Abstract
Cells use feedback to implement a diverse range of regulatory functions. Building synthetic feedback control systems may yield insight into the roles that feedback can play in regulation since it can be introduced independently of native regulation, and alternative control architectures can be compared. We propose a model for microbial biofuel production where a synthetic control system is used to increase cell viability and biofuel yields. Although microbes can be engineered to produce biofuels, the fuels are often toxic to cell growth, creating a negative feedback loop that limits biofuel production. These toxic effects may be mitigated by expressing efflux pumps that export biofuel from the cell. We developed a model for cell growth and biofuel production and used it to compare several genetic control strategies for their ability to improve biofuel yields. We show that controlling efflux pump expression directly with a biofuel-responsive promoter is a straightforward way of improving biofuel production. In addition, a feed forward loop controller is shown to be versatile at dealing with uncertainty in biofuel production rates.
Keywords: Feedback control, Biofuels, Biological control systems, Transcriptional regulation, Efflux pump, Feed forward loop
Introduction
Organisms use feedback to respond to changing conditions, optimize the use of resources, and maintain homeostasis. The versatility of feedback control in gene regulation is evidenced by the frequency with which positive and negative feedback loops appear in regulatory networks (Alon 2007).
Examples of synthetic feedback control systems have been successfully implemented. Applications include a population control circuit (You et al. 2004) and a controllable yeast mating pathway (Bashor et al. 2008). Several studies focus on the modular nature of feedback control (Win and Smolke 2008; Topp and Gallivan 2007; Goldberg et al. 2009). In particular, work on controller design has used toggle switches (Kobayashi et al. 2004; Anesiadis et al. 2008) and synthetic promoters (Farmer and Liao 2000) to control gene expression in response to a sensed signal. A systematic study of the properties of alternative control strategies may lend insight into how different feedback architectures can be used to regulate gene expression.
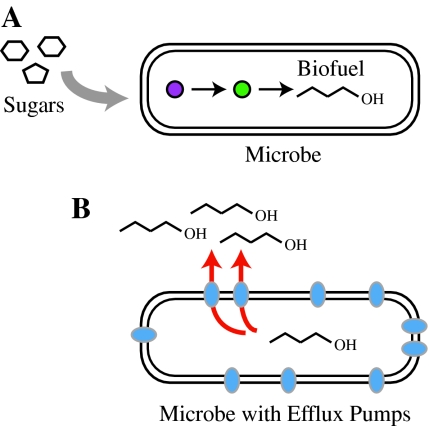
Microbial biofuel production is one area where synthetic feedback regulation has the potential for great impact. Biofuels are a promising form of alternative energy that may replace existing fuel sources such as gasoline, jet fuel, or diesel without requiring engine modifications or additional infrastructure development (Savage et al. 2008; Fortman et al. 2008). Typical biofuel production processes start with cellulose from plant matter. Using enzymes and chemical pretreatment processes, the cellulose is broken down into sugars like glucose or pentose. These sugars are then fed to microorganisms (such as Saccharomyces cerevisiae or Escherichia coli) that convert the sugar into biofuel (Fig. 1a).
Fig. 1.
a Biofuel production using microbes to convert sugar into fuel. b Efflux pumps can be used to export biofuel out of the cell
However, the fuel synthesis stage can be limited by the fact that biofuels are often toxic to microbial growth. Biofuel-producing cells will eventually reach a point where the amount of fuel they produce inhibits their growth, placing a fundamental limit on the amount of biofuel that can be generated (Jones and Woods 1986).
One mechanism for dealing with toxicity is to export the fuel molecules using efflux pumps. These pumps are protein complexes in the cell membrane that recognize toxic substrates and expel them (Fig. 1b) (Paulsen et al. 1996). Once a toxin is sensed, a channel in the membrane opens in an iris-like fashion and the toxin is pushed out using the electrochemical gradient across the cell membrane (Nikaido 1994; Bavro et al. 2008; Symmons et al. 2009). Efflux pumps provide resistance to a wide variety of substrates, but there are certain pumps that are specifically resistant to solvents (Ramos et al. 2002). For example, the toluene tolerance genes from the soil bacterium Pseudomonas putida recognize pentane, hexane, butanol, propanol, toluene, and other solvents (Kieboom et al. 1998).
Efflux pumps provide a mechanism for controlling the level of biofuel within a cell. However, pump overexpression can also be toxic (Wagner et al. 2007). When too many pumps are produced they overload the membrane insertion machinery, change the membrane composition, and inhibit growth (Wagner et al. 2008).
Thus, a genetic feedback loop that controls efflux pump levels to balance toxicity due to biofuel production and toxicity due to pump overexpression may significantly improve biofuel yields.
In this paper we develop a model for cell growth that incorporates the detrimental effects of toxicity from biofuels and pump overexpression. We compare several biologically realistic control strategies for improving fuel production. We find that some control strategies are more robust than others, producing high biofuel yields for a wide range of controller parameters. Controller performance characteristics are also explored, comparing temporal response times and the controllers’ ability to deal with uncertainty in the biofuel production rate. Our findings highlight how ideas from control theory can be used in combination with synthetic control strategies to engineer and design genetic feedback systems. Understanding how feedback architecture design affects gene regulation will extend the set of tools that synthetic biology researchers have at their disposal.
Materials and methods
Butanol toxicity
E. coli strain BW25113  (Baba et al. 2006) was grown overnight in LB medium and then diluted 1:100 in M9 minimal medium (per liter: 30 g Na2HPO4, 15 g KH2PO4, 5 g NH4Cl, 2.5 g NaCl, 15 mg CaCl2, 10 ml 20% glucose, 1 ml 1M MgSO4, 0.1 ml 0.5% thiamine). The culture was aliquoted into a 96-well plate with 100 μl per well and n-butanol was added to the individual wells with each condition tested in triplicate. Optical density (absorbance at 600 nm) readings were collected every 10 min using a plate reader (SpectraMax Plus) over the course of 20 h at 37°C with linear shaking. Data are normalized relative to the maximum optical density of a culture grown without butanol.
(Baba et al. 2006) was grown overnight in LB medium and then diluted 1:100 in M9 minimal medium (per liter: 30 g Na2HPO4, 15 g KH2PO4, 5 g NH4Cl, 2.5 g NaCl, 15 mg CaCl2, 10 ml 20% glucose, 1 ml 1M MgSO4, 0.1 ml 0.5% thiamine). The culture was aliquoted into a 96-well plate with 100 μl per well and n-butanol was added to the individual wells with each condition tested in triplicate. Optical density (absorbance at 600 nm) readings were collected every 10 min using a plate reader (SpectraMax Plus) over the course of 20 h at 37°C with linear shaking. Data are normalized relative to the maximum optical density of a culture grown without butanol.
Efflux pump toxicity
The solvent resistance genes srpABC (Kieboom et al. 1998) from Pseudomonas putida S12 (ATCC 700801) were cloned into the pTYL plasmid (p15A ori, lacI q, kan r) using the forward primer 5′-GTGAGACAGATACGATCCCC-3′ and reverse primer 5′-GTTTTGACTCACGCTCC-3′ with cloning sites appended to both primers. Overnight cultures of E. coli cells containing the plasmid were diluted 1:100 into 5 ml of LB with 30 μg/ml kanamycin and induced with IPTG (isopropyl-1-thio-3-D-galactoside), where full induction of the lacUV5 promoter occurs at 100 μM IPTG. The optical density of the induced cultures was measured after 8 h of growth at 37°C with orbital shaking and normalized relative to growth of a culture containing the empty pTYL vector. For parameter estimation we make the simplifying assumption that pump expression levels vary linearly with IPTG.
Parameter fitting
Model parameters were fitted to experimental data by minimizing the least squares difference between the model and experimental data. For a parameter p
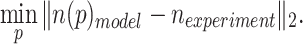 |
Global sensitivity analysis
Global sensitivity analysis was conducted using a variance-based method to find the sensitivity and total sensitivity indices for all controller parameters (Saltelli et al. 2008). The controller parameters were used as inputs and the final biofuel concentration (b e(T); T = 100 h) was used as the output. Monte Carlo distributions for the parameter values were generated by finding the combination of parameters that maximized biofuel yield. For each controller design a gradient ascent algorithm was started from 100 random initial conditions and the maximum and the statistical variation around it were used to generate a set of Monte Carlo points for the sensitivity analysis.
Simulations
All simulations were conducted in Matlab (the MathWorks, Inc.) using the ode45 solver and custom analysis software.
Noise in the biofuel production rate was simulated using an Ornstein-Uhlenbeck process with a log normal distribution with μ = 1, σ = 0.35, and τ = 1 h (Dunlop et al. 2008; Rosenfeld et al. 2005). This noisy signal, η(t), was included in the model by replacing the intracellular biofuel equation with 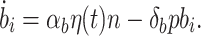 The same noise signal was used to compare the performance of all four controllers. Noise simulations were repeated 5,000 times and results were averaged.
The same noise signal was used to compare the performance of all four controllers. Noise simulations were repeated 5,000 times and results were averaged.
Results
Cell growth and biofuel production model
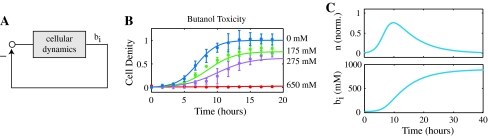
Without any control of biofuel production, the host microbe will make biofuel at the maximum level that the metabolically engineered system allows. When biofuel levels are high enough they will be toxic to cell growth. This negative feedback loop, shown in Fig. 2a, reduces biofuel yields.
Fig. 2.
Biofuel production without export. a Cells produce intracellular biofuel b i, which inhibits their growth, and consequently limits biofuel production. b Experimental growth curves (dots and error bars) are fit to the model in Eq. 1 to estimate the model parameters αn and δn (model fits shown as solid lines). The amount of butanol added to the culture is indicated in the figure, where experimental data is for exogenously added butanol in cells without intracellular butanol production. c Simulation of cell growth and biofuel production. Growth inhibition occurs when biofuel produced by the cells reaches toxic levels. Simulation parameters are αn = 0.66 1/h, δn = 0.91 1/M h, and αb = 0.1 1/h
Cell growth is modeled as in (You et al. 2004) by
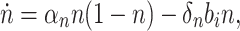 |
1 |
where n is the normalized cell density; a value of n = 1 is the maximum level of cell growth supported by the nutrients supplied in the growth medium, assuming no toxicity. This equation represents the standard growth curve of a microbial culture (Fig. 2b). The parameter αn describes the specific growth rate of the cells. b i is the intracellular biofuel concentration. δn is the biofuel toxicity coefficient, which is biofuel-specific since some compounds are much more toxic than others (Kieboom et al. 1998).
We estimate the parameters αn and δn directly using experimental data. Setting b i = 0 we fit the model in Eq. 1 to the experimental growth curve data without biofuel. αn is estimated to be 0.66 1/h, equivalent to a 1 h cell division time (ln(2)/αn). Figure 2b shows the experimental data and model fits for growth in the absence of biofuel (blue dots and line, respectively).
Experimental data for exogenously added butanol was used to estimate δn. Overall cell growth was inhibited when butanol was added. For the levels of butanol indicated in the figure (the values of b i), δn was estimated by minimizing the difference between experimental data and the modeled system. The model fit is compared to the experimental results in Fig. 2b. In reality, exogenous butanol levels will be higher than the corresponding intracellular levels that cells experience, however, for simplicity we assume these values are the same when estimating δn.
Metabolically engineered microbes can currently produce biofuels like butanol in small quantities (Atsumi et al. 2008; Steen et al. 2008). Although current production levels in E. coli and S. cerevisiae are not toxic to cell growth, as yield improves growth inhibition will become a serious limitation (Jones and Woods 1986).
Initially, we assume biofuel is produced in proportion to cell density:
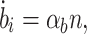 |
where b i is the intracellular level of biofuel and αb is the biofuel production rate. This model makes the simplifying assumption that biofuel cannot diffuse through the cell membrane. Figure 2c shows a simulation of the biofuel-producing cells. The cells begin to produce biofuel, which inhibits their growth, eventually killing the entire population. Similar effects have been observed in the butanol-producing microbe Clostridium acetobutylicum (Van Der Westhuizen et al. 1982; Jones and Woods 1986).
In summary, cell growth and biofuel production are modeled as
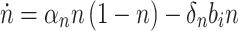 |
2 |
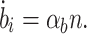 |
3 |
The system is at equilibrium when 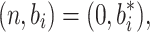 where
where 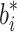 is any value of b
i; the exact value achieved depends on the initial conditions.
is any value of b
i; the exact value achieved depends on the initial conditions.
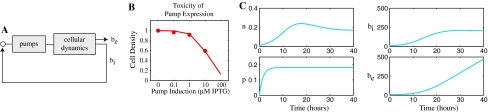
Model for biofuel export using efflux pumps
When efflux pumps are used to export biofuel from the cell the extracellular level of biofuel (b e) will increase, allowing intracellular biofuel levels to remain low (Fig. 3a). However, efflux pump expression is also limited by biological considerations. If pumps are expressed too highly they consume cellular resources that are necessary for cells to function properly and can change the membrane composition (Wagner et al. 2007). Thus, there are two limits on pump expression: levels that are too low will cause biofuel toxicity and kill the cells, while overexpression will also result in cell death. An intermediate level of pump expression can balance these two competing factors.
Fig. 3.
Biofuel production with constant efflux pump expression. a Expression of pumps can reduce intracellular biofuel levels, reducing toxicity. b Overexpressing efflux pumps inhibits cell growth. Experimental data (dots) and a model (line) of this phenomenon for the srpABC efflux pump from Pseudomonas putida S12. γp = 0.14 from the model fits; note that this value is specific to srpABC in E. coli and other pumps and organisms may have different toxicity profiles. c Simulation of cell growth and biofuel production with export using efflux pumps. Simulation parameters are the same as listed in Fig. 2 with δb = 0.5 1/M h, k p = 0.12 1/h, βp = αn, γp = 0.14
The growth model from Eq. 1 is extended to include toxicity due to pump expression:
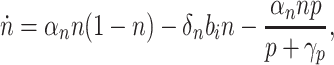 |
4 |
where p is the concentration of efflux pump proteins and γp sets the toxicity threshold.
In the absence of biofuel, measurements of the cell density at stationary phase as pump expression is induced are shown in Fig. 3b. Under these conditions, 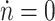 and b
i = 0, thus from Eq. 4 we obtain the relationship
and b
i = 0, thus from Eq. 4 we obtain the relationship
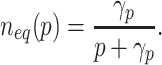 |
γp was estimated by minimizing the error between the experimental data and the model and the results are shown in Fig. 3b.
The biofuel production model is extended to include biofuel export and the intra- and extracellular biofuel levels are modeled as
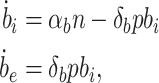 |
where δb is the biofuel export rate per pump.
The pump dynamics per cell are given by
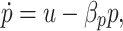 |
where u is the control input, which describes both transcription and translation of the proteins in the efflux pump complexes and is discussed in more detail in the next section. βp is the protein degradation rate, which includes both active protein degradation and dilution due to cell growth. Since there is no evidence that efflux pumps are actively degraded, βp is assumed to be proportional to the growth rate (Rosenfeld et al. 2005).
The simplest form of pump expression places the genes under the control of a constitutive promoter. In this situation pump expression is constant. Figure 3c shows a simulation of the biofuel export system with constant pump expression (u = k p). Note that the number of cells stabilizes at a constant value rather than decaying to zero, as it did without efflux pumps. The levels of intracellular biofuel also stabilize, and extracellular levels of biofuel rise. This constant biofuel production following stationary phase has been observed experimentally (e.g., Supplementary Fig. 2 from Atsumi et al. 2008).
In summary, the following dynamics describe cell growth, biofuel production, and export:
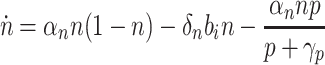 |
5 |
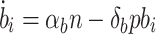 |
6 |
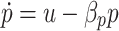 |
7 |
and the output is given by
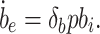 |
8 |
Efflux pump controller models
We investigate several strategies for controlling pump expression to maximize biofuel yield.
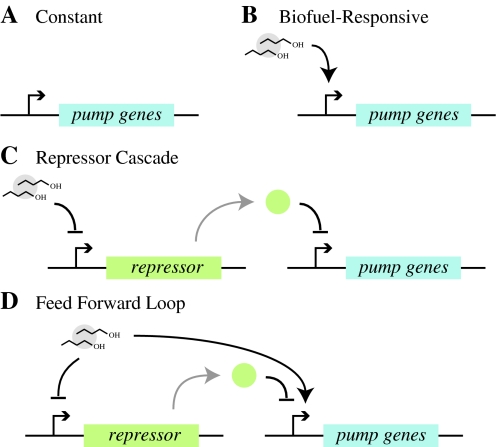
Four alternative controllers are shown in Fig. 4. The simplest controller is a constitutive promoter (Fig. 4a) where pumps are expressed at a constant rate
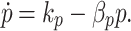 |
The strength of efflux pump expression is represented by k p. It is possible to tune the transcription and translation rates of a gene by choosing promoters with alternative strengths (Ellis et al. 2009) or by modifying the ribosome binding site strength (Salis et al. 2009). Thus, k p can be changed to tune biofuel production. Because the pump expression is not related to biofuel levels we expect this controller will be suboptimal, but it is the easiest system to design and test.
Fig. 4.
Different schemes for controlling efflux pump expression. a With constant control the pump expression is driven by a constitutive promoter. b A promoter regulated by a transcription factor that senses biofuel is used to drive efflux pump expression directly. c In the repressor cascade biofuel represses production of a transcription factor, which in turn represses efflux pump expression. d The feed forward loop uses both biofuel and a transcription factor to control pump expression, while the transcription factor is also controlled by a biofuel-responsive promoter
The second controller actuates pump expression in response to intracellular biofuel levels (Fig. 4b). There are examples of biofuel-responsive promoters that are regulated by transcription factors that sense biofuel, e.g. (Willardson et al. 1998). Such a promoter could be used to drive expression of efflux pump genes. Typical promoters respond linearly within a range, but saturate above a certain threshold of sensed substrate. We model this class of promoters by
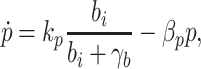 |
where γb defines the threshold level above which the promoter’s response saturates.
Biofuel can also be used indirectly to control pump expression. For example, Fig. 4c shows a controller where a biofuel-sensing repressor regulates expression of a transcription factor that controls efflux pump expression. This repressor cascade strategy has the advantage of using a native regulatory system to control pump expression directly. Efflux pumps are often controlled by a single repressor (Paulsen et al. 1996), where relief of repression leads to efflux pump expression. These native regulatory proteins could be used for control. This type of controller is modeled by
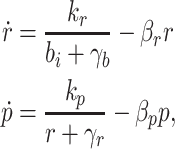 |
where r is the repressor protein concentration, k r and k p are the production rates for the repressor and pump proteins and γb and γr specify the threshold values of biofuel and the repressor protein for the promoters.
The final control strategy we consider is similar to the repressor cascade, but uses a feed forward loop to control pump expression, as shown in Fig. 4d. The feed forward loop discussed here is a network motif that appears frequently in gene regulation (Mangan and Alon 2003). This should be distinguished from the class of feed forward loops used in control theory, where a system is engineered to respond to input signals in a predetermined way based on a model of the dynamics (Astrom and Murray 2008). In the genetic feed forward loop shown in Fig. 4d, biofuel inhibits expression of the pump repressor and simultaneously activates expression of the efflux pump genes. This controller can turn on pump expression to a small extent and then, with a delay, fully commit to expressing the pumps; there is no corresponding delay in turning off gene expression so the process can easily be shut down (Mangan and Alon 2003; Wall et al. 2005). Because pump expression can be toxic, this control strategy may have a distinct advantage since the genes can be turned on at a low level in response to the presence of biofuels and then turned on more completely only if necessary. The controller is modeled by
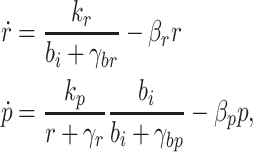 |
where the parameters are the same as defined in the repressor cascade, but γbr and γbp set the thresholds for biofuel sensed at the promoter for the repressor and the pump genes, respectively.
For each of these controllers we calculate the maximum amount of biofuel that can be produced, how sensitive this maximum production level is to changes in the controller parameters (i.e., how precise do the biological parts need to be), and how quickly controller responds to changes.
Controller performance and sensitivity
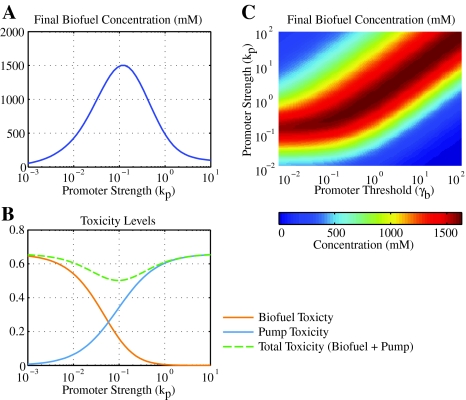
For a constant controller Fig. 5a and b show how the final biofuel yield (b e(T); T = 100 h) varies as a function of the promoter strength. T = 100 h was chosen to allow sufficient time in stationary phase; resulting trends are similar for other values of T. At low levels of pump expression the biofuel is not exported and biofuel toxicity limits yields. When pumps are expressed highly, the toxicity of pump overexpression limits cell growth. Thus, biofuel yield is maximized at an intermediate promoter strength where biofuel and pump toxicity effects are balanced. This maximal level is significantly higher than what could be reached without any biofuel export (Fig. 2c).
Fig. 5.
Toxicity tradeoffs in biofuel production. a Final biofuel concentration (b e(T); T = 100 h) as a function of promoter strength for constitutive pump expression. b Toxicity of biofuel (orange line) and pumps (blue line) at time T is defined by δn b i(T) and αn p(T)/(p(T) + γp), respectively. The sum of the two toxicity curves is also shown (dashed line). Note that the maximum b e(T) in a occurs at the minimum of the total toxicity curve. c Biofuel production as a function of promoter strength and threshold for a biofuel-responsive promoter. Parameters are the same as in Fig. 3 unless noted
For a biofuel-responsive promoter, Fig. 5c shows how the final biofuel yield depends on the parameters k p and γb. Again, there is a trade off between toxicity due to biofuel and toxicity due to pump overexpression. However, the wide plateau of high biofuel production levels suggests that a broad range of biofuel sensors will allow maximal biofuel yield provided the promoter strength can be tuned. This is an encouraging finding since the parameters associated with biofuel sensors (such as the saturation value) can be more complicated to tune than promoter strengths. It is interesting to note that the maximum biofuel yield is not significantly higher with the biofuel-responsive promoter than it is with constitutive expression.
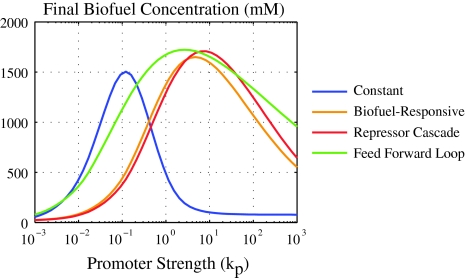
The dependence on k p of the four controllers is compared in Fig. 6. All four controllers are capable of producing similar maximal levels of biofuel, with the three non-constitutive promoters slightly outperforming the constant controller. However, some controllers are more sensitive to the system parameters than others. For example, the constant controller produces high levels of biofuel for only a small set of k p values compared to the other controllers, as evidenced by how “sharp” the peak is. A wider peak indicates that the promoter strength does not need to be tuned as precisely to reach near-maximal biofuel yields. The feed forward loop controller is particularly insensitive to changes in k p, with a large range of promoter strengths giving high biofuel yields.
Fig. 6.
Overall biofuel yield as a function of k p. The controller parameters that give the maximum biofuel yield are: Constant, k p = 0.12; Biofuel-responsive, k p = 4.34, γb = 5.72; Repressor cascade, k p = 7.23, γr = 0.19, k r = 5.01, γb = 0; Feed forward loop, k p = 2.64, γr = 0.016, γbp = 5.89, k r = 0.048, γbr = 0. Other parameters are the same as those listed in Fig. 3
Table 1 shows global sensitivity indices for each of the four controllers’ parameters. The S i sensitivity indices rank which parameters cause the greatest deviation from the maximum b e(T). A larger S i value indicates that a change in this parameter will have a large change on b e(T). The total sensitivity index S Ti quantifies how interactions between this parameter and all other model parameters contribute to changes in b e(T). The constant controller only has one parameter so its sensitivity analysis results are trivial. The biofuel-responsive controller parameters contribute equally to model variation, though when secondary interactions are considered k p plays a greater role. In the repressor cascade controller the promoter strengths, k p and k r, are more sensitive to parameter variation than the thresholds γr and γb. The feed forward loop controller places a more even balance on the role of its five parameters.
Table 1.
Sensitivity and total sensitivity indices for controller parameters
| S i | S Ti | |
|---|---|---|
| Constant | ||
| k p | 1.00 | 1.00 |
| Biofuel-responsive | ||
| k p | 0.26 | 0.76 |
| γb | 0.25 | 0.60 |
| Repressor cascade | ||
| k p | 0.45 | 0.72 |
| γr | 0.07 | 0.11 |
| k r | 0.23 | 0.45 |
| γb | 0.01 | 0.01 |
| Feed forward loop | ||
| k p | 0.09 | 0.31 |
| γr | 0.07 | 0.26 |
| γbp | 0.21 | 0.47 |
| k r | 0.11 | 0.38 |
| γbr | 0.04 | 0.21 |
Controller performance characteristics
The performance characteristics of a controller, such as response time and ability to handle noise, become particularly important when there is uncertainty in the system. Under optimized conditions the four controllers produced similar biofuel yields, but do these results persist when the system deviates from optimal?
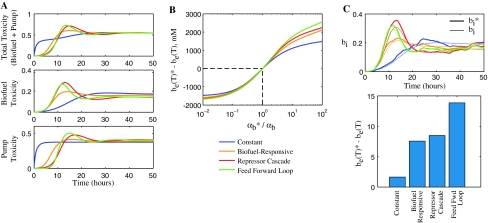
Figure 7a shows toxicity results for the four controllers as a function of time using the parameter values that maximize b e(T). All four controllers ultimately achieve the same total toxicity level, however, they reach this point by different paths. The constant controller turns on pump production immediately, even though there is no biofuel present. This causes an immediate increase in pump toxicity, though the pumps are present as soon as biofuel starts to appear and thus biofuel toxicity is smoothly controlled. The other three controllers all delay pump expression until biofuel is sensed. This gives the cell population a period of time without any toxicity mediating its growth, but pump production must be turned on quickly in response to sensed biofuel. The repressor cascade and feed forward loop controllers produce similar results, but the feed forward loop has a faster temporal response, turning on and then settling to a final expression level more rapidly. Although the biofuel-responsive promoter is not quite as responsive as the more complicated controllers, it still does a good job of responding to biofuel levels and is a good choice for a straightforward control system.
Fig. 7.
Controller performance comparisons. a Toxicity as a function of time. The total toxicity is the sum of the biofuel and pump toxicity, as defined in Fig. 5b. b Difference between the actual final biofuel concentration 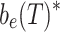 and the nominal b
e(T) given biofuel production rates
and the nominal b
e(T) given biofuel production rates 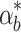 and αb, respectively. c Intracellular biofuel levels given a noisy biofuel production rate.
and αb, respectively. c Intracellular biofuel levels given a noisy biofuel production rate. 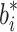 (thick line) and the nominal b
i (thin line) are shown for the four controllers. Lower plot shows the average difference between the actual and nominal final biofuel concentrations. All plots use the nominal parameter values given in Fig. 6 unless otherwise listed
(thick line) and the nominal b
i (thin line) are shown for the four controllers. Lower plot shows the average difference between the actual and nominal final biofuel concentrations. All plots use the nominal parameter values given in Fig. 6 unless otherwise listed
If the actual biofuel production rate 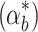 is different from the nominal conditions for which the controller parameters have been optimized (αb), the controller selection becomes more important. Figure 7b shows that the four controllers handle differences in the biofuel production rate to a different extent. When
is different from the nominal conditions for which the controller parameters have been optimized (αb), the controller selection becomes more important. Figure 7b shows that the four controllers handle differences in the biofuel production rate to a different extent. When 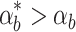 the system produces more biofuel than in the nominal case. Because the constant controller does not sense biofuel levels directly, the cells do not adjust the pump expression levels accordingly and miss out on potential yield. The feed forward loop controller does the best job responding to the changed conditions, exploiting the increase in biofuel production to achieve high biofuel yields. This performance advantage is likely due to the quick response times of the feed forward loop controller. When
the system produces more biofuel than in the nominal case. Because the constant controller does not sense biofuel levels directly, the cells do not adjust the pump expression levels accordingly and miss out on potential yield. The feed forward loop controller does the best job responding to the changed conditions, exploiting the increase in biofuel production to achieve high biofuel yields. This performance advantage is likely due to the quick response times of the feed forward loop controller. When 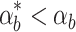 the system produces less biofuel than in the nominal case and none of the systems have good yields because there is less biofuel around.
the system produces less biofuel than in the nominal case and none of the systems have good yields because there is less biofuel around.
These controller performance differences can also matter if there is noise in the system. For example, if 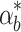 varies over time due to extrinsic or intrinsic noise (Elowitz et al. 2002) in the system a good controller will track relevant changes in internal biofuel levels and adjust pump expression accordingly. For example, Fig. 7c shows the internal biofuel level for the system with four different controllers given the same noisy
varies over time due to extrinsic or intrinsic noise (Elowitz et al. 2002) in the system a good controller will track relevant changes in internal biofuel levels and adjust pump expression accordingly. For example, Fig. 7c shows the internal biofuel level for the system with four different controllers given the same noisy 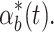 Averages of many noise simulations show that the feed forward loop controller is consistently better at handling noise than the other controllers. These trends are consistent with the results from Fig. 7b. Thus, deviations from the nominal system are handled differently by the four controllers and the constant controller consistently shows the worst performance, while the feed forward loop shows the best.
Averages of many noise simulations show that the feed forward loop controller is consistently better at handling noise than the other controllers. These trends are consistent with the results from Fig. 7b. Thus, deviations from the nominal system are handled differently by the four controllers and the constant controller consistently shows the worst performance, while the feed forward loop shows the best.
Discussion
We have developed a model for microbial biofuel production that includes the toxic effects of both biofuel production and overexpression of export machinery. The model is used to compare alternative strategies for controlling efflux pump expression. Feedback loops can be engineered using synthetic biology to address many of the same problems that are encountered in traditional engineering fields: mitigating uncertainty, tracking changing conditions over time, and altering the fundamental dynamics of a process (Astrom and Murray 2008). Synthetic feedback loops can be inserted independent of native regulatory networks which are often complicated, multi-layer regulatory systems. In addition, the feedback control mechanism can be treated as a biological part and exchanged for another controller if different performance characteristics are required. This work directly compares alternative control strategies in a synthetic circuit design.
For the microbial biofuel production system, we propose a control strategy that uses efflux pumps to export biofuels and mitigate toxicity. All controller designs must balance the competing toxicity from biofuels and efflux pump overexpression. In general we find that there is an intermediate optimum for maximizing biofuel production that balances the two toxic effects.
All controllers tested are capable of producing similar maximal levels of biofuel, however, the controllers that respond to sensed biofuel levels have an advantage, which becomes more pronounced when there is system uncertainty. The biofuel-responsive controller is the simplest system that senses biofuel and is thus the most straightforward feedback control system to implement. We show that the temporal response characteristics of the feed forward loop controller allow it to achieve high biofuel yields even in the presence of variable biofuel production rates.
Although conceptually promising, the feed forward loop controller has the experimental limitation that a combinatorial promoter that responds to both biofuel-responsive and regulatory transcription factors may be challenging to engineer. An alternative to a transcriptionally regulated feed forward loop is an anti-sense strategy where the biofuel-responsive promoter activates both the pump genes and an anti-sense sequence that degrades pump mRNA (Isaacs et al. 2006). This strategy uses the feed forward loop architecture, but eliminates the need for a combinatorial promoter.
In the future it would also be interesting to explore control strategies that modulate biofuel production in addition to its export. In addition, combining active control strategies with chassis engineering through the use of stoichiometric model predictions (Burgard et al. 2003) may have complementary benefits.
This work highlights how a control theory approach can be used to gain insight into synthetic biology design, considering realistic biological control strategies in combination with classic control theory methods. Controller architectures and corresponding analysis methods should be broadly applicable in synthetic biology design.
Acknowledgments
This work was conducted at the Department of Energy Joint BioEnergy Institute (http://www.jbei.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, and through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alon U. An introduction to systems biology: design principles of biological circuits. Boca Raton: Chapman & Hall/CRC; 2007. [Google Scholar]
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10(5):255–266. doi: 10.1016/j.ymben.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Astrom KJ, Murray RM. Feedback systems: an introduction for scientists and engineers. Princeton: Princeton University Press; 2008. [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–U13. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, Article number 2006.0008. doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed]
- Bashor CJ, Helman NC, Yan SD, Lim WA. Using engineered scaffold interactions to reshape map kinase pathway signaling dynamics. Science. 2008;319(5869):1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- Bavro VN, Pietras Z, Furnham N, Perez-Cano L, Fernandez-Recio J, Pei XY, Misra R, Luisi B. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30(1):114–121. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD. OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Dunlop MJ, Cox RS, Levine JH, Murray RM, Elowitz MB. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat Genet. 2008;40(12):1493–1498. doi: 10.1038/ng.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27(5):465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18(5):533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- Fortman JL, Chhabra S, Mukhopadhyay A, Chou H, Lee TS, Steen E, Keasling JD. Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 2008;26(7):375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Goldberg SD, Derr P, DeGrado WF, Goulian M. Engineered single- and multi-cell chemotaxis pathways in E. coli. Mol Syst Biol. 2009;5:283. doi: 10.1038/msb.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24(5):545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50(4):484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom J, Dennis JJ, de Bont JAM, Zylstra GJ. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273(1):85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101(22):8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100(21):11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Prevention of drug access to bacterial targets—permeability barriers and active efflux. Science. 1994;264(5157):382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60(4):575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Gallegos M-T, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307(5717):1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27(10):946–950, October, ISSN 1087-0156. doi:10.1038/nbt.1568. URL 10.1038/nbt.1568 [DOI] [PMC free article] [PubMed]
- Saltelli A, Ratto M, Andres T, Campolongo F, Cariboni J, Gatelli D, Saisana M, Tarantola S. Global sensitivity analysis: the primer. Chichester: Wiley; 2008. [Google Scholar]
- Savage DF, Way J, Silver PA. Defossiling fuel: how synthetic biology can transform biofuel production. ACS Chem Biol. 2008;3(1):13–16. doi: 10.1021/cb700259j. [DOI] [PubMed] [Google Scholar]
- Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Factories. 2008;7:36. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA. 2009;106(17):7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129(21):6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Westhuizen A, Jones DT, Woods DR. Autolytic activity and butanol tolerance of Clostridium acetobutylicum. Appl Environ Microbiol. 1982;44(6):1277–1281. doi: 10.1128/aem.44.6.1277-1281.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Baars L, Ytterberg AJ, Klussmeier A, Wagner CS, Nord O, Nygren PA, van Wijk KJ, de Gier JW. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteomics. 2007;6(9):1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Hogbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci USA. 2008;105(38):14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Dunlop MJ, Hlavacek WS. Multiple functions of a feed-forward-loop gene circuit. J Mol Biol. 2005;349(3):501–514. doi: 10.1016/j.jmb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Willardson BM, Wilkins JF, Rand TA, Schupp JM, Hill KK, Keim P, Jackson PJ. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64(3):1006–1012. doi: 10.1128/aem.64.3.1006-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322(5900):456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LC, Cox RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428(6985):868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]