV particles when expressed in quail cells (Young et al., 1990).
Specific recruitment
In the presence of both MLV and HIV-1 cores, competition studies demonstrated a preferential production of MLV cores with wildtype MLV Env that supersedes the ability of HIV-1 to productively pseudotype with wildtype MLV Env. Although not known, we speculate that MLV cores may sequester available MLV Env from HIV-1 cores. However, in the absence of the MLV Env CTD, infectious particles of MLV and HIV-1 were produced in equal proportion to one another. As a result, we propose that the CTD of MLV Env facilitates selective incorporation into MLV particles.
A reasonable model for specific CTD-dependent recruitment would support a direct interaction between the MLV structural protein Gag and the CTD of Env. Substantial evidence from other retroviruses suggests that the MA domain of Gag directly interacts with the cytoplasmic tail of native Env glycoproteins. In one such example, mutations in HIV-1 Env that block incorporation into HIV-1 particles can be circumvented by compensatory mutations in HIV-1 MA (Murakami and Freed, 2000). The R peptide of MLV Env has been shown to predominantly associate with immature particles, suggesting an R peptide-facilitated interaction between Env and the assembling viral particle (Andersen et al., 2006). Furthermore, wildtype HIV-1 Env becomes fully fusogenic only after maturation of HIV-1 Gag, suggesting a direct interaction between HIV-1 Gag and Env (Jiang and Aiken, 2006; Wyma et al., 2004). Although a direct interaction between MA and Env is the most logical explanation for specific recruitment, other contributing mechanisms cannot be excluded.
Materials and Methods
Plasmid constructs
The ecotropic Friend MLV Env, yellow fluorescent protein (YFP)-tagged MLV Env, and MLV GagPol constructs were kindly provided by Walther Mothes (Yale University) (Sherer et al., 2003). The codon-optimized late domain deficient HIV-1 Gag construct used in SEM images has been described previously (Jorgenson et al., 2009). Truncations of MLV Env were generated by oligonucleotide linker insertion between the ClaI site (33 amino acids upstream of the stop codon) and the EcoRI site (located just downstream of the stop codon). The R479G mutation was introduced by two-step PCR. The MLV reporter construct pQCXIP-TdTomato was described previously (Lucas et al., 2010). The HIV-1 packaging construct CMV ΔR8.2 was obtained from Addgene (plasmid #12263) (Zufferey et al., 1997). The reporter construct pSIN18.cPPT.hEF1a.EGFP.WPRE was designed by Michal Gropp and Benjamin Reubinoff (Gropp et al., 2003).
Cells
The 293FT cell line was obtained from Invitrogen. The 293T mCAT-1 cell line stably expresses the target cellular receptor for ecotropic MLV Env, murine cationic transporter-1, and was obtained, kindly, from Walther Mothes. Cell lines were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM non-essential amino acids, and 0.5 mg/ml G418.
Infectivity assay
Infectivity competition studies were performed by transfecting 293FT cells in 6 well plates with FuGene 6 (Roche). Each MLV Env construct (200 ng) was co-transfected with HIV CMV ΔR8.2 (150 ng), HIV reporter construct pSIN18.cPPT.hEF1a.EGFP.WPRE (150ng), MLV GagPol expression construct (250ng) and the MLV reporter construct pQCXIP-TdTomato (250ng). In experiments where the viruses were expressed individually, the DNA for the second expression construct and reporter was replaced with filler DNA. The media was replaced 24 hours post-transfection to remove residual transfection reagent and viral media was collected ca. 48 h posttransfection, frozen at −80°C for at l east 4 h, thawed, and 1 ml of media was transferred onto 293T mCAT-1 cells with 10 ug/ml polybrene (Invitrogen). Cells were collected 48 hours later, fixed with 4% paraformaldehyde and infectivity was measured as infectious units per ml as determined by flow cytometry with an Accuri flow cytometer.
Surface labeling
For each construct, 293FT cells were transfected with 1000 ng of each YFP-tagged Env. Media was changed 24 h posttransfection to remove residual transfection reagent. At 48 hours posttransfection, cells were collected, labeled with an anti-GFP Alexa Fluor-647 antibody with 1% goat serum for 1 h at 4°C (1:1000, Invitrogen), fixed with 4% paraformaldehyde and analyzed by FACS. Surface expression was calculated as mean channel FL-2 fluorescence to mean channel GFP fluorescence for each construct relative to wildtype MLV Env expression levels.
Syncytia assay
In order to determine fusogenic activity of the MLV Env constructs, 293T mCAT-1 cells were co-transfected with each of the 1000 ng MLV Env constructs along with 20 ng of plasmid expressing mCherry. The mCherry allowed the transfection to be detected by fluorescence microscopy. Cells were fixed ca. 24 hours posttransfection and were treated with Hoechst stain to identify cell nuclei. Cells were imaged via fluorescent microscopy and the number of cells and the number of nuclei were counted per view-field. Syncytia were quantified with the cell fusion index formula as defined by [1-(number of cells/number of nuclei)].
SEM
The distribution of MLV Env and virions on the cell surface was imaged via SEM, as previously described (Jorgenson et al., 2009). Briefly, cells were plated onto coverslips coated with a patterned gold grid and were transfected with a late domain defective HIV-1 Gag expression vector (500 ng) and a YFP-tagged Env expression vector (500 ng) with FuGene 6 according to manufacture’s protocol (Roche). At 20 hours posttransfection, plated cells were fixed with 4% paraformaldehyde and the grid locations of individual transfected cells were recorded. Cells were labeled with primary mouse anti-GFP (1:25, Sigma) and 10- to 12-nm gold conjugated anti-mouse secondary antibody (1:20, Jackson ImmunoResearch). Subsequently, cells were fixed with 2.5% glutaraldehyde, dehydrated in ethanol, critical point dried, coated with carbon, and imaged with a Hitach S-4700 FE-SEM.
Acknowledgments
We thank Walther Mothes and Vineet KewalRemani for reagents. SEM studies were performed at the University of Missouri Electron Microscopy Core facility. This research was supported by U.S. Public Health Service grant AI73098 and the Arnold and Mabel Beckman Foundation Young Investigator Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar HC, Anderson WF, Cannon PM. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R Peptide. Journal of Virology. 2003;77(2):1281–91. doi: 10.1128/JVI.77.2.1281-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KB, Diep HA, Zedeler A. Murine leukemia virus transmembrane protein Rpeptide is found in small virus core-like complexes in cells. Journal of General Virology. 2006;87(Pt 6):1583–8. doi: 10.1099/vir.0.81527-0. [DOI] [PubMed] [Google Scholar]
- Benting J, Rietveld A, Ansorge I, Simons K. Acyl and alkyl chain length of GPI-anchors is critical for raft association in vitro. FEBS Lett. 1999;462(1–2):47–50. doi: 10.1016/s0014-5793(99)01501-x. [DOI] [PubMed] [Google Scholar]
- Blot V, Lopez-Verges S, Breton M, Pique C, Berlioz-Torrent C, Grange MP. The conserved dileucine- and tyrosine-based motifs in MLV and MPMV envelope glycoproteins are both important to regulate a common Env intracellular trafficking. Retrovirology. 2006;3:62. doi: 10.1186/1742-4690-3-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkova M, Stitz J, Engelstadter M, Cichutek K, Buchholz CJ. Identification of Rpeptides in envelope proteins of C-type retroviruses. Journal of General Virology. 2002;83(Pt 9):2241– 6. doi: 10.1099/0022-1317-83-9-2241. [DOI] [PubMed] [Google Scholar]
- Bouard D, Sandrin V, Boson B, Negre D, Thomas G, Granier C, Cosset FL. An acidic cluster of the cytoplasmic tail of the RD114 virus glycoprotein controls assembly of retroviral envelopes. Traffic. 2007;8(7):835–47. doi: 10.1111/j.1600-0854.2007.00581.x. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Wilk T, Fuller SD. Do lipid rafts mediate virus assembly and pseudotyping? J Gen Virol. 2003;84(Pt 4):757–68. doi: 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. Journal of Virology. 2006;80(18):9039–52. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulopoulos I, Cannon PM. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J Virol. 2001;75(9):4129–38. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Dubay JW, Perez LG, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubay JW, Dubay SR, Shin HJ, Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69(8):4675–82. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70(1):341–51. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61(9):2852–6. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman LJ, Kain SR, Firestone GL. Trafficking of wild-type and an endoproteolyticsite mutant of the mouse mammary tumor virus glycoprotein. J Biol Chem. 1993;268(4):2329–36. [PubMed] [Google Scholar]
- Green N, Shinnick TM, Witte O, Ponticelli A, Sutcliffe JG, Lerner RA. Sequencespecific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci U S A. 1981;78(10):6023–7. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp M, Itsykson P, Singer O, Ben-Hur T, Reinhartz E, Galun E, Reubinoff BE. Stable genetic modification of human embryonic stem cells by lentiviral vectors. Mol Ther. 2003;7(2):281–7. doi: 10.1016/s1525-0016(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Guo HG, Veronese FM, Tschachler E, Pal R, Kalyanaraman VS, Gallo RC, Reitz MS., Jr Characterization of an HIV-1 point mutant blocked in envelope glycoprotein cleavage. Virology. 1990;174(1):217–24. doi: 10.1016/0042-6822(90)90070-8. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Sowder R, Copeland TD, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52(2):492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszeski MM, Cannon PM, Chen D, Rozenberg Y, Anderson WF. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71(5):3613–9. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Aiken C. Maturation of the viral core enhances the fusion of HIV-1 particles with primary human T cells and monocyte-derived macrophages. Virology. 2006;346(2):460–8. doi: 10.1016/j.virol.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Jorgenson RL, Vogt VM, Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol. 2009;83(9):4060–7. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369(2):245–62. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Amanuma H. Mutational analysis of the R peptide cleavage site of Moloney murine leukaemia virus envelope protein. J Gen Virol. 2003;84(Pt 8):2253–7. doi: 10.1099/vir.0.19126-0. [DOI] [PubMed] [Google Scholar]
- Kubo Y, Tominaga C, Yoshii H, Kamiyama H, Mitani C, Amanuma H, Yamamoto N. Characterization of R peptide of murine leukemia virus envelope glycoproteins in syncytium formation and entry. Arch Virol. 2007;152(12):2169–82. doi: 10.1007/s00705-007-1054-6. [DOI] [PubMed] [Google Scholar]
- Leung K, Kim JO, Ganesh L, Kabat J, Schwartz O, Nabel GJ. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host & Microbe. 2008;3(5):285–92. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge R, Delamarre L, Lalonde JP, Alvarado J, Sanders DA, Dokhelar MC, Cohen EA, Lemay G. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J Virol. 1997;71(7):5696–702. doi: 10.1128/jvi.71.7.5696-5702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Verges S, Camus G, Blot G, Beauvoir R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving R, Li K, Wallin M, Sjoberg M, Garoff H. R-Peptide cleavage potentiates fusioncontrolling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. Journal of Virology. 2008;82(5):2594–7. doi: 10.1128/JVI.02039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas TM, Lyddon TD, Cannon PM, Johnson MC. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol. 2010;84(6):2666–74. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida CA, Kabat D. Role of partial proteolysis in processing murine leukemia virus membrane envelope glycoproteins to the cell surface. A viral mutant with uncleaved glycoprotein. J Biol Chem. 1982;257(23):14018–22. [PubMed] [Google Scholar]
- Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83(3):1523–31. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger HG. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69(6):3824–30. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53(1):55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Moulard M, Hallenberger S, Garten W, Klenk HD. Processing and routage of HIV glycoproteins by furin to the cell surface. Virus Res. 1999;60(1):55–65. doi: 10.1016/s0168-1702(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74(8):3548–54. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickl WF, Pimentel-Muinos FX, Seed B. Lipid rafts and pseudotyping. J Virol. 2001;75(15):7175–83. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragheb JA, Anderson WF. pH-independent murine leukemia virus ecotropic envelopemediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68(5):3220–31. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. Journal of Virology. 1994;68(3):1773–81. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg-Adler Y, Conner J, Aguilar-Carreno H, Chakraborti S, Dimitrov DS, Anderson WF. Membrane-proximal cytoplasmic domain of Moloney murine leukemia virus envelope tail facilitates fusion. Experimental & Molecular Pathology. 2008;84(1):18–30. doi: 10.1016/j.yexmp.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Sandrin V, Bouard D, Boson B, Negre D, Thomas G, Granier C, Cosset F-L. Interaction of a new sorting motif and its cellular effector controls retroviral envelope assembly. Retroviruses abstract book 2006 [Google Scholar]
- Sandrin V, Cosset FL. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. Journal of Biological Chemistry. 2006;281(1):528–42. doi: 10.1074/jbc.M506070200. [DOI] [PubMed] [Google Scholar]
- Sandrin V, Muriaux D, Darlix JL, Cosset FL. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J Virol. 2004;78(13):7153–64. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire AC, Gallay PA, Bark SJ. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J Proteome Res. 2006;5(3):530–8. doi: 10.1021/pr050276b. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274(4):2038–44. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- Schnierle BS, Stitz J, Bosch V, Nocken F, Merget-Millitzer H, Engelstadter M, Kurth R, Groner B, Cichutek K. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(16):8640–5. doi: 10.1073/pnas.94.16.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4(11):785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- Sjoberg M, Wallin M, Lindqvist B, Garoff H. Furin cleavage potentiates the membrane fusion-controlling intersubunit disulfide bond isomerization activity of leukemia virus Env. J Virol. 2006;80(11):5540–51. doi: 10.1128/JVI.01851-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitz J, Buchholz CJ, Engelstadter M, Uckert W, Bloemer U, Schmitt I, Cichutek K. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology. 2000;273(1):16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- Taylor GM, Sanders DA. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology. 2003;312(2):295–305. doi: 10.1016/s0042-6822(03)00297-6. [DOI] [PubMed] [Google Scholar]
- Thomas A, Gray KD, Roth MJ. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology. 1997;227(2):305–13. doi: 10.1006/viro.1996.8333. [DOI] [PubMed] [Google Scholar]
- Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143(2):162–76. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189(1):167–77. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. Journal of Virology. 2004;78(7):3429–35. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Compans RW. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71(11):8490–6. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79(19):12132–47. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Bates P, Willert K, Varmus HE. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science. 1990;250(4986):1421–3. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- Zavorotinskaya T, Albritton LM. Failure To cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. Journal of Virology. 1999;73(7):5621–9. doi: 10.1128/jvi.73.7.5621-5629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Azam S, Thorpe R. Comparative studies on cellular gene regulation by HIV-1 based vectors: implications for quality control of vector production. Gene Ther. 2005;12(4):311–9. doi: 10.1038/sj.gt.3302414. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nature Biotechnology. 1997;15(9):871–5. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]