Abstract
Dieldrin is a persistent organochlorine pesticide that induces neurotoxicity in the vertebrate central nervous system and impairs reproductive processes in fish. This study examined the molecular events produced by subchronic dietary exposures to 2.95 mg dieldrin/kg feed in the neuroendocrine brain of largemouth bass, an apex predator. Microarrays, proteomics, and pathway analysis were performed to identify genes, proteins, and cell processes altered in the male hypothalamus. Fifty-four genes were induced, and 220 genes were reduced in steady-state levels (p < 0.001; fold change greater than ± 1.5). Functional enrichment analysis revealed that the biological gene ontology categories of stress response, nucleotide base excision repair, response to toxin, and metabolic processes were significantly impacted by dieldrin. Using isobaric tagging for relative and absolute quantitation, 90 proteins in the male hypothalamus were statistically evaluated for changes in protein abundance. Several proteins altered by dieldrin are known to be associated with human neurodegenerative diseases, including apolipoprotein E, microtubule-associated tau protein, enolase 1, stathmin 1a, myelin basic protein, and parvalbumin. Proteins altered by dieldrin were involved in oxidative phosphorylation, differentiation, proliferation, and cell survival. This study demonstrates that a subchronic exposure to dieldrin alters the abundance of messenger RNAs and proteins in the hypothalamus that are associated with cell metabolism, cell stability and integrity, stress, and DNA repair.
Keywords: pesticides, neurodegeneration, biomarkers, neuroendocrine, iTRAQ, proteomics, environment-disease
The cyclodiene insecticide dieldrin is a persistent environmental contaminant that was widely used in the 1960s–1980s for agricultural practices (Jorgenson, 2001). Although applications using dieldrin are restricted or banned, sediment in some areas still contain high concentrations of dieldrin because of heavy agricultural use. Dieldrin is highly lipophilic, and animal tissues such as brain, liver, muscle, and gills accumulate significant amounts of the pesticide (Lamai et al., 1999; Satyanarayan et al., 2005). As a result, there remains continued concern for wildlife and human risks because of bioaccumulation of dieldrin and other organochlorine pesticides (OCPs).
Dieldrin is neuroactive and targets the central nervous system (CNS). Specifically, dieldrin’s mechanism of action is antagonism of gamma-amino-butyric acid (GABAA) receptors, blocking GABA-mediated synaptic transmission in the CNS. Studies in fish have demonstrated that dieldrin is neurotoxic and induces stress in both the developing and adult brain (Martyniuk et al., 2010a; Ton et al., 2006). For example, an acute injection of dieldrin (10 mg/kg) into female largemouth bass (LMB) resulted in transcriptomic responses in the hypothalamus that were indicative of (1) increased ubiquitin-proteasome activity, (2) oxidative stress, (3) inflammation, and (4) DNA damage (Martyniuk et al., 2010a). Experimental in vitro and in vivo studies in mammals suggest that dieldrin exerts adverse effects on the dopaminergic system by inducing apoptosis and oxidative stress (Hatcher et al., 2007; Kanthasamy et al., 2008; Kitazawa et al., 2001, 2004). Both apoptosis and DNA damage are cellular responses common during the progression and manifestation of human neurological diseases, such as Parkinson’s (PD) and Alzheimer’s disease (AD). However, the molecular mechanisms underlying dieldrin-mediated neurotoxicity are not fully characterized.
The hypothalamus is the major neuroendocrine region of the brain that controls pituitary hormone release. The teleostean hypothalamus is a target for dieldrin-induced neurotoxicity because it contains a high density of GABA-producing cells (Martyniuk et al., 2007; Trabucchi et al., 2008). Moreover, GABA concentrations are increased by 25–30% in the hypothalamus of female LMB after a single injection of dieldrin (Martyniuk et al., 2010a), suggesting there are neurochemical changes that occur in this tissue that potentially impact synaptic signaling.
To identify cell processes impacted by dieldrin in the hypothalamus, sexually mature male LMB were fed a diet that contained 2.95 mg dieldrin/kg feed (measured) during a 2-month subchronic feeding study. It has been acknowledged that long-term consequences of chronic low concentration dieldrin exposure are not well studied (Sava et al., 2007), and this study contributes to a better understanding of how environmental exposures may lead to neuronal damage. In this study, LMB were fed a contaminated diet that would result in dieldrin tissue burdens comparable with levels in LMB found in sites with significant sediment contamination.
Genomics (microarrays), proteomics, and bioinformatics approaches were utilized to identify cell processes altered by dieldrin in the LMB male hypothalamus. Proteomic responses to dieldrin were measured using the novel approach, isobaric tagging for relative and absolute quantitation (iTRAQ) (Applied Biosystems, Foster City, CA). This method detects 200–300 proteins within fish tissues (Martyniuk et al., 2009, 2010b) and is becoming a powerful discovery tool for protein biomarker characterization. This study is the first to combine microarrays and iTRAQ to study mechanisms of pesticide neurotoxicity in fish.
MATERIALS AND METHODS
LMB and dieldrin feeding regime.
LMB were purchased from the American Sportfish Hatchery (Montgomery, AL) and maintained at the Aquatic Toxicology Laboratory at the Center for Environmental and Human Toxicology (University of Florida, Gainesville, FL). Fish were acclimated in aerated 147–220-gallon fiberglass tanks under constant conditions of 21°C ± 2°C for a minimum of 1 week prior to exposures. Prior to the feeding regime, dieldrin was added to the diet at a nominal concentration of 3.0 mg/kg by dissolving in 90% ethanol, mixing with menhaden oil, and coating onto a trout diet (Silver Cup, Ogden, UT) using a cement mixer. This increased the oil content of the food by 4.5% (wt/wt). Control pellets were prepared using only 90% ethanol mixed into menhaden oil. Sexually mature male LMB were fed experimental diets beginning at the end of February (27 February 2008) and fed ∼1% body weight per day.
The feed concentration of dieldrin utilized in the current study was designed to target a dieldrin body burden similar to that observed at contaminated sites. Specifically, LMB placed into mesocosms in the OCP-contaminated north shore area of Lake Apopka (Florida) by the St Johns River Water Management District (SJRWMD) contained a mean whole-carcass burden of 0.5 mg dieldrin/kg whole-body wet weight after 4 months (unpublished data). Shad, a preferred LMB prey item, contained 1.2–1.6 mg dieldrin/kg whole-body wet weight. In an early study by Johnson et al. (2007), a concentration of 3.0 mg dieldrin/kg in the feed was sufficient to yield 0.4–0.5 mg dieldrin/kg whole-body wet weight after 30 days of feeding under controlled conditions; similar to what was measured in LMB exposed in the wild at the Lake Apopka mesocosm. Therefore, this study also used 3.0 mg dieldrin/kg feed to obtain whole-carcass body burdens that were comparable with previous data and mimicked environmentally relevant exposures. To verify that the uptake of dieldrin occurred in LMB, muscle, and not whole carcass, was analyzed for dieldrin content.
Natural photoperiod and ambient water temperature (ranging from 18°C–24°C) were maintained throughout the experiment. The experiment was terminated after 57 days (24 April 2008). At termination, the fish were anesthetized with MS-222 (tricaine methanesulfonate) (200 ppm buffered with sodium bicarbonate), blood was collected from the caudal vein with a heparinized syringe, and then fish were euthanized by spinal transection. The hypothalamus was dissected and immediately frozen in liquid nitrogen for messenger RNA (mRNA) and protein analyses. Portions of gonad, liver, and kidney were preserved in 10% buffered formalin for histopathology and reproductive staging. Males used in this study were reproductively mature (> 50% mature sperm in testes). Final body burdens of dieldrin were measured by gas chromatography-mass spectrometry (GC-MS) and were determined to be comparable with levels found in LMB in polluted areas of Lake Apopka (see below). All animals were treated as per the guidelines outlined by University of Florida Institutional Animal Care and Use Committee throughout the experiment.
Determination of dieldrin burden in LMB muscle and feed pellets.
Muscle was excised from LMB carcasses dorsally just posterior to the operculum, and dermal tissue was removed. Muscle from each animal (2.5–5 g) was frozen in liquid nitrogen and pulverized using a BioSpec BioPulverizer cryopulverization device (BioSpec Products, Inc., Bartlesville, OK) and transferred to a Teflon-capped glass extraction vial. One hundred microliters of internal standard solution containing 3 ppm d10-phenanthrene (PHEN; SPEX CertiPrep, Metuchen, NJ) and 9 ppm Ring-13C12-4,4′-dichlorodiphenyldichloroethylene (13C12-DDE; Cambridge Isotope Laboratories, Inc., Andover, MA) in cyclohexane (HPLC grade; Fisher Scientific, Waltham, MA) was added to each vial followed by vortex mixing. Primary organic extraction of muscle samples was performed by the method of Gelsleichter et al. (2005) and repeated twice.
Following preparation by mortar and pestle, 3 μg PHEN was added to each feed sample as an internal standard and organic compounds were extracted using 7.0 ml n-hexane (HEX; ACS Grade, Fisher Scientific). Feed samples were homogenized with a mechanical homogenizer (Tekmar Tissumizer; Tekmar, Cincinnati, OH) and centrifuged for 15 min at 100 × g. The organic phase was transferred to a borosilicate glass tube, and two additional extractions were performed with 3.0 ml of HEX. The final combined extract for both muscle and feed samples was evaporated under nitrogen until solvent free and reconstituted in 2 (muscle) or 3 ml (feed) of acetonitrile (HPLC grade, Fisher Scientific). Further sample cleanup was accomplished by solid-phase extraction chromatography as previously described (Gelsleichter et al., 2005). Final chromatography eluant from muscle and feed samples was evaporated under nitrogen until solvent free and reconstituted in 100 (muscle) or 1000 μl (feed) of cyclohexane.
Analysis of toxicant concentration by GC-MS was performed by comparing sample measurements to a standard curve containing multiple concentrations of dieldrin along with 3 and 9 ppm of the internal standards PHEN and 13C12-DDE, respectively, in cyclohexane. GC separation was accomplished using a 5% phenylmethylpolysiloxane fused silica column with helium as carrier gas (HP-5MS, 30 m × 250 μm inner diameter; Agilent Technologies, Santa Clara, CA). The analytical program utilized a sample injection volume of 1 μl into the splitless inlet, initial oven temperature of 100°C, and final temperature of 290°C (total program time = 27.50 min). Column pressure was maintained at 100 kPa, and initial column flow was 1.4 ml/min, and then reduced to 1.0 ml/min after 2.5 min. Selective ion monitoring was utilized to record ion spectra during set retention windows. Primary and qualifier mass to charge ratios used, respectively, were 262.9 and 260.9 for dieldrin, 188.2 and 80.1 for PHEN, and 258.1 and 330.0 for 13C12-DDE. Data are expressed as mass of dieldrin per wet weight of tissue or feed, respectively. The limit of detection in the reconstituted sample extract (prior to normalization to extracted sample weight) was ∼50 μg/kg muscle samples and 300 μg/kg feed. Dieldrin detected in muscle (prior to tissue normalization) was ∼10- to 30-fold higher than the limit of detection of the method. The R2 value of the standard curve was 1.0 using quadratic regression.
Microarray analysis in the male hypothalamus.
Microarray analysis was performed using a reference design with a pool of control hypothalamic RNA from male LMB (n = 4 biological replicates) and independent biological replicates of dieldrin-treated hypothalamic RNA from male LMB (n = 4). Total RNA for microarray analysis was extracted from LMB hypothalamus (20–40 mg wet weight) using RNA STAT-60 reagent (Friendswood, TX). RNA quantity for microarray analysis was measured using the NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was evaluated using the Agilent 2100 BioAnaylzer. RNA integrity values were > 8.0 for all samples used in microarray analysis.
Details of the LMB 4 × 44K microarray construction and validation have been published elsewhere (Garcia-Reyero et al., 2008). Microarray hybridizations were performed according to the Agilent Two-Color Microarray-Based Gene Expression Analysis protocol using Cyanine 5 (Cy5) (dieldrin-fed) and Cyanine 3 (Cy3) (control) using 1 μg total RNA for each sample. Details on LMB microarray processing using the Agilent two-color protocol (Garcia-Reyero et al., 2008) and scanning of the LMB microarrays (Martyniuk et al., 2010a) have been published in detail elsewhere.
Raw expression data were imported into JMP Genomics v3.2 (SAS, Cary, NC). Raw intensity data for each microarray were normalized using Loess Normalization with a smoothing factor of 0.2. Data quality analysis after normalization verified that microarray signal intensities of Cy5 and Cy3 were sufficiently normalized based on signal histogram and box plots of each microarray. Differentially regulated transcripts were identified using a one-way analysis of variance (false discovery rate [FDR] = 5%). Differentially regulated transcripts p < 0.01; fold change greater than ± 1.5 were subjected to hierarchal clustering. Distance calculations were performed using the program Cluster (Eisen et al., 1998) and visualized using the Java TreeView program (Saldanha, 2004). Raw microarray data for this experiment have been deposited into the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (GSE16402; LMB platform GPL6527).
Functional enrichment of regulated transcripts (p < 0.05) was performed using FatiGO in Babelomics (Al-Shahrour et al., 2005). LMB transcripts on this microarray are annotated with gene ontology (GO) terms for use in functional enrichment analyses. GO level analysis was inclusive, annotation was propagated to upper levels, and the GO levels included 3–9. Functional enrichment was performed for biological processes, molecular function, and cellular components.
Pathway Studio (v5.0) (Ariadne Genomics, Rockville, MD) (Nikitin et al. 2003) was used to probe which disease processes were most impacted by regulated transcripts. Human homologs (NCBI) for LMB genes were obtained (RefSeq in NCBI) and Entrez Gene identifiers retrieved using the ID mapping service in Pathway Studio from MD Anderson GeneLink (University of Texas, Houston, TX). Disease pathways were built by selecting the shortest pathways among entities (genes). A minimum of six genes needed to be associated with a disease for inclusion in the final pathway. In addition, the user-defined connectivity was set at 1500 and those diseases that did not meet that level of connectivity were removed.
Real-time PCR validation.
Gene and protein nomenclature for LMB follows that of recommendations for zebrafish (ZFIN). Nomenclature for mammalian protein follows mammalian protein nomenclature rules. Transcripts investigated further using real-time PCR included ar, esr-beta b (AY211021), gst (AY335905), gpx (FJ030930), hsp70 (FJ751227), and npc2 (FJ751228). LMB hsp70 is most closely homologous to the hsp cognate 70 found in other teleost fish. All transcripts chosen for real-time PCR analysis were identified as differentially expressed by microarray analysis (p < 0.05). In addition, steady-state mRNA levels for these transcripts were previously shown to be altered by dieldrin in the LMB hypothalamus (Martyniuk et al., 2010a) and, in the case of steroid receptors, are also altered by dieldrin in the LMB liver and gonad (Garcia-Reyero et al., 2006). Primer sequences are provided in Table 1. Complementary DNA (cDNA) synthesis protocol for real-time PCR (Superscript-II; Invitrogen, Carlsbad, CA) is described in detail in Martyniuk et al. (2009, 2010a). Real-time PCR reactions were performed using 1X iQ SYBR Green Supermix (Bio-Rad), 1 μl of each gene-specific primer (10mM), and ∼100 ng first-strand cDNA derived from DNase-treated RNA samples. LMB rps18 was used to normalize gene expression (IQ Supermix; Bio-Rad).
TABLE 1.
Real-Time PCR Primers Used to Measure mRNA Steady-State Levels in the LMB Hypothalamusa
| Gene title | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon size (bp) |
| Androgen receptor | CAC CAC AGA GAA TGT GCC TGA | CAG GTG AGT GCG CCG TAA | 66 |
| Estrogen receptor beta b | CCG ACA CCG CCG TGG TGG ACT C | AGC GGG GCA AGG GGA GCC TCA A | 96 |
| Glutathione S-transferase | AAC TTT TCG CTG GCT GAT GT | TCT TGT CCC TGT GGG TTC TC | 173 |
| Heat-shock cognate 70 | CAG TGA TGA AGA CAA GCA GAA GA | GCC ACC AGC ACT CTG ATA CA | 163 |
| Niemann-Pick disease type C2 | GAT GGC TGC AAG TCT GGA AT | ACT GGG AAC CTG ATG CAG AA | 164 |
| Ribosomal 18S | CGG CTA CCA CAT CCA AGG AA | CCT GTA TTG TTA TTT TTC GTC ACT ACC T | 86 |
Note. aThe forward primer is provided in the 5′–3′ orientation in relation to the 5′–3′ coding strand (sense) deposited in GenBank, and the reverse primer is provided in the 5′–3′ direction in relation to the complementary strand (antisense).
Real-time PCR reactions were assayed on an iCycler Thermal Cycler (Bio-Rad). Standard curves, constructed using serial dilutions of estimated copy number, and real-time PCR assay conditions for transcripts are outlined in Martyniuk et al. (2010a). Each real-time reaction plate contained negative control samples that included pooled RNA samples (n = 2) that received no reverse transcriptase during cDNA synthesis to assess efficiency of DNase treatment. In addition, negative control samples that included water instead of template cDNA were used to assess genomic contamination in real-time PCR reagents. Reaction efficiencies for real-time PCR reactions in this study ranged between 95–105% and R2 > 0.990. Relative gene copy numbers among individuals in the control and treatment groups were determined to be non-normally distributed by a Shapiro-Wilk W-test. Therefore, a Mann-Whitney U-test was used to determine if transcript steady-state levels were significantly different between control and dieldrin-fed LMB (p < 0.05). All analyses were performed in SPSS Statistics v17.0 (SPSS, Inc., Chicago, IL).
Quantitative proteomics in the male LMB hypothalamus.
Proteins were acetone precipitated from the phenol layer of STAT-60 by adding six times the volume of each sample (ie, 200 μl sample and 1.2 ml acetone) (control, n = 3; dieldrin, n = 3). Therefore, both genomic and proteomic analyses were conducted in the same individuals. Three separate iTRAQ labeling reactions were each processed according to the manufacturer’s protocol (Applied Biosystems) using label 116 (treatment = 3) and label 117 (control = 3). Each protein sample was acetone precipitated and resolubilized in dissolution buffer (iTRAQ reagent; 0.5M trimethylammonium bicarbonate, pH = 8.5). Each sample was adjusted to 100 μg total protein prior to peptide labeling. Peptide labeling, peptide separation and fractionation, and MS parameters for QSTAR XL (Applied Biosystems) peptide analysis have been described in detail previously (Martyniuk et al., 2009, 2010b).
Tandem mass spectra were extracted by Analyst (v1.1; Applied Biosystems). In order to better assign peptides to proteins, a ray-finned fish database containing trypsin-digested peptides (Martyniuk et al., 2009) was searched using MS/MS data interpretation algorithms within Protein Pilot (Paragon algorithm, v 2.0; Applied Biosystems). Search parameters and modifications are identical to that outlined in Martyniuk et al. (2009, 2010b). To calculate an FDR for peptide-protein assignments, Proteomics System Performance Evaluation Pipeline (ProteomicS PEP; Applied Biosystems) in Protein Pilot was used to create a reversed ray-finned fish database. The methods for the generation of differential ratios for protein quantitation and spectral quality are described in Martyniuk et al. (2010b) and follow methodology used in Protein Pilot. Each protein that was quantified was done so with a minimum of two spectra.
RESULTS
Dieldrin Accumulates in LMB Muscle
The concentration of dieldrin was normalized to tissue wet weight or feed weight for each sample. In the muscle, dieldrin was not detectable in control LMB via GC-MS, whereas the concentration quantified in animals fed a diet of 2.95 mg dieldrin/kg feed for 2 months was 0.036 ± 0.006 mg dieldrin/kg wet weight of muscle (mean ± SEM) (Supplementary figure 1). The range of dieldrin in the tissue was between 0.02 and 0.06 mg dieldrin/kg of muscle. Muscle burden data for dieldrin in LMB were not lipid normalized. We point out that the muscle burden is approximately an order of magnitude lower than whole-carcass measurements obtained by the SJRWMD (unpublished data). This is expected because of low lipid content of muscle relative to other tissues, such as ovary and liver. However, it has been demonstrated in the study by Johnson et al. (2007) that 3.0 mg dieldrin/kg feed will achieve body burdens comparable with field data.
Gene Expression Analysis in the Hypothalamus after Dieldrin Feeding Exposure
Microarray analysis identified 54 genes that were induced and 221 genes that were reduced in steady-state abundance at p < 0.001, fold change greater than ± 1.5 (Supplementary appendix 1). When clustering probe intensity for genes showing significance (p < 0.01; fold change greater than ± 1.5), both control and dieldrin-fed animals separated into two distinct expression clusters (Supplementary figure 2). Differentially regulated transcripts identified in this study (cutoff p < 0.05) were also compared with those affected by an acute exposure to dieldrin in the study by Martyniuk et al. (2010a) using a Venn diagram (Oliveros, 2007) to determine whether there were common transcripts that are regulated by dieldrin by both short- and long-term exposure routes (Supplementary figure 3). In this study, 3135 transcripts were differentially expressed in the male hypothalamus at a significance level of p < 0.05 after feeding exposure. In our previous acute study with females, 1406 transcripts were differentially expressed in the hypothalamus after injection. Approximately 10% (237) of the transcripts were common between the two studies (Supplementary figure 2) despite the fact that the sex of the animal, route of exposure, concentration, and duration of the experiment were quite different. Transcripts affected by dieldrin (p < 0.01) in both studies included npc2, esr-beta b, slc25a20, gabrr3, and gltp.
Functional enrichment analysis revealed that biological processes overrepresented by regulated genes included response to stress and base excision repair (Table 2). The biological process of protein transport and protein targeting to mitochondria were also significantly impacted by dieldrin. Genes that encoded proteins found in the golgi apparatus were also preferentially affected by dieldrin and are also related to protein transport and targeting.
TABLE 2.
Functional Enrichment for Biological Process, Molecular Function, and Cellular Process for Regulated Transcripts (p < 0.05)
| GO category | p Value | |
| Biological processes | Response to stress (GO:0006950) | 0.011 |
| Base excision repair (GO:0006284) | 0.012 | |
| Intracellular protein transport (GO:0006886) | 0.017 | |
| Protein transport (GO:0015031) | 0.025 | |
| Protein targeting to mitochondria (GO:0006626) | 0.031 | |
| Response to toxin (GO:0009636) | 0.04 | |
| Protein import to mitochondria (GO:0045039) | 0.044 | |
| Metabolic processes (GO:0008152) | 0.046 | |
| ER to Golgi-mediated transport (GO:0006888) | 0.047 | |
| Aromatic compound catabolism (GO:0019439) | 0.064 | |
| Molecular functions | Serine carboxypeptidase activity (GO:0004185) | 0.041 |
| Intracellular transporter activity (GO:0005478) | 0.041 | |
| Protein carrier activity (GO:0008320) | 0.041 | |
| Manganese ion binding (GO:0030145) | 0.061 | |
| Epoxide hydrolase activity (GO:0004301) | 0.065 | |
| Rho GTPase activator activity (GO:0005100) | 0.065 | |
| Cellular compartments | Integral to membrane (GO:0016021) | 0.014 |
| Myosin complex (GO:0016459) | 0.032 | |
| Golgi apparatus (GO:0005794) | 0.042 | |
| Mitochondrial intermembrane protein transporter complex (GO:0042719) | 0.046 | |
| Integral to plasma membrane (GO:0005887) | 0.07 |
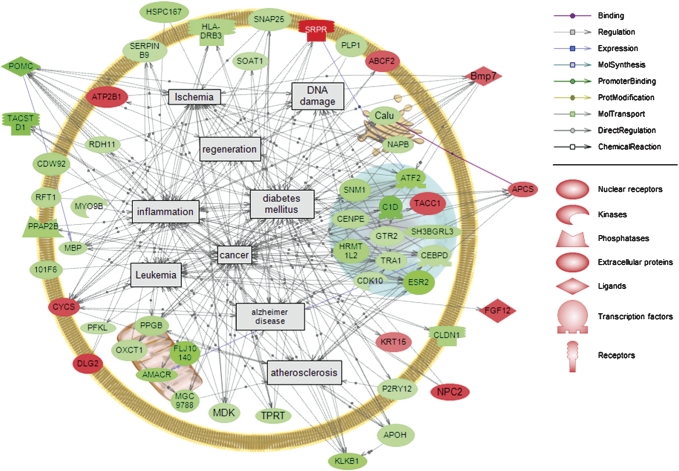
Highly regulated dieldrin transcripts were associated with a number of human disease processes (Fig. 1). All disease associations that did not meet criteria of high connectivity (> 2000) and more than five transcripts associated with the disease were removed to reduce the complexity of the interactions and focus on the processes most likely affected within the hypothalamus. The diseases and disease-related processes present in the final pathway are those most associated with the genes regulated by dieldrin (connectivity > 2000). Noteworthy is that AD, inflammation, DNA damage, and ischemia were also identified as diseases and cellular events affected by dieldrin in the previous acute exposure study (Martyniuk et al., 2010a).
FIG. 1.
Disease processes that involve genes (p < 0.01) regulated by dieldrin. Green indicates a downregulation in the mRNA and red indicates an induction in mRNA level. Proteins in figure follow nomenclature rules for mammalian proteins because pathways are based on mammalian data. 101F6, cytochrome b-561 domain containing 2; ABCF2, ATP-binding cassette, subfamily F (GCN20), member 2 (predicted); AMACR, alpha-methylacyl-CoA racemase; APCS, serum amyloid P-component; APOH, apolipoprotein H; ATF2, activating transcription factor 2; ATP2B1, ATPase, Ca++ transporting, plasma membrane 1; Bmp7, bone morphogenetic protein 7; C1D, nuclear DNA-binding protein; Calu, calumenin; CDK10, cyclin-dependent kinase (CDC2-like) 10; CDW92, CDW92 antigen; CEBPD, CCAAT/enhancer-binding protein (C/EBP), delta; CENPE, centromere protein E (predicted); CLDN1, claudin 1; CYCS, cytochrome c, somatic; DLG2, discs, large homolog 2 (Drosophila); ESR2, estrogen receptor 2 beta; FGF12, fibroblast growth factor 12; FLJ10140, tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (predicted); GTR2, Ras-related GTP binding C (predicted); HLA-DRB3, major histocompatibility complex, class II, DR beta 3; HRMT1L2, heterogeneous nuclear ribonucleoprotein methyltransferase-like 2 (Saccharomyces cerevisiae); HSPC167, CDK5 regulatory subunit associated protein 1; KLKB1, kallikrein B, plasma 1; KRT15, type I keratin KA15; MDK, midkine; MGC9788, 3-hydroxybutyrate dehydrogenase, type 1; MYO9B, myosin Ixb; NAPB, similar to N-ethylmaleimide-sensitive fusion protein attachment protein beta; NPC2, Niemann-Pick type C2; OXCT1, similar to 3-oxoacid CoA transferase 1; P2RY12, purinergic receptor P2Y, G-protein coupled 12; PFKL, phosphofructokinase, liver, B-type; PLP1, proteolipid protein; POMC, pro-opiomelanocortin; PPAP2B, phosphatidic acid phosphatase type 2B; PPGB, protective protein for beta-galactosidase; RDH11, retinol dehydrogenase 11; RFT1, RFT1 homolog (predicted); SERPINB9, similar to SPI6; SH3BGRL3, SH3 domain binding glutamic acid–rich protein-like 3 (predicted); SNAP25, synaptosome-associated protein 25 kDa; SNM1, DNA cross-link repair 1A, PSO2 homolog (S. cerevisiae) (predicted); SOAT1, sterol O-acyltransferase 1; SRPR, hypothetical LOC569063; TACC1, transforming, acidic coiled-coil containing protein 1; TACSTD1, tumor-associated calcium signal transducer 1; TPRT, prenyl (solanesyl) diphosphate synthase, subunit 1.
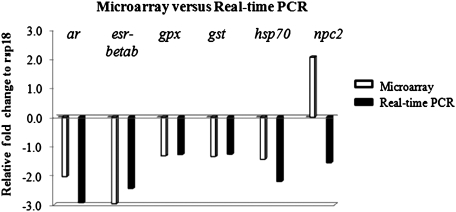
Microarray data in general corresponded to fold changes measured by real-time PCR. There was a decrease in steady-state mRNA levels of ar, gpx, gst, and hsp70 (Fig. 2). Real-time PCR verified microarray data indicating that esr-beta b mRNA levels were significantly decreased in the hypothalamus after treatment with dieldrin (p = 0.043). LMB npc2 transcript levels were increased according to microarray and significantly decreased according to real-time PCR (p = 0.029). This discrepancy between microarray and real-time PCR for npc2 was also observed in the previous acute exposure study with dieldrin (Martyniuk et al., 2010a), and it is hypothesized that the probe on the microarray is not entirely specific for npc2.
FIG. 2.
Relative fold changes for genes analyzed by microarray (white bars; n = 4) and real-time PCR (black bars; n = 4) in the male hypothalamus. Abbreviations are as follows: ar = androgen receptor; esrβb = estrogen receptor beta b; gpx = glutathione peroxidase; gst = glutathione-s-transferase; hsp70= heat-shock protein 70; npc2 = Niemann-Pick Disease C2.
Effects of Dieldrin on the Male Hypothalamic Proteome
There were 7 proteins that were significantly decreased and 11 proteins that were significantly increased in the dieldrin-fed group (Table 3) in the proteomics experiment. Eno1 showed the largest induction of the hypothalamic proteins (16-fold). Two proteins, beta cytoplasmic actin (Actb) (Psetta maxima) (gi|116488218) and beta actin (Oncorhynchus mykiss) (gi|8886013), were identified as being decreased and increased by dieldrin, respectively, and these two proteins may represent different beta actin isoforms in the LMB hypothalamus.
TABLE 3.
Differentially Expressed Proteins Identified by iTRAQ and LC MS/MS (QSTAR) after Dieldrin Treatment.a Quantitative Data for All Proteins Are Provided in Supplementary Appendix 2
| NCBI sequence identifier | Protein | Number of iTRAQb | % Coverage (Protein)c | Number of spectra usedd | Fold change | p Value |
| Downregulated | ||||||
| gi|125812189 | Microtubule-associated protein tau (Mapt) | 1 | 8.8 | 5 | −2.30 | 0.005 |
| gi|62955673 | Stathmin 1a (Stmn1a) | 3 | 31.5 | 17 | −2.30 | 0.010 |
| gi|47222845 | Dynactin 2 (p50) (Dctn2) | 1 | 8.5 | 6 | −1.99 | 0.009 |
| gi|116488218 | Beta cytoplasmic actin (Actb) | 1 | 43.3 | 3 | −1.98 | 0.007 |
| gi|52218952 | Calcineurin subunit B type 1 (protein phosphatase 2B regulatory subunit 1) (Ppp3r1) | 2 | 34.7 | 8 | −1.92 | 0.042 |
| gi|99122203 | Alpha-2 globin (Hba2) | 1 | 28.0 | 3 | −1.35 | 0.073 |
| gi|47224014 | Myelin basic protein (Mpb) | 3 | 36.9 | 128 | −1.22 | < 0.001 |
| gi|94733617 | Parvalbumin (Pvalb) | 2 | 57.8 | 26 | −1.16 | 0.031 |
| Upregulated | ||||||
| gi|98979415 | Enolase A (Eno1) | 3 | 49.1 | 4 | 16.94 | 0.034 |
| gi|1170175 | Hemoglobin subunit beta-C (Hbbc) | 1 | 41.8 | 5 | 3.95 | 0.022 |
| gi|47218629 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide (Atp5b) | 3 | 44.7 | 18 | 2.60 | 0.012 |
| gi|47207317 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 (Atp5a1) | 3 | 33.9 | 14 | 2.21 | 0.014 |
| gi|82264543 | Cytochrome C (Cytc) | 2 | 20.2 | 10 | 1.98 | 0.099 |
| gi|110589604 | Hemoglobin alpha chain (Hba1) | 3 | 55.6 | 20 | 1.95 | 0.063 |
| gi|47216921 | Collagen, type I, alpha 1 (Col1a1) | 2 | 27.6 | 3 | 1.78 | 0.050 |
| gi|68438153 | Histone cluster 1, H2bb (H2bb) | 3 | 50.0 | 18 | 1.56 | < 0.001 |
| gi|37778988 | Apolipoprotein E (Apoe) | 1 | 36.6 | 7 | 1.54 | 0.010 |
| gi|56207279 | Lactate dehydrogenase B4 (Ldhb4) | 3 | 25.1 | 25 | 1.49 | 0.077 |
| gi|27884149 | Mitochondrial isoleucine tRNA synthetase (Iars) | 3 | 5.8 | 8 | 1.48 | 0.053 |
| gi|22002420 | Adult alpha-type globin (N/A) | 3 | 40.6 | 18 | 1.48 | 0.001 |
| gi|8886013 | Beta actin (Oncorhynchus mykiss) (Actb) | 3 | 67.5 | 96 | 1.46 | 0.049 |
| gi|78099194 | Calmodulin (Cam) | 3 | 73.8 | 136 | 1.19 | < 0.001 |
| gi|47222711 | Synaptosomal-associated protein 25-A (Snap25) | 3 | 46.8 | 69 | 1.12 | 0.032 |
Proteins that were significantly altered at p < 0.05 (18) and at p < 0.10 (5).
Number of iTRAQ experiments in which peptides for the protein were detected.
Percent protein coverage for all spectra identified for a protein.
Number of high-quality spectra that was used in the quantification of the proteins (includes all peptides with selected modifications, missed cleavages, and all charge states from all three iTRAQ experiments).
Supplementary appendix 2 provides a list of all detected proteins for all three iTRAQ experiments in addition to the protein NCBI accession, the confidence of the peptide-protein identification, the peptide sequence, modifications to the peptide, and missed peptide cleavages. The total number of spectra detected was 5315. The number of distinct peptides was 1523. Of all the spectra, 39.3% could be assigned to a known protein. The number of proteins identified in each iTRAQ experiment was 122, 121, and 120. False positives using a reverse database search were calculated to be 12, 3, and 8, respectively, corresponding to an FDR of 9.8, 2.5, and 6.7% (average FDR =6.3%). The total number of unique proteins identified in the study was 192 (cutoff 1.3; 95%), and 90 proteins were quantified.
DISCUSSION
Bioaccumulation of Dieldrin in LMB Muscle
The solubility of dieldrin in water is between 0.1 and 0.25 mg/l and its log Kow is 5.48 at 20°C, indicating that it has high potential to bioaccumulate in fatty tissues. In the muscle of LMB, dieldrin was undetectable in control animals, whereas fish fed the dieldrin diet exhibited considerable accumulation after 2 months (ranging between 0.02 and 0.06 mg dieldrin/kg wet weight muscle). Dieldrin has also been shown to bioaccumulate in tissues of other teleost species. This is demonstrated in bioconcentration studies with various OCPs, including dieldrin, in carp (Cyprinus carpio), which confirmed that the liver accumulated significantly higher levels compared with intestine, muscle, and gills (Satyanarayan et al., 2005). Previous laboratory and field-based experiments with LMB have demonstrated that this species accumulates dieldrin. Garcia-Reyero et al. (2006) fed male and female LMB 0.4 and 0.81 mg dieldrin/kg feed for 120 days. Whole-carcass measurements of 0.204 ± 0.038 mg dieldrin/kg body weight for the 0.4 mg dieldrin/kg in feed and 0.265 ± 0.050 mg dieldrin/kg body weight for the 0.81 mg dieldrin/kg in feed were observed. Although concentrations of dieldrin in the tissues will depend on the tissue sampled, duration of exposure, and other factors, the accumulation of dieldrin in the muscle of LMB in the present study verified that animals were ingesting significant amounts of the pesticide from the prepared feed pellets and exposures were comparable with that observed in animals inhabiting contaminated sites around Lake Apopka, Florida.
Genomic and Proteomic Responses in LMB and Human Neurological Disease Markers
The genomics and proteomics analysis in the LMB hypothalamus revealed interesting parallels between molecular responses to dieldrin and known biomarkers of human neurodegenerative diseases. This study does not suggest that LMB display symptoms observed with human neurodegenerative diseases but rather identifies putative molecular mechanisms that may be associated with both environmental pollutants and neurological disorders. Epidemiological evidence supports the hypothesis that some pesticides, including OCPs, are contributing factors toward increased incidence of neurodegenerative diseases, such as PD (Brown et al., 2006; Priyadarshi et al., 2000). A recent epidemiological study by Weisskopf et al. (2010) investigated levels of OCPs in human serum and possible associations with the incidence of PD. Of the nine OCPs measured, only dieldrin was found to be significantly associated with PD after confounding variables such as smoking and age were considered. Furthermore, studies in the postmortem human brain have determined that concentrations of dieldrin can be significantly higher in PD patients compared with individuals who do not show symptoms or signs of PD (Corrigan et al., 2000; Fleming et al., 1994;). Thus, there is some associative evidence that dieldrin may be a factor with increased incidence of PD but we and others point out that a causal link can be difficult to ascertain.
There is also interest in using protein biomarkers as indicators for earlier stages of human neurological disease etiology (Kovacech et al., 2009; Shi et al., 2009), and it is plausible that these same biomarkers can be used as indicators for adverse health in fish because of neuroactive toxicants. Teleost models have been used for human degenerative disease research (Flinn et al., 2008; Weinreb and Youdim, 2007), and molecular signaling cascades have been investigated extensively in teleostean neuroendocrine tissues (Popesku et al., 2008). Therefore, there are some good examples for applications of fish models in disease research, and there is promise in using fish models to study the association between environmental pollutants and impaired CNS function.
The quantitative proteomics analysis in the LMB hypothalamus identified proteins that are also associated with neurodegenerative processes. Proteins that were increased in the LMB hypothalamus after dieldrin treatment included Snap25, Cytc, Eno1, Hba1, and histone cluster 1, H2bb, whereas proteins that were decreased in the hypothalamus included Pvalb. A proteomic survey in a mouse model of AD (3xTgAD mice that express mutant presenilin-1, amyloid precursor protein, and tau proteins) identified some of the same proteins as differentially expressed when compared with age-matched controls. Proteins increased in the 3xTgAD mouse cerebral cortex included SNAP25, CYTC (somatic), ENO1, HBA2, and a number of histone proteins, whereas PVALB was significantly lower in stage-matched controls (Martin et al., 2008). In another study, two-dimensional gel electrophoresis was used to quantify proteins in the cerebral cortex of a 14-month-old transgenic mouse model for AD that overexpresses a mutated form of β-amyloid precursor protein in the brain (Sizova et al., 2007). The study identified 35 proteins as being differentially expressed between the AD model and control mice, and the proteins included APOE, tubulin alpha-4, and CaMK-II alpha subunit (CaMKII).
It is pointed out that neurodegenerative diseases have common pathways leading to the disease state, and there is overlap in proteomic signatures for neurological diseases, such as PD and AD (De Iuliis et al., 2005). AD and PD also share common molecular pathways that include energy production, protection from oxidative damage, and synapse integrity (Sowell et al., 2009). Differential proteins summarized by Sowell et al. (2009) as being common to PD and AD and that were affected by dieldrin included Eno1, Actb, Stmn1, Cytc, Ldh, Apoe, Atp5b, and Mbp. The proteomic responses observed in these studies and LMB may be because of common pathways activated by stress or injury in the CNS and may be the result of apoptosis, inflammation, and oxidative damage that may precede neurotoxicity and neural damage.
Dieldrin Preferentially Modulates Transcripts Involved in DNA Repair, Cell Integrity, and Stress
Gene ontology identified DNA repair mechanisms such as base excision repair as being significantly affected by dieldrin. Base excision repair transcripts altered by dieldrin included mpg, FHA-HIT long isoform, ung, nth1, and parp1. Mpg is involved in repairing alkylation base damage, whereas ung removes uracils generated by deamination of cytosine. These transcripts and others involved in mitochondrial DNA damage are implicated in neurodegeneration (de Souza-Pinto et al., 2008). Genomic data in LMB hypothalamus are consistent with a previous study (Martyniuk et al., 2010a) that suggests that dieldrin impacts cell processes that involve DNA repair mechanisms, despite the fact that both studies differed in the genes involved. The difference in the transcripts regulated by dieldrin between both studies may be because of the difference in exposure duration, dose, and route of exposure. This highlights the importance of a broad bioinformatics-based approach to detect major cell processes altered by neuroactive pollutants.
Structural genes and proteins are also altered by dieldrin, and these play a significant role in cell integrity and survival. These included mapt (−2.3-fold), mbp mRNA (−1.7-fold), and Mbp protein (−1.2-fold); actb mRNA (−2.0-fold) and Actb protein (−2.0-fold); and col1a1 mRNA (1.7-fold) and Col1a protein (1.6-fold). The cytoskeleton and associated proteins play major roles that include maintenance of cell structure, cell migration, differentiation, and growth. In AD pathology, MAPT is abnormally hyperphosphorylated, resulting in abnormal microtubule associations and assembly. MAPT becomes misfolded and contributes to the formation of tangles in the CNS (Iqbal et al., 2009). Environmental exposure risks to pesticides may therefore include impacts on the structural integrity of cells, leading to disease pathology. It has been suggested that the process of oligodendrocyte dysfunction and myelin disruption can serve as a hallmark for neurodegenerative disease progression (Wenning et al., 2008). Dieldrin-induced changes in cell structure and stability may underlie early apoptotic events in the CNS.
In mammals, dieldrin has been shown to induce oxidative stress and generate reactive oxygen species (ROS) (Sharma et al., 2010). The generation of ROS is hypothesized to lead to increased mitochondrial outer membrane permeabilization. Indeed, many studies suggest that neurodegeneration is preceded by increased mitochondrial dysfunction (reviewed in Leuner et al., 2007). Dieldrin may be contributing to this process in the LMB hypothalamus. There was a reduction (20–50%) in the mRNA levels of genes involved in the general and oxidative stress response (p < 0.05), suggesting that the hypothalamus may be responding to ROS production. Transcripts included activator of heat-shock 90 kDa protein ATPase homolog 1, hsp14a, gpx4b, gpx8b, atmpk1 14b, gstt1, and gstk1. In a previous study, many stress genes were induced after dieldrin ip injection after 7 days (Martyniuk et al., 2010a), suggesting temporal genomic responses that include an early response of increasing mRNA for genes involved in protection against ROS, followed by a later reduction in the expression of these transcripts. However, genomic responses are complex and it is currently speculative whether there is a reduced oxidative stress response over time in the LMB hypothalamus.
CONCLUSIONS
There are two additional points that are raised by this study. In this study, many transcripts affected by dieldrin were involved in inflammation, DNA damage, stress, and repair as revealed by pathway analysis and functional enrichment analysis. These cellular responses are also corroborated by a previous study in LMB hypothalamus after acute dieldrin treatment (Martyniuk et al., 2010a). Dieldrin directly impacts GABAergic synaptic transmission and indirectly induces stress responses in the CNS. Additional studies may investigate how pesticides that affect GABAergic synaptic signaling may contribute to dysfunction of GABA-mediated processes in the CNS. This is important because GABA dysfunction is also associated with human neurological diseases (Chen and Yung, 2004).
The second point is that, given the importance of GABA in controlling reproduction in fish, disruptions along the hypothalamus-pituitary-gonadal axis may have long-term reproductive consequences. For example, esr-beta b mRNA levels were significantly decreased in the hypothalamus after dieldrin-fed treatments. In LMB ovary and testis, dieldrin also alters estrogen receptor (ER) isoform expression (Garcia-Reyero et al., 2006), decreasing esr-beta b and esr-beta a expression in the ovary and all three ER isoforms in the testis with 0.4 mg dieldrin/kg feed over a 120 day feeding study. Dieldrin has been shown to act as both an antagonist for human ER binding (Lemaire et al., 2006) as well as an agonist for membrane bound ER signaling (Watson et al., 2007). Interestingly, clinical stages of neurodegenerative diseases have been associated with changes in ER abundance in the mammalian brain (Kelly et al., 2008). Additional studies should evaluate how estrogens mediate molecular responses in the brain during environmental toxicant exposures.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health Pathway to Independence Award (K99 ES016767-01A1 to C.J.M.); Superfund Basic Research Program from the National Institute of Environmental Health Sciences (RO1 ES015449 to N.D.D. and D.S.B.).
Supplementary Material
Acknowledgments
We thank collaborators C. Diaz, S. Alvarez, and S. McClung at the Interdisciplinary Center for Biotechnology Research at University of Florida for assistance in developing the iTRAQ method for use in teleost models. The authors have no competing financial or nonfinancial interests to declare.
References
- Al-Shahrour F, Mínguez P, Vaquerizas JM, Conde L, Dopazo J. Babelomics: a suite of web-tools for functional annotation and analysis of group of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TP, Rumsby PC, Capleton AC, Rushton L, Levy LS. Pesticides and Parkinson’s disease—is there a link? Environ. Health Perspect. 2006;114:156–164. doi: 10.1289/ehp.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yung WH. GABAergic neurotransmission in globus pallidus and its involvement in neurologic disorders. Sheng Li Xue Bao. 2004;56:427–435. [PubMed] [Google Scholar]
- Corrigan FM, Wienburg CL, Shore RF, Daniel SE, Mann D. Organochlorine insecticides in substantia nigra in Parkinson’s disease. J. Toxicol. Environ. Health A. 2000;59:229–234. doi: 10.1080/009841000156907. [DOI] [PubMed] [Google Scholar]
- De Iuliis A, Grigoletto J, Recchia A, Giusti P, Arslan P. A proteomic approach in the study of an animal model of Parkinson’s disease. Clin. Chim. Acta. 2005;357:202–209. doi: 10.1016/j.cccn.2005.03.028. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Wilson DM, III, Stevnsner TV, Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair. 2008;7:1098–1109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming L, Mann JB, Bean J, Briggle T, Sanchez-Ramos JR. Parkinson's disease and brain levels of organochlorine pesticides. Ann. Neurol. 1994;36:100–103. doi: 10.1002/ana.410360119. [DOI] [PubMed] [Google Scholar]
- Flinn L, Bretaud S, Lo C, Ingham PW, Bandmann O. Zebrafish as a new animal model for movement disorders. J. Neurochem. 2008;106:1991–1997. doi: 10.1111/j.1471-4159.2008.05463.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N, Barber DS, Gross TS, Johnson KG, Sepúlveda MS, Szabo NJ, Denslow ND. Dietary exposure of largemouth bass to OCPs changes expression of genes important for reproduction. Aquat. Toxicol. 2006;78:358–369. doi: 10.1016/j.aquatox.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Griffitt RJ, Liu L, Kroll KJ, Farmerie WG, Barber DS, Denslow ND. Construction of a robust microarray from a non-model species largemouth bass, Micropterus salmoides (Lacepede), using pyrosequencing technology. J. Fish Biol. 2008;72:2354–2376. doi: 10.1111/j.1095-8649.2008.01904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsleichter J, Manire CA, Szabo NJ, Cortes E, Carlson J, Lombardi-Carlson L. Organochlorine concentrations in bonnethead sharks (Sphyrna tiburo) from four Florida estuaries. Arch. Environ. Contam. Toxicol. 2005;48:474–483. doi: 10.1007/s00244-003-0275-2. [DOI] [PubMed] [Google Scholar]
- Hatcher JM, Richardson JR, Guillot TS, McCormack AL, Di Monte DA, Jones DP, Pennell KD, Miller GW. Dieldrin exposure induces oxidative damage in the mouse nigrostriatal dopamine system. Exp. Neurol. 2007;204:619–630. doi: 10.1016/j.expneurol.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta. Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Muller JK, Price B, Ware A, Sepúlveda MS, Borgert CJ, Gross TS. Influence of seasonality and exposure on the accumulation and reproductive effects of p, p'-dichlorodiphenyldichloroethane and dieldrin in largemouth bass. Environ. Toxicol. Chem. 2007;26:927–934. doi: 10.1897/06-336r1.1. [DOI] [PubMed] [Google Scholar]
- Jorgenson JL. Aldrin and dieldrin: a review of research on their production, environmental deposition and fate, bioaccumulation, toxicology, and epidemiology in the United States. Environ. Health Perspect. 2001;109:113–139. doi: 10.1289/ehp.01109s1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthasamy AG, Kitazawa M, Yang Y, Anantharam V, Kanthasamy A. Environmental neurotoxin dieldrin induces apoptosis via caspase-3-dependent proteolytic activation of protein kinase C delta (PKCdelta): implications for neurodegeneration in Parkinson’s disease. Mol. Brain. 2008;1:12. doi: 10.1186/1756-6606-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JF, Bienias JL, Shah A, Meeke KA, Schneider JA, Soriano E, Bennett DA. Levels of estrogen receptors alpha and beta in frontal cortex of patients with Alzheimer’s disease: relationship to Mini-Mental State Examination scores. Curr. Alzheimer Res. 2008;5:45–51. doi: 10.2174/156720508783884611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin-induced oxidative stress and neurochemical changes contribute to apoptopic cell death in dopaminergic cells. Free Radic. Biol. Med. 2001;31:1473–1485. doi: 10.1016/s0891-5849(01)00726-2. [DOI] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy A, Kanthasamy AG. Dieldrin promotes proteolytic cleavage of poly(ADP-ribose) polymerase and apoptosis in dopaminergic cells: protective effect of mitochondrial anti-apoptotic protein Bcl-2. Neurotoxicology. 2004;25:589–598. doi: 10.1016/j.neuro.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Kovacech B, Zilka N, Novak M. New age of neuroproteomics in Alzheimer’s disease research. Cell Mol. Neurobiol. 2009;29:799–805. doi: 10.1007/s10571-009-9358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamai SL, Warner GF, Walker CH. Effects of dieldrin on life stages of the African catfish, Clarias gariepinus (Burchell) Ecotoxicol. Environ. Saf. 1999;42:22–29. doi: 10.1006/eesa.1998.1723. [DOI] [PubMed] [Google Scholar]
- Lemaire G, Mnif W, Mauvais P, Balaguer P, Rahmani R. Activation of alpha- and beta-estrogen receptors by persistent pesticides in reporter cell lines. Life Sci. 2006;79:1160–1169. doi: 10.1016/j.lfs.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Leuner K, Hauptmann S, Abdel-Kader R, Scherping I, Keil U, Strosznajder JB, Eckert A, Müller WE. Mitochondrial dysfunction: the first domino in brain aging and Alzheimer’s disease? Antioxid. Redox Signal. 2007;9:1659–1675. doi: 10.1089/ars.2007.1763. [DOI] [PubMed] [Google Scholar]
- Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer’s mice: understanding the interface between physiology and disease. PLoS ONE. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Alvarez S, McClung S, Villeneuve DL, Ankley GT, Denslow ND. Quantitative proteomic profiles of androgen receptor signaling in the liver of fathead minnows (Pimephales promelas) J. Proteome Res. 2009;8:2186–2200. doi: 10.1021/pr800627n. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Awad R, Hurley R, Finger TE, Trudeau VL. Glutamic acid decarboxylase 65, 67, and GABA-transaminase mRNA expression and total enzyme activity in the goldfish (Carassius auratus) brain. Brain Res. 2007;1147:154–166. doi: 10.1016/j.brainres.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Feswick A, Spade DJ, Kroll KJ, Barber DS, Denslow ND. Effects of acute dieldrin exposure on neurotransmitters and global gene transcription in largemouth bass (Micropterus salmoides) hypothalamus. Neurotoxicology. 2010a;31:356–366. doi: 10.1016/j.neuro.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Kroll KJ, Doperalski NJ, Barber DS, Denslow ND. Environmentally relevant exposure to 17α-ethinylestradiol affects the telencephalic proteome of male fathead minnows. Aquat. Toxicol. 2010b;98:344–353. doi: 10.1016/j.aquatox.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn diagrams. 2007 Available at http://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed October 15, 2009. [Google Scholar]
- Popesku JT, Martyniuk CJ, Mennigen J, Xiong H, Zhang D, Xia X, Cossins AR, Trudeau VL. The goldfish (Carassius auratus) as a model for neuroendocrine signaling. Mol. Cell Endocrinol. 2008;293:43–56. doi: 10.1016/j.mce.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Priyadarshi A, Khuder SA, Schaub EA, Shrivastava S. A meta-analysis of Parkinson’s disease and exposure to pesticides. Neurotoxicology. 2000;21:435–440. [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Satyanarayan S, Ramakant, Satyanarayan A. Bioaccumulation studies of organochlorinated pesticides in tissues of Cyprinus carpio. J. Environ. Sci. Health B. 2005;40:397–412. doi: 10.1081/pfc-200047572. [DOI] [PubMed] [Google Scholar]
- Sava V, Velasquez A, Song S, Sanchez-Ramos J. Dieldrin elicits a widespread DNA repair and antioxidative response in mouse brain. J. Biochem. Mol. Toxicol. 2007;21:125–135. doi: 10.1002/jbt.20165. [DOI] [PubMed] [Google Scholar]
- Sharma H, Zhang P, Barber DS, Liu B. Organochlorine pesticides dieldrin and lindane induce cooperative toxicity in dopaminergic neurons: role of oxidative stress. Neurotoxicology. 2010;31:215–222. doi: 10.1016/j.neuro.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Shi M, Caudle WM, Zhang J. Biomarker discovery in neurodegenerative diseases: a proteomic approach. Neurobiol. Dis. 2009;35:157–164. doi: 10.1016/j.nbd.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova D, Charbaut E, Delalande F, Poirier F, High AA, Parker F, Van Dorsselaer A, Duchesne M, Diu-Hercend A. Proteomic analysis of brain tissue from an Alzheimer’s disease mouse model by two-dimensional difference gel electrophoresis. Neurobiol. Aging. 2007;28:357–370. doi: 10.1016/j.neurobiolaging.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Sowell RA, Owen JB, Butterfield DA. Proteomics in animal models of Alzheimer's and Parkinson's diseases. Ageing Res. Rev. 2009;8:1–17. doi: 10.1016/j.arr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton C, Lin Y, Willett C. Zebrafish as a model for developmental neurotoxicity testing. Birth Defects Res. A Clin. Mol. Teratol. 2006;76:553–567. doi: 10.1002/bdra.20281. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Trudeau VL, Drouin G, Tostivint H, Ihrmann I, Vallarino M, Vaudry H. Molecular characterization and comparative localization of the mRNAs encoding two glutamic acid decarboxylases (GAD65 and GAD67) in the brain of the African lungfish, Protopterus annectens. J. Comp. Neurol. 2008;506:979–988. doi: 10.1002/cne.21552. [DOI] [PubMed] [Google Scholar]
- Watson CS, Bulayeva NN, Wozniak AL, Alyea RA. Xenoestrogens are potent activators of nongenomic estrogenic responses. Steroids. 2007;72:124–134. doi: 10.1016/j.steroids.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb O, Youdim MB. A model of MPTP-induced Parkinson’s disease in the goldfish. Nat. Protoc. 2007;2:3016–3021. doi: 10.1038/nprot.2007.393. [DOI] [PubMed] [Google Scholar]
- Weisskopf MG, Knekt P, O'Reilly EJ, Lyytinen J, Reunanen A, Laden F, Altshul L, Ascherio A. Persistent organochlorine pesticides in serum and risk of Parkinson disease. Neurology. 2010;74:1055–1061. doi: 10.1212/WNL.0b013e3181d76a93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenning GK, Stefanova N, Jellinger KA, Poewe W, Schlossmacher MG. Multiple system atrophy: a primary oligodendrogliopathy. Ann. Neurol. 2008;64:239–246. doi: 10.1002/ana.21465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.