Abstract
The astrocytic glutamate transporter GLAST (also known as EAAT1) is a key regulator of extracellular glutamate levels in many regions of vertebrate brains. To identify novel interacting partners that might regulate the localization and function of GLAST in astrocytes, we screened the transporter’s C-terminus (GLAST-CT) against a proteomic array of 96 different PDZ domains. The GLAST-CT robustly and specifically interacted with PDZ domains from two related scaffolding proteins, the Na+/H+ exchanger regulatory factors 1 and 2 (NHERF-1 and NHERF-2). Studies on cultured rat cortical astrocytes revealed that these cells are highly enriched in NHERF-2 relative to NHERF-1. Endogenous GLAST and NHERF-2 from cultured astrocytes were found to robustly co-immunoprecipitate, and further co-immunoprecipitation studies on mutant versions of GLAST expressed in transfected cells revealed the GLAST/NHERF-2 interaction to be dependent on the last amino acid of the GLAST-CT. Knockdown of endogenous NHERF-2 in astrocytes via siRNA treatment resulted in a significant reduction in GLAST activity, which corresponded to significantly reduced total expression of GLAST protein and reduced half-life of GLAST, as assessed in pulse-chase metabolic labeling studies. These findings reveal that NHERF-2 can interact with GLAST in astrocytes to enhance GLAST stability and activity.
Glutamate is the most abundant neurotransmitter in the mammalian central nervous system and the mediator of excitatory neurotransmission at the majority of synapses in the brain. The extracellular concentration of glutamate must be tightly regulated, however, as excessive glutamate signaling can lead to excitotoxic cellular death[4]. Five transporters have been identified as the principal regulators of extracellular glutamate levels and therefore have been named excitatory amino acid transporters EAAT-1 (rat homologue known as GLAST), EAAT-2 (rat homologue known as GLT-1), EAAT-3, EAAT-4, and EAAT-5. Genetic studies have shed light on the functions of the individual transporter sub-types, revealing a dominant role for GLAST and GLT-1 in controlling synaptic glutamate levels[16, 21]. Interestingly, GLAST and GLT-1 are exclusively expressed in astrocytes[3, 12], thereby speaking to the importance of astrocytes in clearing synaptic glutamate. Moreover, mutations in GLAST are linked with a variety of disease states, including epilepsy, stroke, Alzheimer’s Disease, Parkinson’s Disease, and schizophrenia[2], further highlighting the importance of understanding GLAST function.
We set out to identify novel interacting partners that might regulate the selective localization and function of GLAST in astrocytes. Since the C-terminus (CT) of GLAST possesses a consensus motif for association with PDZ domains, a family of protein-protein interaction domains named after the first 3 proteins in which they were identified (post-synaptic density protein of 95 kDa, discs-large, and zona-occludens 1), we screened the GLAST-CT against a proteomic array of PDZ domains in order to identify novel GLAST interacting partners.
Overlays of the PDZ domain array were performed as previously described[6, 8]. Briefly, 1 µg of His- and S-tagged PDZ domain fusion proteins were spotted onto nitrocellulose, dried overnight, and overlaid with GST-alone (control) or GST-GLASTCT. Membranes were washed and incubated with an HRP-coupled anti-GST monoclonal antibody (Amersham Pharmacia Biotech) and binding was visualized using enhanced chemiluminescence.
HEK-293 (ATCC) cells were cultured in Dulbecco’s modified Eagle medium containing GlutaMAX™, 10% fetal bovine serum (Atlanta Biologicals) and 1% penicillin/streptomycin, and were maintained at 37°C in an atmosphere of 95% air/5% CO2. Transfections were performed with Lipofectamine 2000, as previously described[10]. DNA constructs used were rat GLAST in pcDNA3 (kindly provided by Jeff Rothstein, Johns Hopkins), human FLAG-GLAST (kindly provided by Susan Amara, Univ. Pittsburgh), and rabbit HA-NHERF-2 in pBK-CMV.
Purified primary astrocyte cultures were prepared by the method of McCarthy and de Vellis[14]. All animal work was performed under the guidance of the National Institute of Health Guide for the Care and Use of Laboratory Animals. Briefly, neocortices were dissected from E18–19 Sprague Dawley rat embryos and dissociated in medium by trituration. The cells were re-suspended in GlutaMAX™ DMEM (10% FBS and 1% Pen/Strep) on poly-D-lysine coated tissue culture flasks. The medium was changed the next day and then every 48 hours. After 6–8 days, cells were shaken overnight (280–310 rpm) to remove microglia and oligodendrocytes and passaged 48 hours later. Immunostaining with the astrocytic marker GFAP revealed that this culturing method results in at least 90% purified GFAP-positive astrocyte cultures (data not shown).
Overlay assays, immunoprecipitation experiments, and Western blotting were performed as previously described[10]. For overlay experiments, 1 µg of purified protein was separated by SDS-PAGE gel electrophoresis, transferred onto nitrocellulose, and overlaid with purified His/S-tagged NHERF-2-PDZ domain 2 fusion protein (prepared using a pET30A construct and corresponding to amino acids 149–337 of human NHERF- 2) in increasing concentrations. Binding of His/S-tagged NHERF-2 to GLAST was visualized with S-protein HRP and quantified via densitometry. Numbers were expressed as percentage of maximum binding to generate binding curves for affinity estimates.
Immunoprecipitation experiments were performed using anti-FLAG M2 affinity gel (Sigma), or NHERF-2 antibody bound to Protein A/G agarose beads (Pierce). Astrocyte lysates were also incubated with purified NHERF-2 or GST-alone bound to beads for pull-downs. For all experiments, cells were lysed with a buffer containing 50mM NaCl, 20mM Hepes, 5mM EDTA, 1 protease inhibitor cocktail tablet (Roche Diagnostics), 1% Triton X-100, pH 7.4. To detect endogenous and/or recombinant NHERF-1, NHERF-2, and GLAST, the reagents used were S-protein HRP conjugate (1:4,000, Novagen), polyclonal guinea pig anti-GLAST (1:10,000, Chemicon), rabbit polyclonal anti-GLAST (H-50, Santa Cruz), rabbit anti-NHERF-1 (1:7,000) and rabbit anti-NHERF-2 (1:5000)[24].
Mutagenesis of the last amino acid of the GLAST (Amara construct) from a methionine to an alanine (M542A) was performed using the QuikChange Site-directed mutagenesis kit (Stratagene). Primers used for PCR were 5’- GACAGCGAAACCAAGGCGTAG-3’ and its reverse complement. Sequencing was performed to confirm mutation of the target bases. Knockdown of NHERF-2 was accomplished using TransIT-siQuest transfection reagent (Mirus). NHERF-2 siRNA was Silencer pre-designed siRNA (ID# 55838; Ambion). Control siRNA was Silencer Negative Control #1 siRNA (Ambion).
For the amino acid uptake experiments, astrocytes were incubated at 37°C in assay buffer containing 1 mM of the GLT-1-selective inhibitor dihydrokainate (DHK) (Tocris), followed by increasing concentrations of [3H]aspartate for 6 minutes in a 37°C water bath. Uptake was stopped with three washes of ice-cold buffer containing choline chloride in replacement of sodium chloride. Cells were lysed with 0.5 M NaOH and samples were counted in 30% Scintisafe (Fisher).
Pulse-chase assays were performed as previously described[1]. Briefly, primary astrocytes that had been treated with either control siRNA or NHERF-2 siRNA were incubated in methionine-free DMEM for thirty minutes, then 60 µCi of Redivue L-[35S]- methionine (Amersham Biosciences) was added to each plate to incubate at 37 °C for an additional 30 minutes. Cells were washed and immediately chased with 3 mM cold L-methionine for various time points (0, 2, 4, 8, 12, 24, 48, and 72 hrs). Samples were adjusted to normalize for protein concentration, GLAST was immunoprecipitated using guinea pig anti-GLAST antibody, and the amount of 35S incorporated into the GLAST protein was determined.
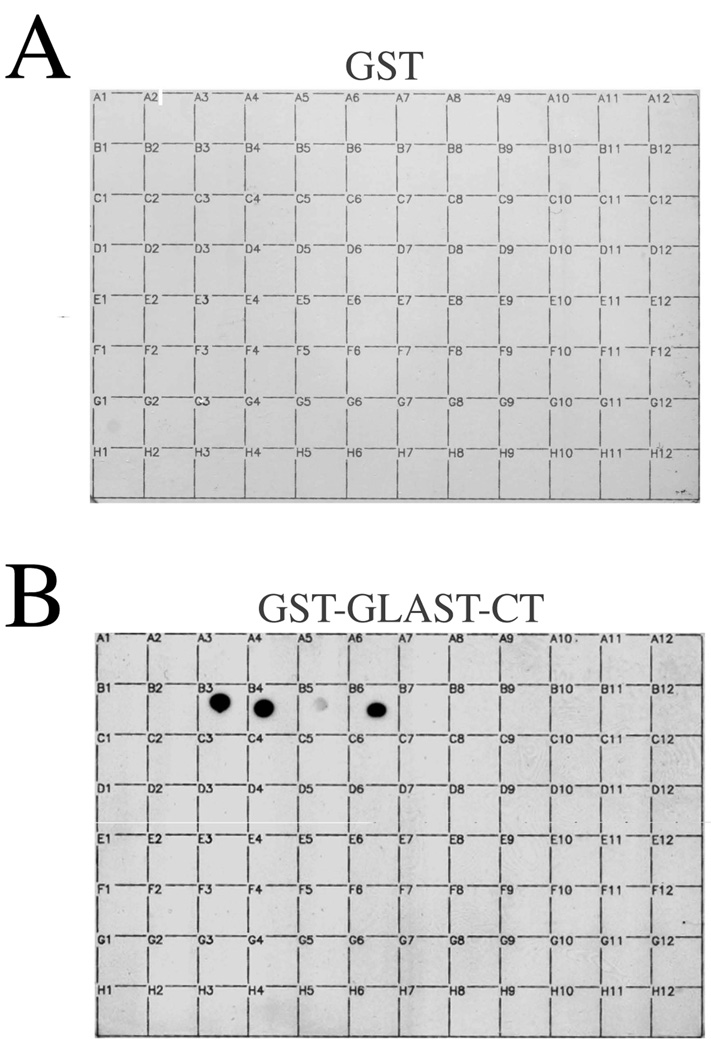
In order to gain a panoramic view of potential PDZ interactions for GLAST, we screened a GST fusion protein of the GLAST-CT against a previously-described PDZ proteomic array containing 96 distinct PDZ domains (Fig. 1). The GLAST-CT did not detectably bind to the vast majority of PDZ domains on the array. However, strong binding of the GLAST-CT was observed to both PDZ domains of the Na+/H+ exchanger regulatory factor (NHERF-1), as well as to PDZ domain 2 of the related protein NHERF-2 (Fig. 1). Weaker binding was also observed to the first PDZ domain of NHERF-2. The interaction of GLAST with NHERF-1 has been reported previously[11], but the association of GLAST with NHERF-2 is novel. Thus, we characterized the GLAST/NHERF-2 interaction in further detail.
Figure 1.
The C-terminus of GLAST binds selectively to NHERF PDZ domains. Control GST (A) or a GST-fusion protein comprising the last 25 amino acids of GLAST (B) were overlaid at 100 nM onto a proteomic array containing 96 distinct PDZ domains (n = 3). A complete list of the PDZ proteins on this array has been described previously by He et al.[8].
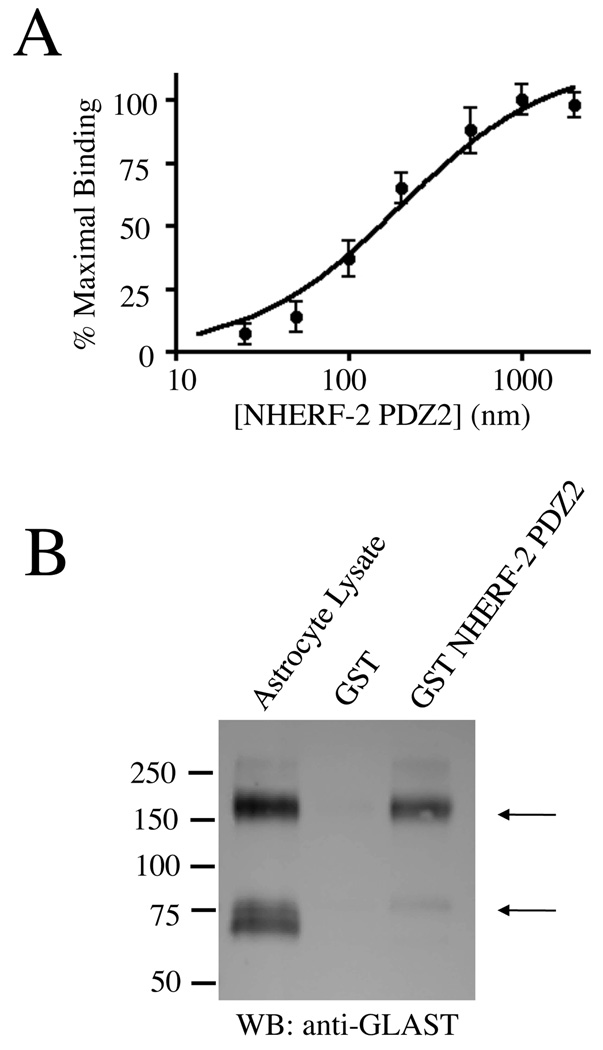
Saturation binding assays overlaying His-tagged NHERF-2 PDZ2 onto purified GLAST-CT-GST revealed the KD of the GLAST/NHERF-2 PDZ2 interaction to be 196 nM (Fig. 2A). This affinity is in the range of other PDZ domain-mediated associations that are known to be physiologically relevant[18]. Furthermore, full-length GLAST from astrocyte lysates was robustly pulled down by GST-NHERF-2 (Fig. 2B). NHERF-2 seemed to preferentially interact with oligomers of GLAST in these experiments, but it should be noted that it is unclear whether oligomers of GLAST observed in SDS-PAGE gels correspond to oligomers of GLAST in living cells.
Figure 2.
In vitro association between GLAST and PDZ2 of NHERF-2. A) Purified GST-GLAST-CT was transferred to nitrocellulose and overlaid with increasing concentrations of purified His/S-tagged NHERF-2-PDZ2. Bound NHERF-2 was visualized using S-protein HRP and quantified via densitometry (n = 4). B) Full-length endogenous GLAST from astrocyte lysates was pulled down with purified GST-NHERF-2 (lane 3; arrows indicate GLAST monomers and oligomers) but not GST alone (lane 2; n = 5).
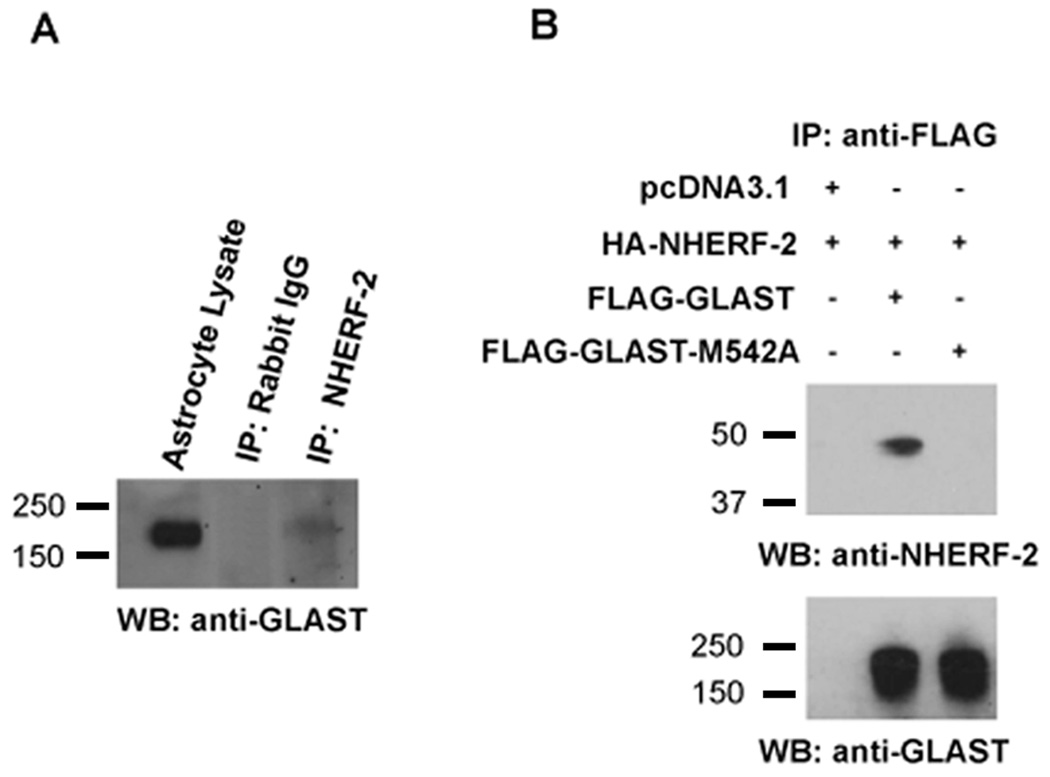
In order to assess whether GLAST and NHERF-2 might associate in a cellular context, thereby validating the initial screen, co-immunoprecipitation experiments were performed. Immunoprecipitation of endogenous NHERF-2 from cultured astrocyte lysates resulted in a robust co-immunoprecipitation of endogenous oligomers of GLAST (Fig. 3A). We then examined whether GLAST/NHERF-2 cellular association was dependent on the PDZ-binding motif by mutating the last amino acid of GLAST. Immunoprecipitation experiments revealed that mutant GLAST-M542A was unable to bind to NHERF-2 (Fig. 3B) or NHERF-1 (data not shown).
Figure 3.
Association of NHERF-2 with GLAST in primary astrocytes and transfected cells via the GLAST C-terminus. A) Immunoprecipitation of endogenous NHERF-2 from cultured primary rat astrocyte lysates resulted in the specific co-immunoprecipitation of GLAST, as detected with guinea pig anti-GLAST antibody (lane 3; n = 6). No GLAST co-immunoprecipitation was observed with an irrelevant rabbit antibody bound to A/G beads (lane 2). B) Mutation of the last amino acid in the GLASTCT (methionine 542 to an alanine) disrupted cellular association of GLAST with NHERF-2 in HEK-293 cells transfected with HA-NHERF-2 and either wild-type FLAGGLAST or FLAG-GLAST-M542A mutant. The amount of co-immunoprecipitated NHERF-2 is shown in the top panel, while equal amounts of immunoprecipitated FLAG-GLAST and FLAG-GLAST-M542A, detected with rabbit anti-GLAST antibody, are shown in the lower panel (oligomer bands only shown; n = 3–5).
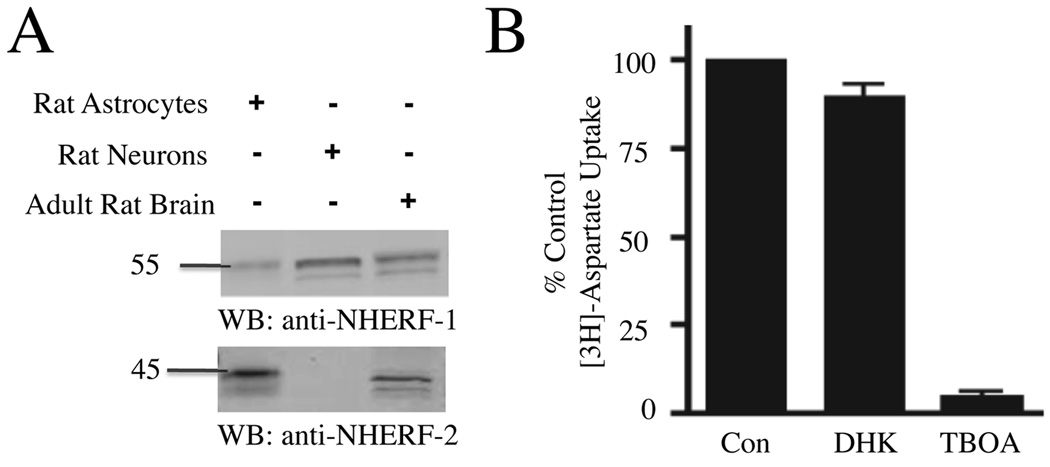
We next examined the effects of GLAST/NHERF interactions on GLAST function in cultured astrocytes. Western blot studies revealed that our cultured astrocytes were highly enriched in NHERF-2 relative to total brain tissue, but in contrast expressed levels of NHERF-1 much lower than total brain tissue (Fig. 4A). We then examined which glutamate transporter was primarily responsible for aspartate uptake by these cells. Treatment of astrocytes with TBOA (100 µM), a non-selective glutamate transporter inhibitor, resulted in nearly complete inhibition of [3H]-aspartate transport, while treatment with DHK (1mM), a GLT-1 selective blocker, only modestly prevented [3H]- aspartate transport (Fig. 4B). Consistent with previous observations[20], these studies revealed that GLAST was the predominant functional glutamate transporter in our cultured astrocytes, with only minor contributions resulting from GLT-1 and potentially other transporters (such as EAAC1/EAAT3).
Figure 4.
Cultured astrocytes are enriched in NHERF-2 and express functional GLAST. A) Equal protein levels of lysates from cultured rat astrocytes, cultured rat neurons, and total adult rat brain were analyzed by Western blot, revealing that the cultured astrocytes were enriched in NHERF-2 (lane 1 versus lane 3), while the cultured neurons preferentially expressed NHERF-1, relative to total brain expression (lane 2 versus lane 3; n = 4). B) The uptake of [3H]-aspartate was measured in cultured astrocytes in the absence and presence of the non-selective glutamate transporter inhibitor TBOA (100 µM) and the GLT-1-selective blocker DHK (1mM; n = 3).
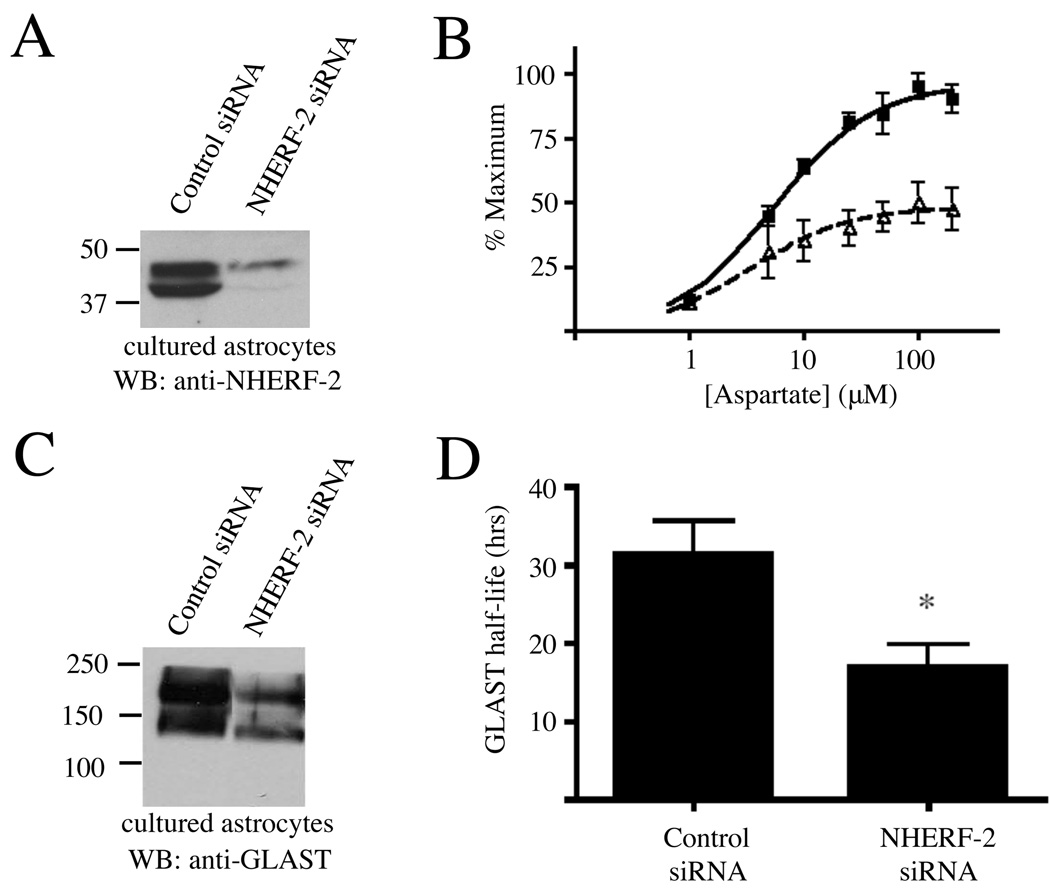
Given the high expression of GLAST and NHERF-2 in our cultured astrocytes, we then examined if knockdown of NHERF-2 might alter GLAST function. Treatment with siRNA resulted in a >90% knockdown of NHERF-2 in the cultured astrocytes (Fig. 5A). Strikingly, this reduction in NHERF-2 expression resulted in approximately a 50% decrease in the maximum total uptake of [3H]-aspartate by the NHERF-2 siRNA-treated astrocytes relative to control siRNA-treated astrocytes (Fig. 5B; paired t-test, p < 0.001).
Figure 5.
Knockdown of endogenous NHERF-2 in astrocytes reduces GLAST activity and protein levels. A) Treatment of astrocyte cultures with NHERF-2 siRNA (lane 2) versus scrambled control siRNA (lane 1) resulted in the specific knockdown of endogenous NHERF-2 (n = 4). B) GLAST activity was defined in cultured astrocytes by measuring uptake of different concentrations of [3H]-aspartate in the presence of the GLT-1 inhibitor DHK (1mM). Data were normalized within each experiment and expressed as a percentage of the maximal uptake observed for either condition. Cultured astrocytes treated with NHERF-2 siRNA (open triangles, dashed line, Vmax = 6.4 ± 0.7 nmol/mg/min) transported nearly 50% less maximal [3H] aspartate in comparison to control siRNA-treated astrocytes (black squares, solid line, Vmax = 12.5 ± 1.0 nmol/mg/min) containing normal endogenous levels of NHERF-2 (paired t-test, p < 0.001; n = 4). C) Cultured astrocytes were treated with control or NHERF-2 siRNA and subsequently harvested to check for GLAST expression via Western blot using the guinea pig anti-GLAST antibody (n = 4). D) Pulse-chase metabolic labeling studies revealed that knockdown of NHERF-2 in cultured primary astrocytes resulted in decreased half-life of the GLAST protein (n = 3; *, p < 0.05).
Since the functional effects of NHERF-2 knockdown in cultured astrocytes were on the maximal activity of GLAST (not apparent affinity for substrate), we assessed GLAST expression levels in the absence and presence of NHERF-2 interactions. Western blots revealed that siRNA knockdown of NHERF-2 in cultured astrocytes resulted in significantly reduced expression of GLAST (Fig. 5C). Pulse-chase metabolic labeling studies revealed that the half-life of the GLAST protein was reduced by 46% in NHERF-2 siRNA-treated astrocytes relative to control siRNA-treated astrocytes (Fig. 5D). Taken together, these studies indicate that NHERF-2 interactions with GLAST are important for GLAST stability and expression.
The studies described here have identified NHERF-2 as a novel interacting partner for the astrocytic glutamate transporter GLAST. Previously, the related protein NHERF-1 was identified as a GLAST binding partner by Lee et al.[11]. Our findings expand on this earlier report in several important ways. First, Lee et al. examined only the specific interaction of GLAST and NHERF-1, but did not screen for potential GLAST interactions with other PDZ scaffolds. In our studies, we screened the GLAST-CT for potential binding to 96 distinct PDZ domains and established a high specificity of binding to the PDZ domains from NHERF-1 and NHERF-2.
Secondly, Lee et al. discounted the possibility that GLAST might interact with NHERF-2 on the basis of light microscopy studies showing a high concentration of NHERF-2 labeling around CNS blood vessels, which they interpreted to indicate NHERF-2 expression in endothelial cells but not neurons or astrocytes[11]. However, in electron microscopy studies that were published contemporaneously with the studies by Lee et al., we showed that NHERF-2 expression in the brain is actually highest in astrocytes[15]. The perivascular astrocytes surrounding blood vessels express particularly high levels of NHERF-2[15], which suggests an explanation for the observations made at the light microscopy level by Lee et al. Since there is substantial functional redundancy between the NHERF proteins[23], our finding that GLAST interacts with both of the NHERF proteins reveals that it will be important to study double NHERF-1/NHERF-2 knockout mice in order to fully assess the effects of PDZ scaffold regulation of GLAST function in vivo.
A third way that our studies expand on the earlier findings by Lee et al. is in providing evidence that GLAST/NHERF interactions have effects on GLAST functional activity in astrocytes. The work by Lee et al. solidly established the GLAST/NHERF-1 interaction[11], and a follow-up study revealed that knockdown of NHERF-1 expression in COS-7 cells resulted in reduced activity of transfected GLAST[19], but this work did not address the physiological importance of endogenous GLAST/NHERF associations in astrocytes. Our studies reported here demonstrate that disruption of the GLAST/NHERF-2 interaction in primary astrocytes results in decreased GLAST activity due to impaired stability of the GLAST polypeptide. These findings are analogous to the effects of PDZ scaffold interactions with certain other neurotransmitter transporters. For example, the associations of EAAT4 with GTRAP48[9], the dopamine transporter DAT with PICK1[22], and the GLT-1 splice variant GLT-1b with PSD-95[7] result in enhanced surface expression and activity of the transporters. The detailed molecular mechanism(s) by which PDZ scaffold interactions can enhance transporter stability and function are not presently understood, but will be an interesting area for future investigation.
In summary, we have found a novel interaction between GLAST and NHERF-2 that modulates GLAST functional activity in astrocytes. GLAST and other glutamate transporters are considered to be prime targets for the development of future therapeutics aimed at reducing neurodegeneration in stroke and other pathological conditions[2, 17]. Thus, scaffolds that modulate glutamate transporter function may also be excellent targets for new therapeutics. Given the short peptide motifs with which PDZ domains interact, these domains are attractive candidates for therapeutic intervention[5], and in fact small peptidomimetic molecules that bind to and specifically regulate the NHERF PDZ domains are already being developed[13]. Future work on GLAST/NHERF interactions may shed light on the mechanisms controlling GLAST function in vivo and also provide opportunities for the development of novel therapeutics.
Acknowledgements
We thank Erin Garcia and Stacy Schaefer for outstanding technical assistance. This work was supported by NIH grants RO1-NS055179 (R.A.H.) and RO1-DK061418 (C.C.Y.). S.L.R. was supported by NIDA training grant T32-DA015040.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balasubramanian S, Fam SR, Hall RA. GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J Biol Chem. 2007;282:4162–4171. doi: 10.1074/jbc.M607695200. [DOI] [PubMed] [Google Scholar]
- 2.Beart PM, O'Shea RD. Transporters for L-glutamate: an update on their molecular pharmacology and pathological involvement. Br J Pharmacol. 2007;150:5–17. doi: 10.1038/sj.bjp.0706949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhry FA, Lehre KP, van Lookeren Campagne M, Ottersen OP, Danbolt NC, Storm-Mathisen J. Glutamate transporters in glial plasma membranes: highly differentiated localizations revealed by quantitative ultrastructural immunocytochemistry. Neuron. 1995;15:711–720. doi: 10.1016/0896-6273(95)90158-2. [DOI] [PubMed] [Google Scholar]
- 4.Choi DW. Excitotoxic cell death. J Neurobiol. 1992;23:1261–1276. doi: 10.1002/neu.480230915. [DOI] [PubMed] [Google Scholar]
- 5.Dev KK. Making protein interactions druggable: targeting PDZ domains. Nat Rev Drug Discov. 2004;3:1047–1056. doi: 10.1038/nrd1578. [DOI] [PubMed] [Google Scholar]
- 6.Fam SR, Paquet M, Castleberry AM, Oller H, Lee CJ, Traynelis SF, Smith Y, Yun CC, Hall RA. P2Y1 receptor signaling is controlled by interaction with the PDZ scaffold NHERF-2. Proc Natl Acad Sci U S A. 2005;102:8042–8047. doi: 10.1073/pnas.0408818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Gonzalez IM, Garcia-Tardon N, Cubelos B, Gimenez C, Zafra F. The glutamate transporter GLT1b interacts with the scaffold protein PSD-95. J Neurochem. 2008;105:1834–1848. doi: 10.1111/j.1471-4159.2008.05281.x. [DOI] [PubMed] [Google Scholar]
- 8.He J, Bellini M, Inuzuka H, Xu J, Xiong Y, Yang X, Castleberry AM, Hall RA. Proteomic analysis of beta1-adrenergic receptor interactions with PDZ scaffold proteins. J Biol Chem. 2006;281:2820–2827. doi: 10.1074/jbc.M509503200. [DOI] [PubMed] [Google Scholar]
- 9.Jackson M, Song W, Liu MY, Jin L, Dykes-Hoberg M, Lin CI, Bowers WJ, Federoff HJ, Sternweis PC, Rothstein JD. Modulation of the neuronal glutamate transporter EAAT4 by two interacting proteins. Nature. 2001;410:89–93. doi: 10.1038/35065091. [DOI] [PubMed] [Google Scholar]
- 10.Lau AG, Hall RA. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry. 2001;40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- 11.Lee A, Rayfield A, Hryciw DH, Ma TA, Wang D, Pow D, Broer S, Yun C, Poronnik P. Na+-H+ exchanger regulatory factor 1 is a PDZ scaffold for the astroglial glutamate transporter GLAST. Glia. 2007;55:119–129. doi: 10.1002/glia.20439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehre KP, Levy LM, Ottersen OP, Storm-Mathisen J, Danbolt NC. Differential expression of two glial glutamate transporters in the rat brain: quantitative and immunocytochemical observations. J Neurosci. 1995;15:1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayasundari A, Ferreira AM, He L, Mahindroo N, Bashford D, Fujii N. Rational design of the first small-molecule antagonists of NHERF1/EBP50 PDZ domains. Bioorg Med Chem Lett. 2008;18:942–945. doi: 10.1016/j.bmcl.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paquet M, Kuwajima M, Yun CC, Smith Y, Hall RA. Astrocytic and neuronal localization of the scaffold protein Na+/H+ exchanger regulatory factor 2 (NHERF-2) in mouse brain. J Comp Neurol. 2006;494:752–762. doi: 10.1002/cne.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, Kanai Y, Hediger MA, Wang Y, Schielke JP, Welty DF. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 17.Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, Jin L, Dykes Hoberg M, Vidensky S, Chung DS, Toan SV, Bruijn LI, Su ZZ, Gupta P, Fisher PB. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 18.Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan SM, Lee A, Bjorkman ST, Miller SM, Sullivan RK, Poronnik P, Colditz PB, Pow DV. Cytoskeletal anchoring of GLAST determines susceptibility to brain damage: an identified role for GFAP. J Biol Chem. 2007;282:29414–29423. doi: 10.1074/jbc.M704152200. [DOI] [PubMed] [Google Scholar]
- 20.Swanson RA, Liu J, Miller JW, Rothstein JD, Farrell K, Stein BA, Longuemare MC. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, Okuyama S, Kawashima N, Hori S, Takimoto M, Wada K. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 22.Torres GE, Yao WD, Mohn AR, Quan H, Kim KM, Levey AI, Staudinger J, Caron MG. Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron. 2001;30:121–134. doi: 10.1016/s0896-6273(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 23.Weinman EJ, Hall RA, Friedman PA, Liu-Chen LY, Shenolikar S. The association of NHERF adaptor proteins with G protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- 24.Yun CH, Lamprecht G, Forster DV, Sidor A. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J Biol Chem. 1998;273:25856–25863. doi: 10.1074/jbc.273.40.25856. [DOI] [PubMed] [Google Scholar]