Abstract
The alphavirus nsP2 protease is essential for correct processing of the alphavirus nonstructural polyprotein (nsP1234) and replication of the viral genome. We have combined molecular dynamics simulations with our structural studies to reveal features of the nsP2 protease catalytic site and S1’-S4 subsites that regulate the specificity of the protease. The catalytic mechanism of the nsP2 protease appears similar to the papain-like cysteine proteases, with the conserved catalytic dyad forming a thiolate-imidazolium ion pair in the nsP2-activated state. Substrate binding likely stabilizes this ion pair. Analysis of bimolecular complexes of Venezuelan equine encephalitis virus (VEEV) nsP2 protease with each of the nsP1234 cleavage sites identified protease residues His510, Ser511, His546, and Lys706 as critical for cleavage site recognition. Homology modelling and molecular dynamics simulations of diverse alphaviruses and their cognate cleavage site sequences revealed general features of substrate recognition that operate across alphavirus strains as well as strain specific covariance between binding site and cleavage site residues. For instance, compensatory changes occurred in the P3 and S3 subsite residues to maintain energetically favourable complementary binding surfaces. These results help explain how alphavirus nsP2 proteases recognize different cleavage sites within the non-structural polyprotein and discriminate between closely related cleavage targets.
Keywords: alphavirus, nsP2 protease, protease specificity
1. Introduction
Alphaviruses are enveloped arthropod borne (+)-sense RNA viruses that cause a variety of human and animal diseases worldwide.[1] Many human diseases resulting from alphavirus infections are non-lethal and characterized by fever, rash, and intense joint pain.[1] These diseases are typically associated with “Old World” alphaviruses, such as Ross River virus (RRV) in Africa, Barmah Forest virus (BFV) in Australia, Semliki Forest virus (SFV) in Asia, and Sindbis virus (SINV) in Europe, Africa, Asia, and Australia. The recent Chikungunya virus epidemic in India and the Indian Ocean islands infected more than a million people, with infection rates approaching 45% in some areas.[2, 3] In contrast, “New World” alphaviruses such as Eastern, Western, and Venezuelan equine encephalitis viruses, typically cause severe flu-like symptoms, encephalitis, and death. Mortality rates range from ∼1% for Venezuelan equine encephalitis virus (VEEV) to upwards of 80% for Eastern equine encephalitis virus (EEEV).[4] No vaccines or therapeutics are approved to combat alphavirus infections in humans.
Following infection, the alphavirus genome is released into the host cell cytoplasm and, depending on the alphavirus strain, translated into either one or two nonstructural polyproteins (nsPs). Strains possessing an opal codon between nsP3 and nsP4 produce two nonstructural polyproteins; nsP123 is produced when translation terminates at the opal codon and nsP1234 is produced by opal codon readthrough.[1] Strains lacking the opal codon only produce the nsP1234 polyprotein. The alphavirus polyprotein forms the initial virus replication complex, which is cleaved into the individual nonstructural proteins by nsP2 to enable replication to proceed productively. The necessary role of nsP2 protease in alphavirus replication makes it an appealing target for antiviral drug design.
Biochemical studies of the nsP2 protease showed considerable differences in cleavage efficiency at the nsP12, nsP23, and nsP34 polyprotein cleavage sites,[5–7] with the substrate preference of both full-length nsP2 and the isolated protease domain being nsP34 > nsP12 ≫ nsP23.[7] In vitro protease activity was enhanced when the peptide substrate contained at least ten prime side residues and five non-prime side residues (Schechter and Berger nomenclature[8]). Even though 15 residues flanking the scissile bond were required for optimal activity, in controlled biochemical studies only substrate residues P1’-P4 specifically impacted substrate recognition by the nsP2 protease.[5] The P2 position is a conserved glycine residue in all alphavirus nsP2 substrates. The remaining P1’-P4 substrate residues diverge both within and between alphavirus strains, although the P1 and P3 sites show less variability than the P1’ and P4 sites.
The recently determined crystal structure of the VEEV nsP2 protease identified likely interactions between the protease binding site and substrate residues.[9] However, that preliminary analysis did not investigate the molecular basis for the observed substrate preferences and how each alphavirus protease recognizes at least three different substrates. Here we provide a detailed analysis of alphavirus nsP2 protease substrate specificity and how variable target substrates are recognized. In addition, amino acid sequence covariation between the protease and its substrates across different alphaviruses is identified. These studies define the nsP2 protease residues that participate in substrate recognition and explain the observed variation of polyprotein target sequences between alphavirus strains.
2. Methods
2.1. Protein/peptide models
All three-dimensional models were constructed with PyMOL using VEEV nsP2 protease coordinates (Protein Data Bank identifier 2HWK[9]). Modelled peptide substrates were positioned by aligning the Cα and Cβ atoms of nsP2 protease residues Cys477, His546, and Trp547 with the corresponding residues in the Ulp1-SUMO complex (PDB identifier 1EUV[10]). Following alignment, Ulp1-SUMO substrate residues were converted to the corresponding nsP2 substrate residues using the PyMOL mutagenesis function. Appropriate residues were added to the peptide ends using PyMOL builder, and the substrate manually docked. These protease/peptide complexes were subsequently refined using energy minimization and molecular dynamics (MD) simulations within the program CHARMM.[11]
2.2. Simulations
To perform MD simulations of each nsP2 protease/peptide substrate complex, non-carbon hydrogens were added to each complex using the HBUILD[12] module in CHARMM. Histidine residues, including the catalytic His546, were uncharged. The nsP2 protease/peptide substrate complex was parameterized using the CHARMM 22 force field.[13] Simulations were performed in vaccuo. Energy minimizations were initially performed with the protease fixed and the substrate unconstrained, followed by minimization with both the protease and substrate unconstrained. MD simulations were performed with 1 fs steps for 1 ns with no atomic positional constraints. The final 500 ps of each MD simulation were used to calculate average structures, which were subjected to a final energy minimization and used to analyze protease-substrate contacts.
Two-dimensional contact diagrams of the nsP2pro/substrate complex were generated using LIGPLOT[14] with a 4 Å cutoff distance for displayed contacts. Protease substrate contacts were also evaluated using CSU software[15] at the LPC CSU server (bip.weizmann.ac.il/oca-bin/lpccsu). Active site pKa values were calculated using the PROPKA web interface (http://nova.colombo58.unimi.it/propka.htm[16]).
2.3. Sequences
Representative alphavirus sequences were obtained from the National Center for Biotechnology Information (NCBI) Protein sequence database as follows: Aura virus (AURAV), NCBI ID: NP_819011; Barmah Forest virus (BFV), NCBI ID: NP_054023; O'nyong-nyong virus (ONNV), NCBI ID: AAC97204; Ross River virus (RRV), NCBI ID: NP_740679; salmon pancreas disease virus (SPDV), NCBI ID: Q8JJX1; Semliki Forest Virus (SFV) NCBI ID: NP_740666; Sindbis virus (SINV), NCBI ID: AAM10974; sleeping disease virus (SDV), NCBI ID: NP_598184; Venezuelan equine encephalitis virus (VEEV), NCBI ID: AAB02516; and Western equine encephalitis virus (WEEV), NCBI ID: CAA52868.
3. Results and discussion
3.1. Overview of the VEEV nsP2 protease structure and catalytic mechanism
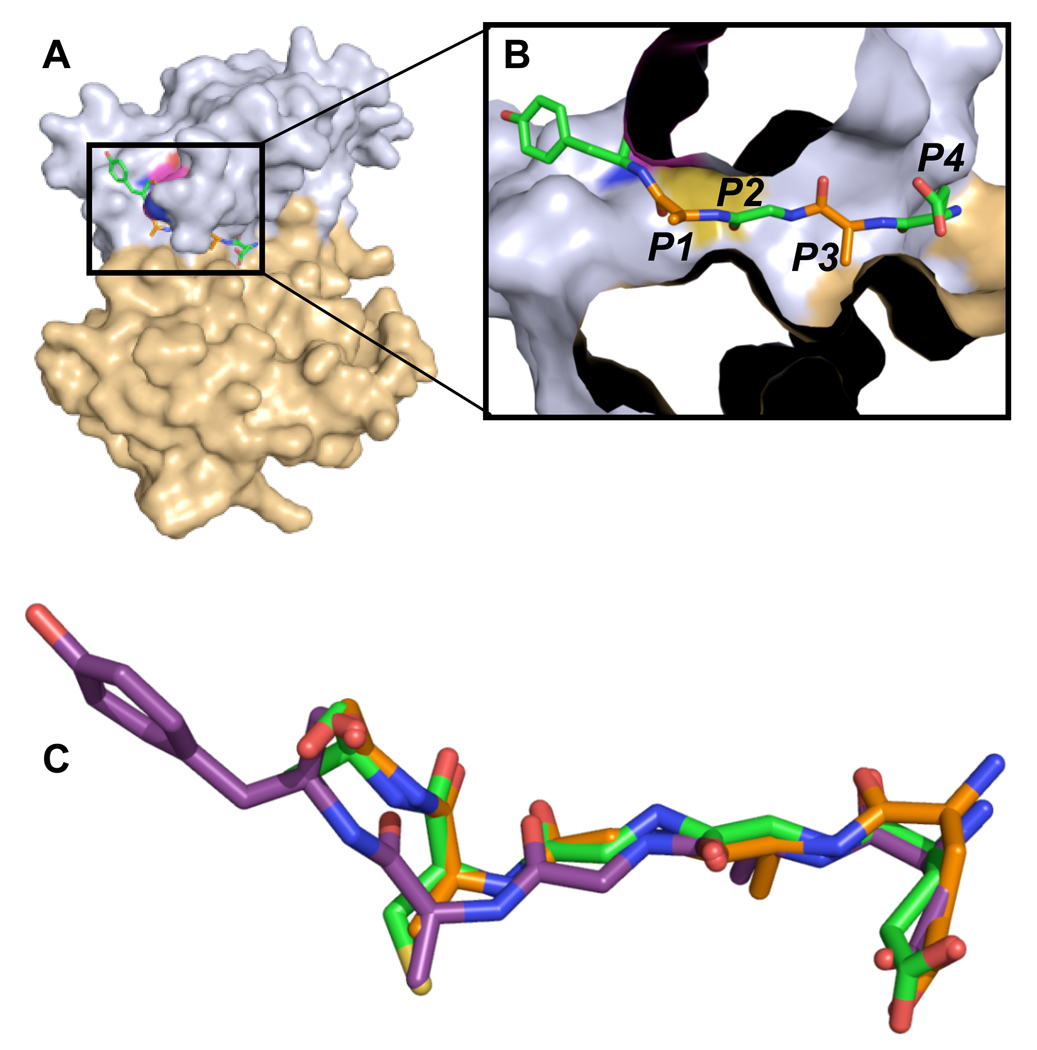
The protease domain of VEEV nsP2 is organized into two domains (Fig. 1). The N-terminal domain is composed of residues 468 to 603 and contains the catalytic dyad residues Cys477 and His546.[9] The N-terminal domain also contains the majority of the residues involved in substrate binding. The C-terminal domain, composed of residues 604 to 793, contains several residues involved in substrate binding. However, these residues do not form part of the central catalytic site. Asn544 and Asn545 form the turn at the end of a highly dynamic β-hairpin that extends towards the C-terminal domain, possibly restricting access of substrate to the binding groove and contributing to substrate binding. The substrate-binding groove of the protease can accommodate a five residue peptide in an extended conformation. In modelling studies, longer peptides extended beyond the binding groove.
Figure 1. nsP2pro with modeled nsP34 substrate peptide.
(a) The N-terminal domain is colored light blue and the C-terminal domain is colored light brown. Substrate peptide, derived from the cleavage site between nsP1 and nsP2, is colored by atom type, with carbon, oxygen, and nitrogen atoms colored green, red, and blue, respectively. (b) Boxed region in panel A is magnified, rotated slightly, and the surface of nsP2 protease cropped to provide a clearer image of the relationship between the substrate residues P1’-P4 and the protease binding subsites S1’-S4. The protease is colored as in panel A, and the coloring of the substrate residues is alternated. P1’, P2, and P4 are colored by atom as in panel A with carbon atoms in green. P1 and P3 are colored the same as in panel A with carbon in orange. (c) Overlay of models of peptides representing VEEV nonstructural polyprotein cleavage substrates nsP12, nsP23, and nsP34. Peptides are colored with nsP12 carbon atoms in orange, nsP23 carbon atoms in green, and nsP34 carbon atoms in purple. Peptides extend left to right from P1’ to P5.
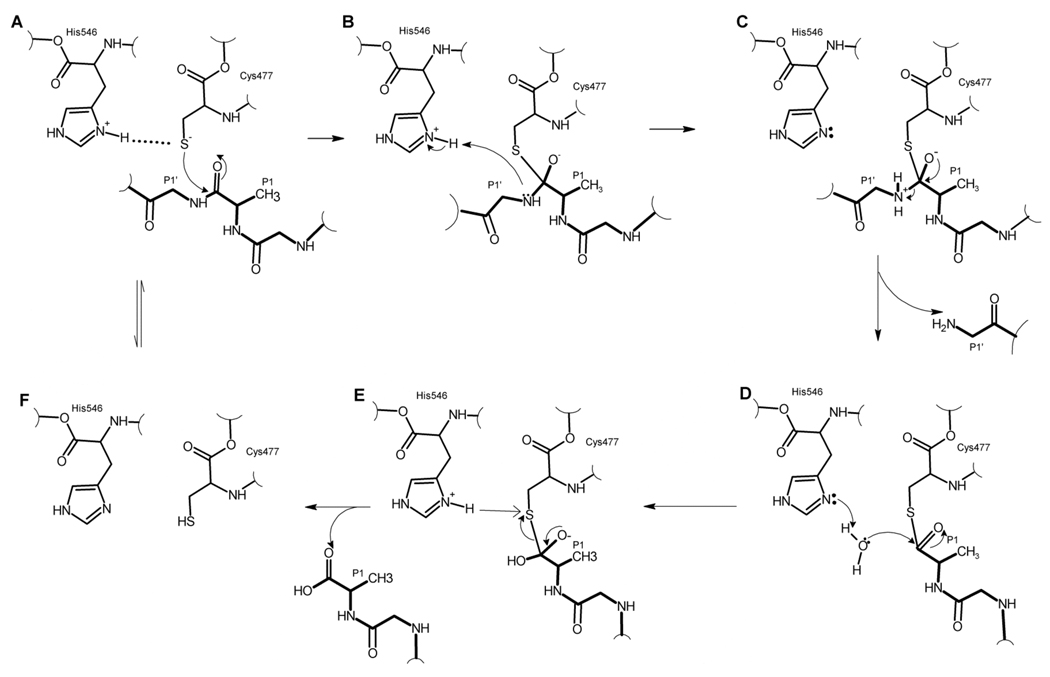
The VEEV protease structure, together with the reaction mechanism proposed for related papain and papain-like proteases,[17, 18] provided insights into the catalytic mechanism of the VEEV nsP2 protease (Fig. 2). In its activated substrate-bound state, the nsP2 protease catalytic dyad likely exists as a Cys477 thiolate-His546 imidazolium ion pair.[18] In the apo nsP2 protease crystal structure, the His546 rotamer was not positioned to participate in the ion pair necessary for catalysis. However, electron density for the His546 side chain is weak and poorly defined, indicating this residue is conformationally flexible. A simple torsion of His546 positioned a rotomer to form the catalytically active thiolate-imidazolium ion pair with Cys477 (Fig. 2f). This His546 rotomer did not affect the backbone conformation and was energetically favourable. It is probable that binding of substrate (Fig. 2a) stabilizes His546 in the active form of the catalytic dyad, thus allowing the thiolate anion to attack the scissile carbonyl carbon and the reaction to proceed as presented in Fig. 2.
Figure 2. Proposed catalytic cycle of VEEV nsP2 protease.
(a) Catalytic dyad is in the ionized state represented by a thiolate/imidazolium ion pair. Substrate (indicated by boldface bonds) binds to the active site, and the P1 carbonyl carbon is attacked by the deprotonated thiol. (b) The negatively charged tetrahedral intermediate is formed and the P1’ amine is protonated by the imidazolium ion. (c) The unstable intermediate breaks down, releasing the C-terminal product and leaving an acylated thiol. (d) and (e). The acyl thiol undergoes hydrolysis releasing the N-terminal product and free enzyme with active ionized catalytic dyad or the inactive uncharged dyad (f).
Structure-based predictions of side chain pKa values for the catalytic dyad residues provide additional support for a thiolate-imidazolium ion pair catalytic mechanism for VEEV nsP2 protease. In papain, the pKa values for the catalytic dyad cysteine and histidine residues were measured to be 3.3 and 8.5, respectively.[19] These values are substantially shifted from free amino acid pKa values of 8.4 for cysteine and 6.0 for histidine.[20] The pKa values for Cys477 and His546 within the nsP2 protease active conformation structure were calculated (PROPKA program) to be 5.1 and 8.0, respectively. These pKa values are consistent with the existence of a thiolate-imidazolium ion pair at physiological pH in VEEV nsP2 protease.
3.2. VEEV S1' subsite
Simulated annealing of the protease-substrate complexes was performed with molecular dynamic simulations and energy minimization to improve understanding of the molecular requirements for polyprotein processing and identify interactions between nsP2pro and its peptide substrates. In the following discussion we use the convention of Schechter and Berger,[8] where S and P prefixes refer to complementary protease binding sites and peptide substrate residues, respectively. In this convention, substrate cleavage occurs between the P1’ and P1 residues.
Limited intermolecular interactions between the S1' and P1' subsites are observed in the VEEV nsP2pro/peptide complex (Fig. 3, Table 1). The P1' residue of the VEEV nsP12, nsP23, and nsP34 polyprotein cleavage substrates is glycine, alanine, and tyrosine, respectively. The S1' subsite within VEEV nsP2pro is defined by residues Ala474, Asn475, Cys477, and His546. Interactions between nsP2pro and the nsP12 and nsP23 cleavage sites are similar, and consist of van der Waals contacts between Asn475 and Cys477 and the P1’ residue, and a hydrogen bond between His546 and the P1’ amide nitrogen (Fig. 3). In addition, Ala474 interacts with the alanine Cβ methyl group of the nsP23 P1’ (Fig. 3, Table 1). Although these contacts are also observed between nsP2pro and the P1’ tyrosine of the nsP34 cleavage site, an additional interaction between Lys480 and the P1’ tyrosine of nsP34 occurs. This interaction can be considered as either a hydrogen bond between the lysine ε-amino and tyrosine hydroxyl groups or a cation-π interaction between lysine and the tyrosine ring. This additional interaction between the nsP34 cleavage site and the S1’ is likely one of the dominant factors influencing the preference of the nsP2 protease for nsP34-based substrate peptides.[6, 7] Other than the side chain interaction with Lys480, the observed contacts between nsP2pro and the P1’ tyrosine residue are consistent with the other two nonstructural polyprotein cleavage sites. This is largely because the S1’ subsite of VEEV nsP2pro is shallow and solvent exposed. Thus, contacts between protease and the substrate P1’ residue are mostly through substrate backbone atoms.
Figure 3. Schematic of the nsP2 protease-binding site with modeled substrates.
(a) nsP12 substrate. (b) nsP23 substrate. (c) nsP34 substrate. Substrate peptide bonds are purple and nsP2 protease bonds are brown. Substrate residues labels are blue. Numbering for peptide substrates is according to the sequence of VEEV nonstructural polyprotein. Schematics were produced with LIGPLOT.[14]
Table 1.
Interactions between VEEV nsP2 protease and peptide substrates.
| nsP12 | Substrate nsP23 |
nsP34 | Enzyme | nsP12 | Contact nsP23 |
nsP34 | |
|---|---|---|---|---|---|---|---|
| Ala474 | + | + | |||||
| Asn475 | + | + | + | ||||
| P1’ | Gly536 | Ala1330 | Tyr1867 | Cys477 | + | + | + |
| Lys480 | ±‡ | ||||||
| His546 | ± | ± | ± | ||||
| Asn475 | + | + | + | ||||
| Cys477 | + | ± | + | ||||
| Trp478 | ± | ||||||
| P1 | Ala535 | Cys1329 | Ala1866 | Ala509 | + | + | + |
| Asn545 | ± | ± | |||||
| His546 | + | + | + | ||||
| Leu665 | + | + | + | ||||
| Cys477 | + | + | |||||
| Trp478 | + | ||||||
| P2 | Gly534 | Gly1328 | Gly1865 | Ala509 | + | + | + |
| His510 | ± | ± | ± | ||||
| Trp547 | + | + | + | ||||
| His510 | + | + | + | ||||
| Ile542 | + | + | |||||
| P3 | Ala533 | Ala1327 | Ala1864 | Trp547 | + | + | + |
| Ile698 | + | + | + | ||||
| Met702 | + | + | + | ||||
| Ser511 | ± | ± | ± | ||||
| Glu513 | ± | ||||||
| P4 | Glu532 | Glu1326 | Asp1863 | Trp547 | + | + | + |
| Met702 | + | ||||||
| Lys706 | ± | ± | ± |
3.3. VEEV S1 subsite
The P1 residue of VEEV polyprotein substrates binds to the S1 subsite of nsP2pro. For each modeled polyprotein substrate, the catalytic thiol of Cys477 and the carbonyl carbon of the substrate scissile bond are correctly positioned for nucleophilic attack to occur (Fig. 2, Fig. 3). To facilitate this reaction, Cys477 is deprotonated and stabilized by a thiolate-imidazolium ion pair with His546. For all cleavage sites within the VEEV nonstructural polyprotein, the P1 residue is either alanine or cysteine (Table 2). The nsP2 protease S1 subsite contains residues that make polar and non-polar contacts to the P1 substrate residue (Table 1, Fig. 3). In the nsP23 substrate, the P1 carbonyl oxygen orientation relative to the Cys477 backbone amide is consistent with hydrogen bonding between these residues; in this case the Cys477 amide nitrogen serves as an oxyanion hole for the hydrolysis reaction. In contrast, molecular modelling of the nsP12 substrate suggested that the indole nitrogen of Trp478 might also function as part of the oxyanion hole. Molecular modelling of the nsP34 substrate revealed the P1 carbonyl oxygen is oriented such that conformational rearrangements expected to occur when the tetrahedral reaction intermediate is formed would position Cys477 and/or Trp478 to serve as the oxyanion hole during the hydrolysis reaction.
Table 2.
Alphavirus sequence covariance for protease subsite residues S1’-S4 and polyprotein cleavage substrate residues P1’-P4.
| Virus | S1’ subsite residues | P1’ residue | ||||||
|---|---|---|---|---|---|---|---|---|
| nsP12 | nsP23 | nsP34 | ||||||
| VEEV | A474 | N475 | C477 | K480 | H546 | G | A | Y |
| WEEV | V | - | - | - | - | - | - | - |
| EEEV | - | - | - | - | - | - | - | - |
| RRV | - | K | - | - | - | - | - | - |
| SFV | - | - | - | - | - | - | - | - |
| ONNV | - | - | - | - | - | - | - | - |
| BFV | V | - | - | - | - | - | - | - |
| SINV | T | - | - | - | - | A | - | - |
| AURAV | V | - | - | - | - | A | - | - |
| SPDV | R | - | - | V | - | - | S | - |
| SDV | R | - | - | V | - | - | S | - |
| Virus | S1 subsite residues | P1 residue | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| nsP12 | nsP23 | nsP34 | ||||||||
| VEEV | N475 | C477 | W478 | A509 | N545 | H546 | L665 | A | C | A |
| WEEV | - | - | - | - | Q | - | - | - | R | - |
| EEEV | - | - | - | - | E | - | - | - | R | - |
| RRV | K | - | - | - | - | - | - | - | - | - |
| SFV | - | - | - | - | - | - | - | - | - | - |
| ONNV | - | - | - | - | - | - | - | - | - | G |
| BFV | - | - | - | - | D | - | - | E | S | G |
| SINV | - | - | - | P | A | - | F | - | A | G |
| AURAV | - | - | - | P | A | - | F | - | A | G |
| SPDV | - | - | - | P | S | - | - | - | A | G |
| SDV | - | - | - | P | S | - | - | S | A | G |
| Virus | S2 subsite residues | P2 residue | ||||||
|---|---|---|---|---|---|---|---|---|
| nsP12 | nsP23 | nsP34 | ||||||
| VEEV | W478 | C477 | A509 | H510 | W547 | G | G | G |
| WEEV | - | - | - | Y | - | - | - | - |
| EEEV | - | - | - | F | - | - | - | - |
| RRV | - | - | - | Y | - | - | - | - |
| SFV | - | - | - | Y | - | - | - | - |
| ONNV | - | - | - | Y | - | - | - | - |
| BFV | - | - | - | Y | - | - | - | - |
| SINV | - | - | P | - | - | - | - | - |
| AURAV | - | - | P | Y | - | - | - | - |
| SPDV | - | - | P | - | - | - | - | - |
| SDV | - | - | P | - | - | - | - | - |
| Virus | S3 subsite residues | P3 residue | ||||||
|---|---|---|---|---|---|---|---|---|
| nsP12 | nsP23 | nsP34 | ||||||
| VEEV | H510 | I542 | W547 | I698 | M702 | A | A | A |
| WEEV | Y | Y | - | - | - | - | - | - |
| EEEV | F | Y | - | - | - | - | - | - |
| RRV | Y | Y | - | M | - | - | - | - |
| SFV | Y | Y | - | M | - | - | - | - |
| ONNV | Y | Y | - | M | - | - | - | - |
| BFV | Y | Y | - | M | - | - | - | - |
| SINV | - | Y | - | A | T | I | V | V |
| AURAV | Y | F | - | G | T | - | S | S |
| SPDV | - | L | - | V | A | - | V | V |
| SDV | - | L | - | V | A | V | V | V |
| Virus | S4 subsite residues | P4 residue | ||||||
|---|---|---|---|---|---|---|---|---|
| nsP12 | nsP23 | nsP34 | ||||||
| VEEV | S511 | E513 | W547 | M702 | K706 | E | E | D |
| WEEV | - | - | - | - | - | - | - | E |
| EEEV | - | - | - | - | - | - | - | E |
| RRV | - | - | - | - | D | Y | T | R |
| SFV | - | V | - | - | D | Y | T | R |
| ONNV | - | - | - | - | D | D | R | R |
| BFV | - | - | - | - | D | - | P | R |
| SINV | - | V | - | T | S | A | G | G |
| AURAV | - | M | - | T | S | D | G | G |
| SPDV | - | L | - | A | G | D | M | G |
| SDV | - | L | - | A | G | D | M | G |
3.4. VEEV S2 subsite
Molecular modelling and molecular dynamic simulations of the binding between nsP2 protease and its varied polyprotein substrates identified crucial interactions between Trp547 in the S2 subsite and the P2 residue. In all examined alphavirus sequences, nsP2 Trp547 is completely conserved and the P2 glycine is invariant in the nonstructural polyprotein cleavage targets. The orientation of Trp547 in the model restricts the volume of the S2 subsite such that it can only accommodate a P2 glycine residue. Modelling other residues at the P2 position, including alanine, resulted in energetically unfavourable steric clashes between the P2 residues and Trp547. These results provide a structure-based explanation of the correlation noted by Golubtsov and co-workers that cysteine proteases that cleaved substrates containing a P2 glycine had bulky aromatic residues immediately following the active site histidine.[21] This glycine specificity motif (GSM) was speculated to be involved in alphavirus nsP2 substrate recognition.[21]
The S2 subsite is not defined exclusively by steric constraints imposed by Trp547. Several additional residues (e.g., Cys477, Ala509) contribute van der Waals interactions, and the backbone carbonyl of His510 forms a hydrogen bond to the amine of P2 glycine (Table 1). Although a non-glycine amino acid at the P2 position could undergo 180° backbone rotation to direct its side chain out of the S2 subsite and into the solvent, such a rotation would be energetically unfavourable since it would disrupt the hydrogen bonding between the cleavage substrate and His510.
3.5. VEEV S3 and S4 subsites
The S3 subsite accommodates a small non-polar residue. In VEEV, each cleavage substrate contains an alanine residue at the P3 position (Table 1). Contacts between S3 residues and the P3 amino acid are predominately hydrophobic (Fig. 3, Table 1). With the exception of Trp547, residues that form the S3 subsite are poorly conserved (Table 2).
VEEV cleavage substrates contain either aspartate or glutamate at the P4 position. The S4 subsite is formed from polar and non-polar residues, although polar contacts dominate the interactions between P4 and S4 residues. In all cleavage substrate models, the side chain hydroxyl of Ser511 makes a hydrogen bond to the P4 backbone carbonyl, and appears to be important for correct positioning and orientation of the substrate peptide. In addition, Lys706 forms a salt bridge with the acidic P4 sidechain, and this contact is probably a significant determinant of substrate recognition.
3.6. Covariance of cleavage site (Pn) and binding subsite (Sn) sequences in alphaviruses
3.6.1. S1’ subsite
The majority of P1’ residues in alphavirus nsP12 and nsP23 cleavage substrates are glycine or alanine, although serine occurs at the nsP23 P1’ position in the distantly related salmonid viruses (Table 2). Tyrosine is conserved at the nsP34 cleavage site P1’ position in all examined alphavirus sequences.
Five residues, located at positions 474, 475, 477, 480, and 546, contribute to the S1 subsite. Cys477and His546 are completely conserved in all alphaviruses. Asn475 is highly conserved among alphaviruses (Table 2). Alanine is the predominant residue at position 474, although several differences are observed at this position in other alphavirus strains. Only backbone atoms of Ala474 interacted with the nsP23 and nsP34 P1’ residue. This lack of sidechain involvement in S1’ specificity removes constraints on sidechain identity and enables substitutions at residue 474 to be accommodated. Lys480 is conserved in the non-salmonid alphaviruses and valine replaces Lys480 in the salmonid alphaviruses. Lys480 interacts with the P1’ tyrosine of nsP34 cleavage substrate through either a cation-π interaction with the tyrosine ring or hydrogen bond with the tyrosine hydroxyl and provides a strong interaction that dictates nsP34 specificity. This interaction is absent in the salmoid alphaviruses, since Lys480 is replaced by valine. However, in the salmonid viruses a compensatory change of Ala474 to Arg474 in the S1’ subsite preserves the specific cation-π interaction (Table 2).
3.6.2. S1 subsite
Within and between alphavirus strains, the S1 subsite accommodates diverse P1 residues. In Western equine encephalitis virus (WEEV), the P1 residue is either alanine or arginine; in Barmah Forest virus (BFV) the P1 residue is glutamate, serine, or glycine. The S1 subsite primarily interacts with the Cα and carbonyl carbon atoms of the P1 residue. The P1 backbone orientation enables long flexible sidechains (e.g., arginine) to adopt conformations that position the P1 sidechain beyond the S1 subsite and in solvent. Branched or aromatic sidechains at the P1 position do not have the necessary flexibility to circumvent the steric constraints of the S1 subsite, and thus are not observed at the P1 position of cleavage substrates.
Seven residues, located at positions 475, 477, 478, 509, 545, 546, and 665, form the alphavirus S1 subsite (Table 2), although residue 478 does not interact with nsP23 and nsP34 cleavage substrates. Residues that delineate the S1 subsite are largely conserved among alphaviruses, with substitutions generally restricted to chemically similar residues (Table 2). Asn545 interacts with the P1 residue through hydrogen bonding between their backbone amides; this S1 residue can tolerate diverse substitutions because its sidechain does not participate in substrate binding. Covariant substitutions between the P1 and S1 subsite residues are not readily apparent, implying this site is relatively tolerant of amino acid substitutions.
Lulla and co-workers[5] observed that the SFV nsP2 protease cleaved synthetic substrates containing alanine, serine, and glycine (and presumably cysteine) at the P1 position, whereas substrates containing P1 glutamate and arginine residues were not cleaved. Glutamate is observed at P1 in BFV cleavage substrates, while arginine is observed at P1 in WEEV and EEEV cleavage substrates. Nearly all S1 residues are conserved among these four alphavirus strains. The notable exception is Asn545 whose contributions to the S1 pocket are through backbone atoms as discussed previously. Interestingly, our molecular modelling studies do not provide a structure-based explanation as to why the SFV nsP2 protease was observed to be inactive against substrates with P1 arginine or glutamate, whereas other alphavirus strains with >95% sequence identity in the S1 subsite are active against these substrates. Future work will investigate the thermodynamic factors that may be responsible for the observed substrate specificities.
3.6.3. S2 subsite
The P2 residue in all alphavirus polyprotein cleavage substrates is glycine, which interacts with the S2 subsite formed in part by conserved residues Cys477, Trp478, and Trp547. Residues 509 and 510 also contribute to the S2 subsite, with alanine or proline residues found at position 509, and histidine, tyrosine or phenylalanine residues found at position 510. The conservative substitutions observed at positions 509 and 510 do not significantly affect the binding characteristics or geometry of the S2 subsite. The Cα atom of Ala509 makes hydrophobic contacts with the Cα atom of the P2 glycine; this interaction is preserved when proline is substituted at this position. Residue 510 interacts with the P2 glycine through backbone hydrogen bonds, with the planar ring sidechain buried within the interface between nsP2pro domains. Thus, substitution of His510 with tyrosine or phenylalanine maintains the local structure of the S2 subsite, does not disrupt backbone interactions with the P2 residue, and preserves substrate specificity.
3.6.4. S3 subsite
Within each alphavirus strain, the P3 residue is almost completely conserved among the nsP12, nsP23, and nsP34 polyprotein cleavage substrates. Alanine occupies the P3 position of the polyprotein cleavage substrates in the majority of alphavirus strains examined. However, in Sindbis, Aura, and salmonid alphaviruses, valine, isoleucine, and serine are found at the P3 position. These substitutions increase sidechain volume, and result in steric clashes between the P3 and S3 residues unless there are compensatory substitutions within the S3 subsite (Fig. 4).
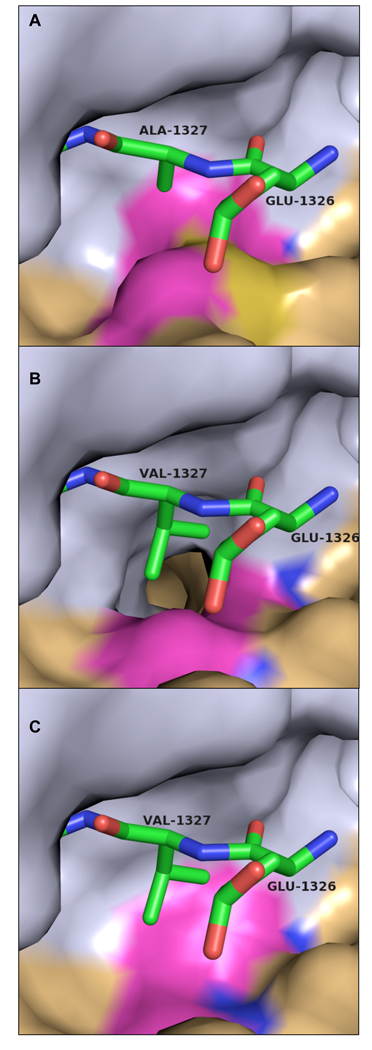
Figure 4. Covariance of amino acid substitutions in different alphaviruses strains at the S3 subsite and the P3 cleavage substrate.
(a) VEEV nsP2 protease/nsP23 substrate complex with P3 alanine (Ala1327). S3 residues Met702 and Ile698 are highlighted. (b) Complex as modeled in the Aura virus with a P3 valine, threonine at position 702, and glycine at position 698. (c) Complex as modeled in the salmonid alphaviruses (salmon pancreas disease virus and sleeping disease virus) with P3 valine, alanine at position 702, and valine at position 698. Substrate peptide is shown as sticks. Carbon atoms are green, nitrogen blue, and oxygen red. The nsP2 protease surface is shown with the N-terminal domain colored gray and the C-terminal domain colored tan. S3 subsite residues exhibiting significant variation are highlighted in magenta. All residue numbers correspond to VEEV numbering convention.
Residues 510, 542, 547, 698, and 702 form the S3 subsite. Trp547 is invariant, with conservative substitutions occurring for His510 and Ile698 (Table 2). Variations in S3 residues are highly correlated with changes at P3 residue. In Sindbis, Aura, and salmonid viruses, the P3 valine or serine residues are >50 Å3 larger than the canonical Ala residue. In Sindbis and Aura virus nsP2 proteases, threonine replaces the canonical Met702, resulting in ∼50 Å3 increase in the volume of the S3 subsite relative to the VEEV S3 subsite (Fig. 4b). In addition, Ile698 is replaced by small hydrophobic sidechains in the Sindbis and Aura viruses, resulting in S3 volume increases of >80 Å3 relative to Ile698 (Fig. 4b). In the salmonid alphaviruses, increases in the volume of the S3 subsite are achieved through substitution of alanine for Met702 and valine for Ile698 (Table 2, Fig. 4c). In each case, the increased volume of the S3 subsite exceeds the volume increase of the P3 sidechain. Thus, residue substitutions that increase the size of the P3 sidechain are complemented with covariant S3 substitutions that increase the volume of the S3 subsite.
3.6.5. S4 subsite
Diverse residues occur at the P4 position of the polyprotein cleavage substrates both within and between alphaviruses. Residues 511, 513, 547, 702, and 706 form the S4 subsite. Interestingly, subsite residues Ser511 and Trp547 are conserved, although the P4 residue is highly variable. Little correlation is observed between most S4 residues and the P4 residue. However, variations in S4 subsite residue 706 and the P4 substrate residue are highly correlated, implying this interaction is a key determinant of substrate specificity. In the equine encephalitis viruses, residue 706 is lysine and the nsP34 P4 residue is acidic, allowing these residues to interact through a salt bridge (Fig. 3). In Ross River, Semliki forest, O’nyong-nyong and Barmah Forest viruses, residue 706 is aspartic acid and the nsP34 P4 residue is arginine, which maintains the salt bridge between these residues. In the nsP12 and nsP23 cleavage substrates, covariance between the P4 residue and residue 706 largely preserves the salt-bridge and hydrogen-bond interactions between these residues.
4. Conclusion
Proteolytic processing of alphavirus nonstructural polyproteins requires the nsP2 protease to differentiate between three similar cleavage sites within the nonstructural polyprotein so that polyprotein cleavage occurs in the correct order and at the correct time during viral replication cycle. . This work elucidates structural features of the nsP2 protease domain/substrate interaction that contribute to the catalytic mechanism, substrate specificity, and discrimination between the nonstructural polyprotein cleavage sites and indicates how specific mutations to the cleavage sites and substrates might affect the protease-substrate interaction. Work by Lulla and co-workers[5] suggested that major determinants of specificity between cleavage sites within the SFV nonstructural polyprotein reside in substrate residues P1’ through P4, with the influence of the non-prime residues dominating specificity. They implicated P4 as a major determinant of substrate specificity in SFV. The covariance between S4 subsite residue 706 and the P4 substrate residue, which maintains a salt bridge in the majority of alphavirus strains, provides a structural basis for the observed specificity. In addition, the P1’ residue contributes to substrate specificity due to cation-π interactions between the invariant P1’ tyrosine of the nsP34 cleavage site and conserved basic residues in the S1’subsite; this interaction likely accounts for the preferential cleavage of this substrate.
We identified several structural determinants of substrate specificity that recognize features common to all three cleavage sites within the viral polyprotein. These determinants are strongly conserved among alphaviruses. Examination of contacts between S2/P2 and S3/P3 residues illustrate preserved binding motifs among nonstructural polyprotein cleavage sites such as the ubiquitous P2 glycine interaction with Trp547 and hydrogen bonding between P2 and His510. Important substrate binding interactions involving P3 are preserved by covariance of residues that form the S3 subsite; compensatory residue changes maintain similar volumes for the P3 sidechain and S3 subsite. These observations benefit drug discovery efforts by identifying target sites and interactions on the nsP2 protease that might lead to either broad-spectrum anti-alphaviral drugs or highly specific treatments for individual alphavirus diseases.
Ackowledgements
We thank Dr. M. Karplus for kindly providing the CHARMM molecular dynamic software. This work was supported in part by NIAID/NIH AI53551 (SJW). MAW was supported by a grant from the Sealy and Smith Foundation to the Sealy Center for Structural Biology and Molecular Biophysics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahariya C, Pradhan SK. Emergence of chikungunya virus in Indian subcontinent after 32 years: A review. J Vector Borne Dis. 2006;43:151–160. [PubMed] [Google Scholar]
- 3.N/A. Outbreak news. Chikungunya. India. Wkly Epidemiol Rec. 2006;81:409–410. [PubMed] [Google Scholar]
- 4.Aguilar PV, Paessler S, Carrara AS, Baron S, Poast J, Wang E, Moncayo AC, Anishchenko M, Watts D, Tesh RB, Weaver SC. Variation in interferon sensitivity and induction among strains of eastern equine encephalitis virus. J Virol. 2005;79:11300–11310. doi: 10.1128/JVI.79.17.11300-11310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lulla A, Lulla V, Tints K, Ahola T, Merits A. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol. 2006;80:5413–5422. doi: 10.1128/JVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kaariainen L, Ahola T. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem. 2003;278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 7.Vasiljeva L, Valmu L, Kaariainen L, Merits A. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J Biol Chem. 2001;276:30786–30793. doi: 10.1074/jbc.M104786200. [DOI] [PubMed] [Google Scholar]
- 8.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 9.Russo AT, White MA, Watowich SJ. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure. 2006;14:1449–1458. doi: 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 11.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunger AT, Karplus M. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988;4:148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- 13.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins, Ć. The Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 14.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 15.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 17.Beveridge AJ. A theoretical study of the active sites of papain and S195C rat trypsin: implications for the low reactivity of mutant serine proteinases. Protein Sci. 1996;5:1355–1365. doi: 10.1002/pro.5560050714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polgar L. Mercaptide-imidazolium ion-pair: the reactive nucleophile in papain catalysis. FEBS Lett. 1974;47:15–18. doi: 10.1016/0014-5793(74)80415-1. [DOI] [PubMed] [Google Scholar]
- 19.Lewis SD, Johnson FA, Shafer JA. Potentiometric determination of ionizations at the active site of papain. Biochemistry. 1976;15:5009–5017. doi: 10.1021/bi00668a010. [DOI] [PubMed] [Google Scholar]
- 20.Dawson RMC. 3 ed. Oxford: Clarendon Press; 1986. [Google Scholar]
- 21.Golubtsov A, Kaariainen L, Caldentey J. Characterization of the cysteine protease domain of Semliki Forest virus replicase protein nsP2 by in vitro mutagenesis. FEBS Lett. 2006;580:1502–1508. doi: 10.1016/j.febslet.2006.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]