Abstract
Comprehensive identification of all functional elements encoded in the human genome is a fundamental need in biomedical research. Here, we present a comparative analysis of the human, mouse, rat and dog genomes to create a systematic catalogue of common regulatory motifs in promoters and 3′ untranslated regions (3 ′ UTRs). The promoter analysis yields 174 candidate motifs, including most previously known transcription-factor binding sites and 105 new motifs. The 3′-UTR analysis yields 106 motifs likely to be involved in post-transcriptional regulation. Nearly one-half are associated with microRNAs (miRNAs), leading to the discovery of many new miRNA genes and their likely target genes. Our results suggest that previous estimates of the number of human miRNA genes were low, and that miRNAs regulate at least 20% of human genes. The overall results provide a systematic view of gene regulation in the human, which will be refined as additional mammalian genomes become available.
Comparative genomics provides a powerful approach for the systematic discovery of functional elements in the human genome1–8, by virtue of their evolutionary conservation across related species. Recent studies have revealed that most functional elements are non-protein-coding; these are likely to include regulatory signals, RNA genes and structural elements. Although the largest and most conserved elements are readily identified9, the vast majority of non-coding functional elements remain unknown. It has been particularly difficult to recognize short regulatory sequences, such as the DNA binding sites for transcription factors.
Our goal here is to create a catalogue of ‘common regulatory motifs’, by which we mean short, functional sequences (typically, 6–10 bases) that are used many times in a genome. Although reliably recognizing each individual occurrence of a motif requires evolutionary comparison with many species10, it should be possible to define the motifs themselves with many fewer species based on their multiple conserved occurrences in the genome. Recent studies of four related yeast species have shown the potential power of this approach in a relatively small genome11,12.
To apply this approach to the vastly larger human genome, we focused on two limited subsets that are likely to be highly enriched for regulatory elements: promoter regions and 3′ UTRs of protein-coding genes. We aligned these regions across the human, mouse, rat and dog genomes13–15, and searched for highly conserved, frequently occurring sequence patterns. In promoter regions, we discovered 174 candidate motifs, including most previously known transcription-factor binding sites and 105 new motifs that seem to be functional based on multiple criteria. In 3′ UTRs, we discovered 106 motifs likely to be involved in post-transcriptional regulation. Nearly one-half of these are associated with miRNAs, demonstrating the extraordinary importance of this recently discovered regulatory mechanism16. Our results suggest that previous estimates of human miRNA number were low17, and that at least 20% of human genes are probably regulated by miRNAs. More broadly, our results suggest that it will be possible to construct a comprehensive catalogue of common regulatory motifs in the human genome as more species become available.
Alignment of promoters and 3′ UTRs
We first constructed genome-wide alignments for the four mammalian species in promoter regions and 3′ UTRs. We studied a total of ~17,700 well-annotated genes from the RefSeq database18, with a single reference messenger RNA selected for each gene. Each promoter region was defined as the non-coding sequence contained within a 4-kilobase (kb) window centred at the annotated transcriptional start site (TSS), and each 3′ UTR was defined based on the annotation of the reference mRNA (see Methods). The promoter set contains ~68 megabases (Mb) of sequence, whereas the 3′-UTR set comprises ~15 Mb. In addition, we defined a control set comprising ~123 Mb of intronic sequence consisting of the last two introns from the genes; these sequences were used as a control because terminal introns are thought to contain relatively fewer regulatory elements.
The human sequence in each set was aligned to the other three mammalian genomes19,20. Some segments could not be aligned in all four species, primarily because they represent new sequence recently inserted by transposons, ancestral sequence deleted in one of the other species, or regions still missing from the draft genome sequences (see Supplementary Information). The proportion of bases that could be aligned across all four species was ~51% for promoters (44% upstream and 58% downstream of the TSS) and ~73% for the 3′UTRs. These proportions are much higher than for the control intronic regions (34%) or for the whole genome (28%), presumably reflecting the presence of important conserved elements (see Supplementary Information).
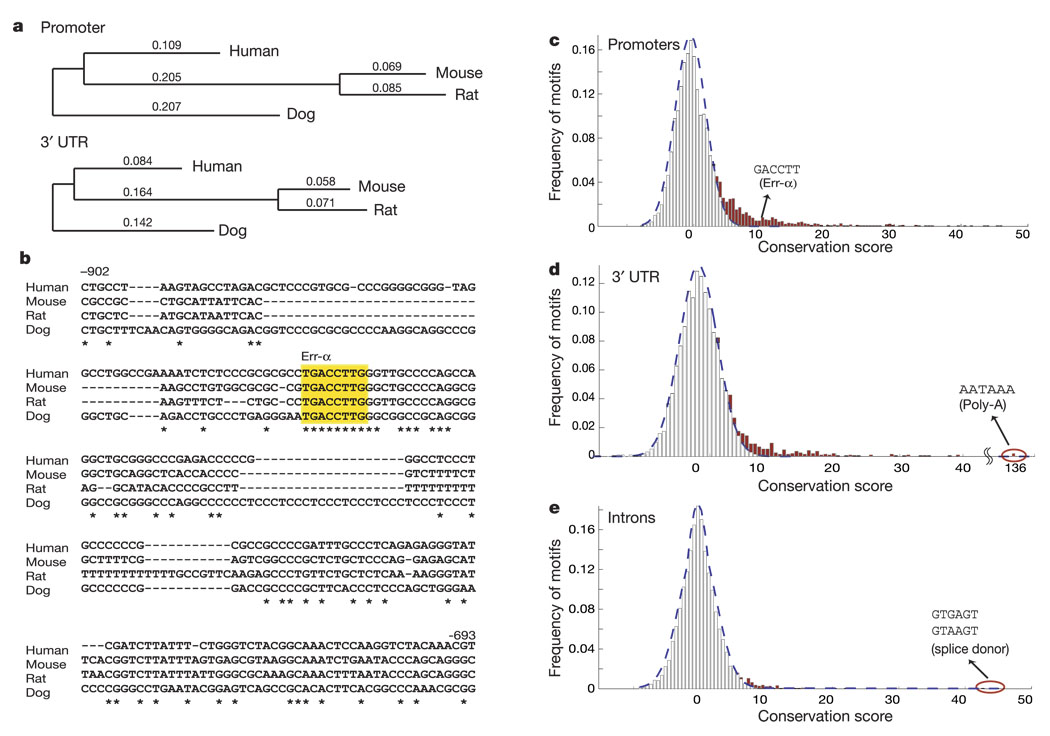
The total number of aligned bases is ~35 Mb for the promoters and ~11 Mb for the 3′ UTRs, which is comparable to the size of yeast genomes. Additionally, the evolutionary tree (Fig. 1a) of the four mammals has a total branch length similar to that used for comparative analysis of Saccharomyces species11 (0.68 substitutions per base in human promoters versus 0.83 in yeast intergenic regions; see Supplementary Information).
Figure 1.
Conservation properties in human promoter regions and 3′ UTRs. a, Evolutionary tree relating the four mammalian species. Branch lengths denote number of substitutions per site. Average nucleotide per cent identity to human is 62% for mouse, 60% for rat and 69% for dog in promoter regions, and respectively 68%, 67% and 76% in 3′ UTRs. b, Conservation in GABPA promoter region reveals functional Err-α motif. Asterisks denote conserved bases. The yellow box marks the experimentally validated Err-α-binding site. c–e, Excess conservation in promoter and 3′-UTR regions reveals short sequences under evolutionary selection. Motif conservation score (MCS) distribution is shown for all 6-mer motifs in aligned promoters (c), 3′-UTR regions (d) and introns (e). The dashed curve shows fit to gaussian distribution. Excess conservation relative to this distribution is shown in red.
Conservation properties of regulatory motifs
Our comparative analysis can be illustrated by considering a known regulatory motif. The 8-mer TGACCTTG is known to be a binding site of the Err-α protein and to occur in the promoters of many genes induced during mitochondrial biogenesis21,22. The promoter of the GABPA gene contains a well-studied Err-α-binding site, which is conserved across all four species, and stands out from the non-conserved flanking sequence (Fig. 1b). More generally, the Err-α motif occurs 434 times in human promoter regions and 162 of these occurrences are conserved across all four species; the conservation rate is thus 37%. By contrast, a random 8-mer motif shows a markedly lower conservation rate in promoters (6.8%). Moreover, the high conservation of the Err-α motif is specific to promoter regions; it shows a much lower conservation rate in introns (6.2%).
We quantified the extent of excess conservation by using a motif conservation score (MCS), which essentially represents the number of standard deviations (s.d.) by which the observed conservation rate of a motif exceeds the expected conservation rate for comparable random motifs (see Methods). A ‘conserved occurrence’ of a (possibly degenerate) motif is an instance in which an exact match to the motif is found in all four species; the ‘conservation rate’ of a motif is defined to be the ratio of conserved occurrences to total occurrences in the human genome. Comparable random motifs are generated by sampling human genomic sequence in promoters (divided according to high or low CpG content) or 3′ UTRs in order to ensure equivalent sequence composition (see Methods). For the Err-α-binding motif, the MCS is 25.2 s.d. (the binomial probability of observing 162 conserved instances out of 434, given an expected rate of conservation of 6.8%).
We examined the conservation properties of known motifs in mammalian promoters using the TRANSFAC database of reported transcription-factor binding sites23. The 446 motifs in TRANSFAC were clustered on the basis of sequence similarity, resulting in 123 motif clusters (see Methods). Most of these 123 motifs had high MCS, with 63% having MCS > 3 and nearly one-half having MCS > 5. By contrast, a control set of random motifs has only 1.6% with MCS > 3 and none with MCS above 5. The TRANSFAC motifs with low conservation scores have three likely explanations: they may be erroneous or over-specified (there is no uniform standard of evidence for inclusion in the database), they may have diverged between the species compared, or they may have too few biologically functional occurrences for the conserved instances to stand out with sufficient statistical significance.
For 3′ UTRs, few common motifs have been defined and there is no database analogous to TRANSFAC. One well-known example is the polyadenylation signal (AATAAA). This motif indeed shows a high conservation rate, with 6,617 out of 14,266 (46%) occurrences in 3 ′ UTRs being conserved (conservation rate = 46%, MCS = 135). Control random motifs show a conservation rate of only 10% in 3′ UTRs, and the polyadenylation signal shows a conservation rate of only 6% in the intronic controls.
Discovery of new motifs
We next explored the general conservation properties of short sequences within promoters and 3′ UTRs. Specifically, we measured the conservation rate of all 6-mer sequences. For both regions, there is a large excess of 6-mers with high MCS scores (Fig. 1c, d). The distribution of MCS scores suggests that a significant proportion of 6-mers are under purifying selection (13% in promoters and 8.6% in 3′ UTRs, indicated by the shaded regions in Fig. 1c, d). By contrast, intronic regions have a largely symmetric distribution (Fig. 1e) with only a modest number of outliers. (The two most extreme cases correspond to known mRNA splice donor signals, GTGAGT and GTAAGT.)
On the basis of these properties, we set out to systematically discover common regulatory motifs in promoters and 3′ UTRs. We evaluated motifs containing 6–18 bases, with degeneracy allowed at each position; a computational algorithm was developed to make it feasible to screen ~1012 possible motifs across the genomic regions (see Methods). Motifs with high conservation scores (MCS > 6) were identified, similar motifs were clustered together and the motif with the highest MCS in each cluster was selected (see Methods). This resulted in a small number of motifs with MCS > 6, which we refer to as ‘highly conserved motifs’
Motifs in promoters
In promoters, we found 174 highly conserved motifs. Among these, some could be immediately recognized as known regulatory elements. For example, the three strongest motifs correspond to binding sites for the oncogene regulator ELK-1, the cell-cycle regulator Myc and the mitochondrial respiratory chain regulator NRF-1. Overall, 59 discovered motifs showed strong matches to known motifs and 10 showed weaker matches (see Supplementary Information), together accounting for 72% of the 123 previously known motifs that we assembled from the TRANSFAC database (Supplementary Information). (By contrast, a comparable collection of random motifs would match only ~5% of the TRANSFAC motifs; see Supplementary Information.)
The remaining 105 discovered motifs represent potentially new regulatory elements. The list includes numerous notable examples, some with conservation scores much higher than for most known motifs. For example, the newly discovered motif M4 (ACTAYRNNNCCCR) occurs 520 times in human promoter regions, of which 317 (61%) are conserved, and the new nearpalindromic motif M8 (TMTCGCGANR) occurs 368 times, of which 236 (64%) are conserved.
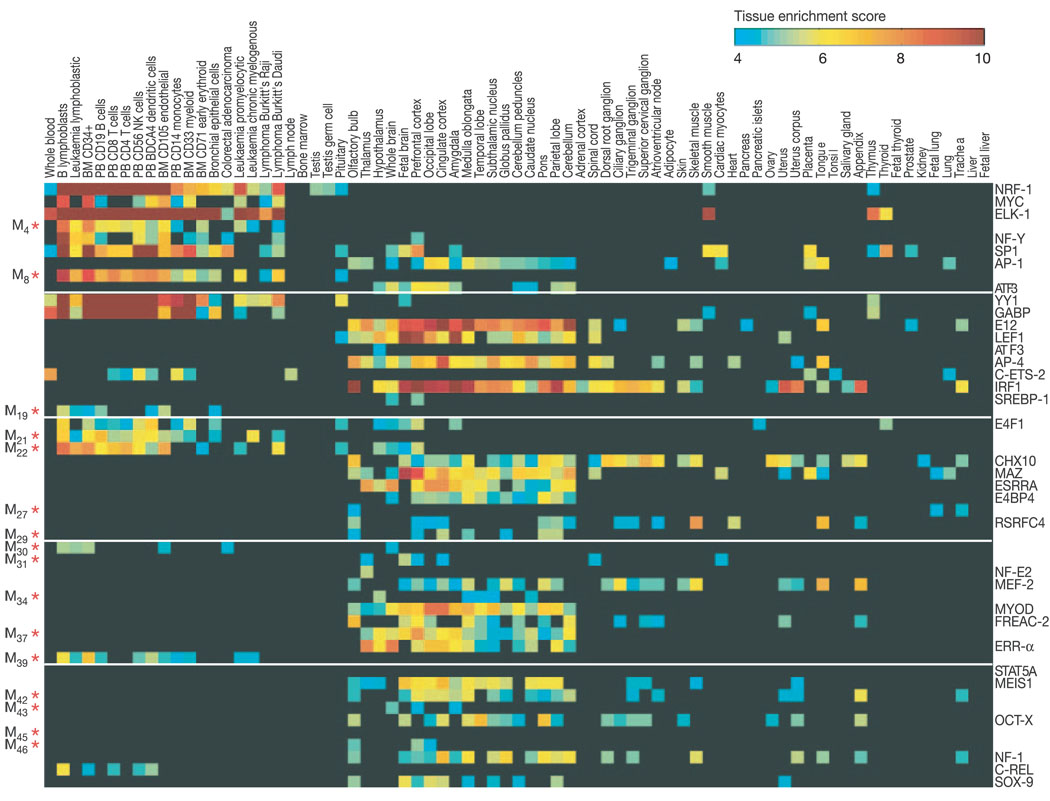
In the absence of specific information about the role of these putative motifs, we used two approaches to demonstrate that most of the new motifs are likely to be biologically meaningful. First, we correlated the presence of motifs with the tissue specificity of gene expression. We reasoned that the genes controlled by a common regulator would often (although not always) show enriched expression in specific sets of tissues. For each motif, we defined the set S1 of genes with conserved occurrences of the motif and an equal-sized control set S2 of genes in which the motif occurs in the human genome but is not conserved (see Methods). Using gene expression data from 75 human tissues24, significant enrichment in one or more tissues (z-score > 4.0) was seen for 59 of the 69 (86%) known motifs and 53 of the 105 (50%) new motifs (Table 1 and Fig. 2). In contrast, the control sets show little or no enrichment across the same tissues (Supplementary Fig. S2). For example, the best-conserved new motifs M4 and M8 show enrichment in haematopoietic cells, motif M27 (TGGN6KCCAR) is enriched in trachea and lung, and motif M29 (CTTTAAR) is enriched in brain-related tissues such as pons, parietal lobe and cingular cortex.
Table 1.
Top 50 of 174 discovered motifs in human promoters
| No. | Discovered motif | MCS | Known factor* | Conservation rate† | Tissue enrichment‡ | Position bias§ |
|---|---|---|---|---|---|---|
| 1 | RCGCAnGCGY | 107.8 | NRF-1 | 0.49 | 15.0 | −62 |
| 2 | CACGTG | 85.3 | MYC | 0.47 | 8.8 | −62 |
| 3 | SCGGAAGY | 80.4 | ELK-1 | 0.44 | 22.4 | −24 |
| 4 | ACTAYRnnnCCCR | 69.5 | – | 0.61 | 8.1 | −89 |
| 5 | GATTGGY | 64.6 | NF-Y | 0.51 | 9.8 | −63 |
| 6 | GGGCGGR | 63.9 | SP1 | 0.21 | 11.4 | −63 |
| 7 | TGAnTCA | 62.8 | AP-1 | 0.38 | 6.5 | – |
| 8 | TMTCGCGAnR | 55.7 | – | 0.64 | 9.4 | −62 |
| 9 | TGAYRTCA | 55.7 | ATF3 | 0.50 | 6.1 | −66 |
| 10 | GCCATnTTG | 54.7 | YY1 | 0.72 | 12.2 | – |
| 11 | MGGAAGTG | 51.6 | GABP | 0.43 | 13.9 | −23 |
| 12 | CAGGTG | 47.6 | E12 | 0.26 | 9.9 | – |
| 13 | CTTTGT | 46.0 | LEF1 | 0.42 | 13.6 | – |
| 14 | TGACGTCA | 44.8 | ATF3 | 0.44 | 4.2 | −22 |
| 15 | CAGCTG | 43.9 | AP-4 | 0.27 | 8.9 | – |
| 16 | RYTTCCTG | 43.0 | C-ETS-2 | 0.32 | 7.4 | −24 |
| 17 | AACTTT | 42.1 | IRF1 | 0.43 | 11.1 | – |
| 18 | TCAnnTGAY | 40.4 | SREBP-1 | 0.47 | 4.9 | −64 |
| 19 | GKCGCn(7)TGAYG | 40.1 | – | 0.35 | 5.6 | −62 |
| 20 | GTGACGY | 38.4 | E4F1 | 0.34 | 6.6 | −56 |
| 21 | GGAAnCGGAAnY | 37.7 | – | 0.68 | 7.0 | −33 |
| 22 | TGCGCAnK | 37.4 | – | 0.24 | 8.2 | −17 |
| 23 | TAATTA | 37.3 | CHX10 | 0.29 | 7.1 | – |
| 24 | GGGAGGRR | 33.5 | MAZ | 0.16 | 9.4 | – |
| 25 | TGACCTY | 33.4 | ESRRA | 0.30 | 7.7 | – |
| 26 | TTAYRTAA | 32.6 | E4BP4 | 0.34 | 6.1 | – |
| 27 | TGGn(6)KCCAR | 32.3 | – | 0.27 | 4.5 | – |
| 28 | CTAWWWATA | 32.3 | RSRFC4 | 0.36 | 7.6 | – |
| 29 | CTTTAAR | 30.8 | – | 0.43 | 5.4 | – |
| 30 | YGCGYRCGC | 30.5 | – | 0.19 | 5.2 | −31 |
| 31 | GGGYGTGnY | 30.0 | – | 0.24 | 5.4 | −63 |
| 32 | TGASTMAGC | 27.2 | NF-E2 | 0.39 | 5.4 | −66 |
| 33 | YTATTTTnR | 26.4 | MEF-2 | 0.21 | 7.1 | – |
| 34 | CYTAGCAAY | 26.1 | – | 0.50 | 5.2 | −142 |
| 35 | GCAnCTGnY | 25.7 | MYOD | 0.25 | 8.2 | – |
| 36 | RTAAACA | 25.6 | FREAC-2 | 0.46 | 7.0 | – |
| 37 | GTTRYCATRR | 25.3 | – | 0.54 | 7.6 | −56 |
| 38 | TGACCTTG | 25.2 | ERR-α | 0.37 | 8.1 | – |
| 39 | TCCCRnnRTGC | 24.3 | – | 0.30 | 6.8 | −60 |
| 40 | TTCYnRGAA | 24.3 | STAT5A | 0.19 | – | – |
| 41 | TGACAGnY | 24.1 | MEIS1 | 0.27 | 6.9 | – |
| 42 | TGACATY | 23.8 | – | 0.23 | 5.8 | – |
| 43 | GTTGnYnnRGnAAC | 23.7 | – | 0.47 | 4.7 | −57 |
| 44 | YATGnWAAT | 23.5 | OCT-X | 0.53 | 6.9 | – |
| 45 | CCAnnAGRKGGC | 23.4 | – | 0.47 | – | −101 |
| 46 | WTTGKCTG | 23.0 | – | 0.25 | 5.0 | −63 |
| 47 | TGCCAAR | 22.9 | NF-1 | 0.25 | 7.0 | – |
| 48 | GCGnnAnTTCC | 22.8 | C-REL | 0.30 | 6.0 | −12 |
| 49 | CATTGTYY | 22.5 | SOX-9 | 0.43 | 5.8 | – |
| 50 | RGAGGAARY | 22.4 | PU.1 | 0.22 | 4.0 | – |
Name of the best-matching motif in TRANSFAC database, if any. Weak matches are indicated in italics.
The percentage of human motif occurrences that match the motif consensus across all four species.
A measure of the maximum enrichment of conserved motif occurrences upstream of genes expressed in a compendium of 75 human tissues.
For motifs with strong positional bias (score > 5 s.d.), the mode of the distance distribution upstream of the TSS.
Figure 2.
Tissue specificity of expression for genes containing discovered motifs. For each of the 174 motifs, we defined the set of genes whose promoters contain conserved occurrences of the motif, and tested for enriched expression in 75 human tissues. The enrichment score (see Methods) is represented in pseudo-colour, with only scores greater than 4 shown. Motifs are ordered by MCS; asterisks denote new motifs (left); factor names are shown for known motifs (right). Only the top 50 motifs are shown. The maximum enrichment score across all tissues is reported in Table 1. Control gene sets were also constructed for each motif, consisting of an equal-sized set of genes in which the motif occurs but is not conserved; these control sets show little or no enrichment across the same tissues (see Supplementary Fig. S2).
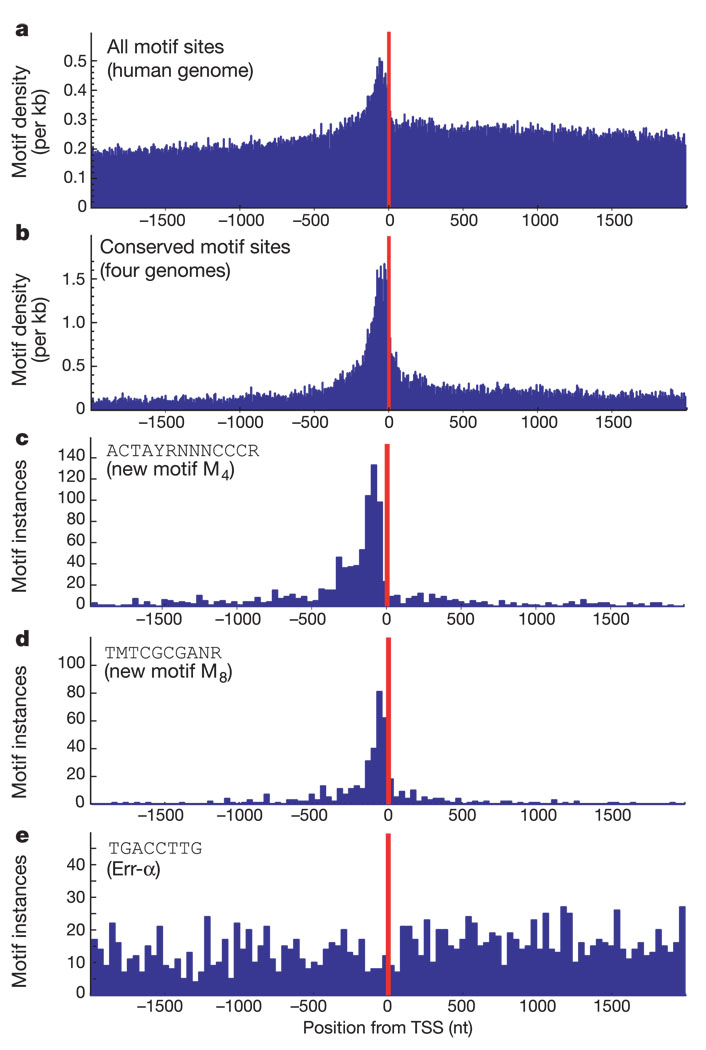
Second, we examined positional bias of the motifs relative to the TSS. Although the analysis covered a 4-kb region surrounding the TSS, the discovered motifs preferentially occur in the human genome within ~100 bases of the TSS, and their conserved occurrences across all four species show an even more marked enrichment in this region (Fig. 3a, b), consistent with the motifs being involved in transcription initiation. Within this overall trend, certain motifs show distinctive positional preferences. Approximately 28% of the known motifs and 35% of the new motifs show significant positional preference (see Methods). For example, the two strongest new motifs (M4 and M8) tend to occur at distances centred around −89 and −62 bp upstream of the TSS, respectively (Table 1 and Fig. 3c, d). Overall, 89% of the known motifs and 69% of the new motifs show tissue specificity, positional bias or both. Taken together, these results strongly suggest that the new promoter motifs are likely to be biologically meaningful.
Figure 3.
Discovered promoter motifs show positional bias with respect to transcriptional start site (TSS). a, Distribution of distance from TSS for all occurrences in human genome peaks within 100 bp before TSS. b, Distribution for conserved occurrences shows an even stronger peak. c, d, New motifs M4 and M8 peak at −81 and −69 respectively. e, Some motifs do not show specific peaks, including the known Err-α motif. nt, nucleotides.
In addition, several of the motifs tend to appear in multiple copies in the promoter. For example, 17.5% of genes containing motif M4 have more than one copy of M4 within 200 bp of each other (27-fold increase compared with 0.66% expected at random), and 10% of genes containing motif M8 have multiple copies of this motif within 200 bp (compared with 0.26% expected by chance). Such clustering is a common feature of several known regulatory motifs.
Motifs in 3′ UTRs
We next turned to motif discovery in 3′-UTR regions25. Using the same approach as for promoter regions, we discovered 106 highly conserved motifs in 3′ UTRs (MCS > 6). Because 3′-UTR motifs have not been extensively studied, we could not compare the discovered motifs to a large collection of previously known motifs; however, the motifs show two unusual properties that give insight into their nature.
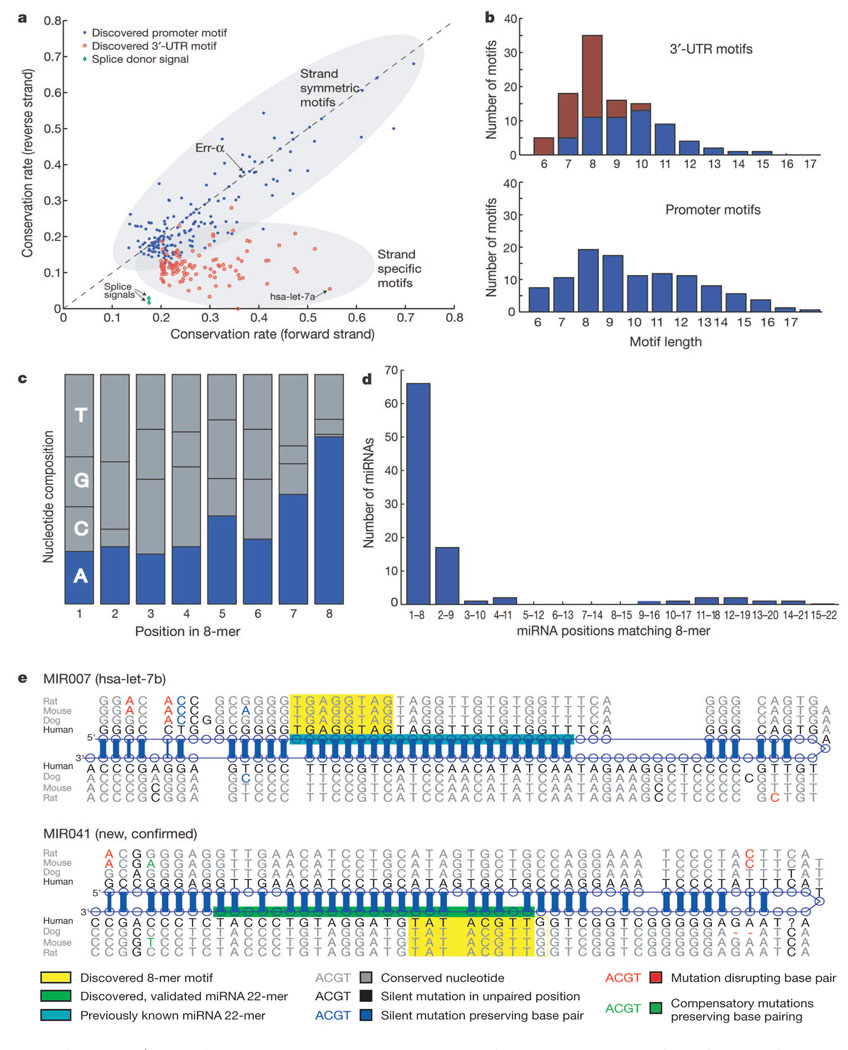
First, the 3′-UTR motifs show a strong directional bias with respect to DNA strand. Whereas promoter motifs tend to have similar conservation rates on the coding and non-coding strands, the 3′-UTR motifs are preferentially conserved in only one strand (Fig. 4a). The strand specificity is consistent with the 3′-UTR motifs acting at the level of RNA rather than DNA and thus having a role in post-transcriptional regulation.
Figure 4.
Properties of discovered 3′-UTR motifs and corresponding miRNA genes. a, Directionality of 3′-UTR motifs revealed by comparing conservation on forward and reverse strands. Strand preference is also seen for splice signals, but conservation of promoter motifs is largely symmetric. hsa-let-7a, Homo sapiens let-7a. b, Length distribution of 3′-UTR motifs shows abundance of motifs of length 8, but no such preference is seen for promoter motifs. Motifs overlapping conserved 8-mers (red) account for the length bias. c, A total of 72% of 8-mer motifs end with nucleotide A, suggesting complementarity with mature miRNA genes, frequently starting with U. d, Ninety-five per cent of discovered 8-mers match known miRNA genes at position 1–8 or 2–9. e, Alignments of known and new miRNA stem-loop structures, identified by their complementarity to a discovered 8-mer motif. New ~22-bp mature miRNA products predicted based on the observation shown in d and validated experimentally.
Second, the 3′-UTR motifs show an unusual length distribution. They have a strong peak at an 8-base length, whereas no such bias was found for promoter motifs (Fig. 4b). Moreover, the motifs of length 8 have a strong tendency to end with the nucleotide ‘A’ (Fig. 4c). These properties are reminiscent of a feature of miRNA16: many mature miRNAs start with a ‘U’ base followed by a 7-base ‘seed’ complementary to a site in the 3′ UTR of target mRNAs17,26,27; binding at such sites can guide degradation or repress translation of the mRNA. We reasoned that many of the highly conserved 8-mer motifs discovered by our unbiased procedure might be binding sites for conserved miRNAs.
Relationship with miRNAs
To investigate the relationship with miRNAs, the motif discovery procedure was repeated using only contiguous, non-degenerate 8-mers. We identified the subset of 8-mers with conservation rate > 18% (compared with the rate for a random 8-mer of 7.6%) and clustered these 8-mers into motifs defined as sets of similar 8-mers (see Supplementary Information). We refer to these as ‘highly conserved 8-mer motifs’.We obtained 72 highly conserved 8-mer motifs, which match 46% of the full set of 3′-UTR motifs (Fig. 4b).
We then searched for complementary matches of the 8-mer motifs to the 207 distinct human miRNAs (encoded by 222 miRNA genes) listed in the current registry28. Roughly 43.5% of the known miRNAs can match through Watson–Crick pairing to the highly conserved 8-mer motifs (versus 2% for an equal number of random 8-mers). When we relax the Watson–Crick base pairing to allow one mismatch, an additional 27 miRNAs can be identified (including four miRNAs containing T–G pairing and ten miRNAs whose first 5′ base was paired unselectively to ‘A’ in the conserved 8-mers). Moreover, the matches begin at nucleotide 1 or 2 of the miRNA gene in more than 95% of cases (Fig. 4d). These results strongly suggest that these 8-mer motifs represent target sites for miRNAs.
The remainder of the known miRNA genes do not contain complementary matches to any of the highly conserved 8-mer motifs. These miRNAs may bind relatively few targets, form only partial complements with their targets, tend to bind regions outside the 3′ UTR, or have been diverged between the species we are comparing. It is worth noting that, among the known miRNAs, those that do not match highly conserved 8-mers are evolving much more rapidly; they show a rate of disruptive mutation that is fivefold higher than for known miRNAs that match the highly conserved 8-mers. This suggests that the miRNA genes that match the highly conserved 8-mers have many more targets and, as a result, are much more constrained in their evolution.
New miRNA genes
We then sought to use the 8-mer motifs to discover new miRNA genes. Towards this end, we searched the four aligned genomes for conserved sequences complementary to any of the 72 discovered 8-mer motifs, extracted the sequence flanking each conserved site, and used the published RNAfold29,30 program to test for a conserved RNA-folding pattern characteristic of miRNA genes (stem-loop structures with calculated folding free energy of at least 25 kcal mol−1 in all four species).
We identified 242 conserved and stable stem-loop sequences, containing conserved instances of most (52 of 72) of the highly conserved 8-mer motifs (see Supplementary Information). These include 113 sequences encoding known miRNAs genes (52% of 222 known miRNA genes) and 129 sequences encoding predicted new miRNAs (Table 2 and Fig. 4e; see also Supplementary Table S8). The predicted miRNA genes show relatively few mutations that would disrupt an inferred base pair in the stem structure; the frequency of such mutations is similar to that seen for the 222 known miRNA genes.
Table 2.
Top 50 conserved 8-mers in 3′ UTRs and corresponding miRNAs
| No. | Motif | Conservation rate | MiRNA* |
|---|---|---|---|
| 1 | GTGCAATA | 0.55 | miR-92, miR-32, miR-137, miR-367, miR-25, miR-217(†), new(12) |
| 2 | GTGCCTTA | 0.54 | miR-124a, miR-224(†), miR-208(†), miR-34b(†), miR-9*(†), miR-34c(†), miR-330(†), new(6) |
| 3 | CTACCTCA | 0.53 | miR-98, let-7i, let-7 g, let-7f, let-7e, let-7c, let-7b, let-7a, let-7d, miR-196b, miR-196a, new(4) |
| 4 | ACCAAAGA | 0.49 | miR-9, new(11) |
| 5 | TGTTTACA | 0.48 | miR-30e-5p, miR-30d, miR-30c, miR-30b, miR-30a-5p, new(4) |
| 6 | GCACTTTA | 0.48 | miR-20, miR-106b, miR-18(†), miR-93, miR-372, miR-17-5p, miR-106a, miR-302d, miR-302c, miR-302b, miR-302a, miR-373, new(4) |
| 7 | TGGTGCTA | 0.43 | miR-29c, miR-29b, miR-29a, miR-107(†), miR-103(†), new(6) |
| 8 | CTATGCAA | 0.42 | miR-153, new(9) |
| 9 | TACTTGAA | 0.42 | miR-26b, miR-26a, new(4) |
| 10 | CGCAAAAA | 0.42 | New(2) |
| 11 | GTGCCAAA | 0.41 | miR-96, miR-182, miR-183, new(16) |
| 12 | GTACTGTA | 0.40 | miR-101, miR-199a‡, miR-144, new(2) |
| 13 | ATACGGGT | 0.40 | miR-99a, miR-100, miR-99b(†) |
| 14 | AAGCACAA | 0.40 | miR-218, new(8) |
| 15 | TTTGCACT | 0.37 | miR-19b, miR-19a, miR-301, miR-130b, miR-130a, miR-152, miR-148b, miR-148a, miR-139, new(10) |
| 16 | TGTACATA | 0.36 | – |
| 17 | AAGCCATA | 0.35 | miR-135b, miR-135a |
| 18 | ACTGTGAA | 0.35 | miR-27b, miR-27a, miR-128b, miR-128a, miR-23b(†), miR-23a(†), new(5) |
| 19 | AGACAATC | 0.33 | miR-219, new(2) |
| 20 | TGCTGCTA | 0.33 | miR-195, miR-16, miR-15b, miR-15a, miR-338(†), miR-424, new(5) |
| 21 | TTTTGTAC | 0.32 | New(1) |
| 22 | ACATTCCA | 0.32 | miR-206, miR-1, miR-122a(†) |
| 23 | TGAATGTA | 0.31 | miR-181b(†), miR-181c, miR-181a |
| 24 | ACGGTACA | 0.30 | – |
| 25 | CAGTATTA | 0.30 | miR-200c, miR-200b, new(1) |
| 26 | TTGCATGT | 0.29 | New(7) |
| 27 | TCGCATGA | 0.29 | New(1) |
| 28 | CTCAGGGA | 0.29 | miR-125b, miR-125a, new(6) |
| 29 | CAAGTGCC | 0.28 | New(2) |
| 30 | ACTACTGA | 0.28 | – |
| 31 | TGGACCAA | 0.28 | miR-133b, miR-133a, new(3) |
| 32 | GTAAATAG | 0.28 | New(1) |
| 33 | TGTAGATA | 0.28 | – |
| 34 | ACACTACA | 0.27 | miR-142-3p, new(3) |
| 35 | GTACAGTT | 0.26 | New(1) |
| 36 | CACCAGCA | 0.26 | miR-138(†), new(4) |
| 37 | GGTACGAA | 0.25 | miR-126(†) |
| 38 | TGTATAGT | 0.24 | miR-381 |
| 39 | AAGGGCTA | 0.24 | New(1) |
| 40 | AGCTTTAA | 0.24 | New(1) |
| 41 | ATTTATCG | 0.23 | – |
| 42 | GGCAGCTA | 0.23 | miR-22(†), new(1) |
| 43 | GCTGTAAA | 0.23 | New(4) |
| 44 | GCACTAAT | 0.22 | – |
| 45 | AAAGGTGC | 0.22 | – |
| 46 | ATGTAGCA | 0.22 | miR-221(†), miR-222(†) |
| 47 | ACACTGGA | 0.21 | miR-199b(†), miR-199a(†), miR-145(†), new(2) |
| 48 | GTATATAG | 0.21 | – |
| 49 | TTTGATAA | 0.21 | miR-361(†) |
| 50 | AAGCATGC | 0.21 | New(1) |
Top 50 of 72 highly conserved 8-mer motifs in 30 UTRs and their corresponding miRNAs.
Known human miRNAs matching complements of each 8-mer and its variants grouped in the same cluster. The dagger symbol (in parentheses) indicates miRNAs with one mismatch. When multiple new miRNAs are discovered using the 8-mer motifs as seeds, their number is indicated in parentheses. (See Supplementary Information for full alignments of known and discovered miRNAs.)
The miRNA mature product comes from the 3′ arm of the stem loop.
A representative set of 12 predicted new miRNA genes was selected for validation according to accepted criteria16,31 (involving purification of small RNAs, ligation of adaptors, polymerase chain reaction (PCR) amplification, cloning and DNA sequencing to verify the precise sequence and junction; see Methods). Lacking prior information about tissue or temporal specificity of expression, we used pooled small RNAs prepared from ten human tissues (breast, pancreas, prostate, colon, stomach, uterus, lung, brain, liver and kidney) (see Methods). This may miss many miRNAs expressed primarily during development or at low levels in the adult, or not expressed in the tissues we tested. Nonetheless, 6 of the 12 (50%) predicted miRNAs genes were found to be clearly expressed in the pooled adult tissues.
We conclude that many of the 129 candidates are likely to be bona fide miRNA genes. Moreover, there are likely to be many additional miRNAs inasmuch as our approach was only designed to find those containing a conserved 8-mer and thus only detects one-half of known miRNAs. This challenges a recent conclusion, based on computational analysis, that nearly all human miRNA genes have already been discovered, and that fewer than 40 remain to be found27.
Targets of miRNAs
The properties of the conserved motifs also allow inferences about the prevalence of regulation by miRNAs. Roughly 40% of human 3′ UTRs contain a conserved occurrence of one of the miRNA-associated 8-mer motifs, whereas only ~25% contain conserved occurrences of a comparable control set. This suggests that at least 20% of 3′ UTRs may be targets for conserved miRNA-based regulation at the 8-mer motifs (see Methods). This greatly expands previous estimates of the number of targets of miRNA genes17. With sequence from more mammalian genomes, it should be possible to distinguish the conserved target sites with high specificity and sensitivity10. There are likely to be additional miRNA target sites in the human genome that are not conserved across all mammals; it should be possible to find most of these by genomic comparison with closer relatives such as primates.
The results thus provide an unbiased assessment of the relative importance of miRNA-based regulation in the human genome, consistent with recent independent studies32–34. Overall, ~45% of the 108 highly conserved motifs in 3′ UTRs appear to be related to miRNA regulation and ~5,000 human genes (~20% of the genome) are likely to be regulated by miRNAs through these conserved motifs.
Remaining motifs in 3′ UTRs
When the miRNA-related motifs are removed, there remains a list of 60 3′-UTR motifs with a length distribution similar to that seen for promoters (Fig. 4b). Although there is no systematic procedure for evaluating these motifs, the list contains several interesting features. It includes the known polyadenylation signal and various (A + T)-rich elements35, which are believed to be involved in controlling mRNA stability and degradation. It also contains 11 motifs with a TGTA core sequence, flanked by different variants of an (A +T)-rich element (the most conserved being TGTANATA). Recent work in yeast has identified such motifs as binding sites for the Puf family of RNA-binding proteins; they may represent binding sites for the homologous mammalian proteins (such as PUM1 and PUM2 in human36).
Conclusion
Our comparative genomic analysis of four mammalian species has provided an initial systematic catalogue of human regulatory motifs in promoters and 3′ UTRs. In promoters, the approach automatically rediscovered many known motifs and discovered many new ones that appear to be functional based on multiple criteria; many of these show tissue-specific expression and distance constraints with respect to the TSS. In 3′ UTRs, the approach provided a first view of common regulatory motifs. Notably, roughly one-half of the discovered motifs in 3′ UTRs appear to be related to miRNAs, showing the extraordinary importance of this recently discovered regulatory mechanism. Moreover, these motifs also led to the identification of numerous new miRNA genes. The remaining motifs may be targets of RNA-binding proteins, with a few matching known motifs. The next challenge will be to develop systematic methods to discern the specific functions of these motifs in a genome-wide fashion.
The results here are, of course, only a step towards a comprehensive inventory of human regulatory motifs to serve as a foundation for understanding cellular circuitry and its role in health and disease. Our analysis used stringent thresholds to focus on the most abundant and most conserved motifs. With sequence from a few dozen additional mammals37, it should be possible to create a complete dictionary of such common functional elements.
Methods
Motif conservation score
Regulatory motifs are represented as consensus sequences (profiles), over an alphabet of 11 characters, consisting of A, C, G, T and N, and the six twofold degenerate characters. A conserved occurrence of a motif m matches the motif consensus in each of the four species. The observed conservation rate p of a motif is the proportion of human occurrences that are conserved across all four species. The expected conservation rate p0 for a motif of given length and redundancy is computed as the average observed conservation rate of 1,000 random motifs of the same length and redundancy, obtained by sampling the human genome in 1,000 loci. The MCS of a motif m is evaluated by comparing its observed conservation rate p to the expected conservation rate p0; it corresponds to the binomial probability of observing k conserved instances out of total n instances given probability p0 of conservation for any one instance.
Motif discovery overview
The MCS is evaluated separately for each type of region (promoters, introns, 3′ UTR), thus matching the specific nucleotide composition of each type of region and eliminating biases from the scoring scheme. Additionally, for promoter motifs, the MCS is evaluated separately for sequences inside CpG islands and those outside CpG islands, thus accounting for their radically different nucleotide compositions. In each type of region motifs are enumerated by hashing and refinement of 6-mer seeds possibly with a central gap. All motifs above the cutoff of MCS > 6 are selected, corresponding to specificity values higher than 98%. The selected motifs are grouped into clusters using genome-wide co-occurrence and sequence similarity: we first merge motif pairs which overlap by more than 80% of their sites, keeping only the motif with the highest MCS score; we then cluster the remaining motifs based on their pairwise sequence similarity, defined as the Pearson correlation of their equivalent position weight matrices.
Motif gene-set enrichment score (MGES)
The enrichment of a motif m in a given tissue is evaluated as the enrichment of its gene set S in the ranked list L for that tissue. The non-randomness of the ranks of S in the list L is evaluated using the Mann–Whitney rank sum statistic; the MGES is reported as the standard deviations corresponding to this statistic. For each motif m, three gene sets are generated: a target gene set S1, and two control gene sets, S2 and S3, with the same number of genes. The target gene set S1 of ‘conserved instances’, consists of all genes whose promoters contain at least one conserved instance of the motif m. The control gene set S2 consists of genes with ‘non-conserved instances’ randomly sampled from genes containing instances of the motif in the human genome, regardless of conservation. The control gene set S3 consists of genes with ‘shuffled conserved instances’, randomly sampled from the union of all conserved gene sets S1, across all motifs.
Identification of new miRNAs
Conserved occurrences of the 3′ UTR 8-mer motifs are identified in the entire human genome by searching both strands for motifs reverse complementary to each 8-mer, and excluding previously annotated functional elements. The neighbourhoods of these alignments are then searched for stable stem-loops over a sliding window of 110 bp every 3 bp, and those with a folding free energy of at least 25 kcal mol−1 in each aligned species are selected using the program RNAfold. These were further evaluated based on several observed features of known miRNAs: higher conservation in the ~22 bp stem, lower conservation in the loop and surrounding regions, and appropriate base-pairing and bulges in the stem region.
Experimental validation of new miRNA genes
A set of 12 new miRNA genes was selected for experimental validation, such that their conservation properties and folding free energy values is representative of the set of all 258 predicted miRNAs. The experimental procedure was carefully designed to ensure that the exact ~22 bp predicted product is specifically expressed. First, three steps of rigorous PAGE purification of small RNA fractions exclude large RNA and genomic DNA. Second, PCR is done with one primer specific to the gene, and one complementary to the artificial adaptors used in miRNA ligation. Finally, the resulting clones are sequenced and the precise expected sequence and junction are verified.
Additional methods
Supplementary Information includes a complete description of the methods for whole-genome alignment, definition of promoter and 3′ UTR databases, rapid motif enumeration and scoring, motif clustering, comparison with TRANSFAC motifs, motif gene-set enrichment analysis, motif positional bias, 3′-UTR 8-mer discovery, identification of new miRNA genes, estimating number of miRNA targets, and experimental verification of miRNA genes.
Supplementary Material
Acknowledgements
We thank B. Birren, M. Kamal, K. O’Neill and A. Subramanian for advice and discussions. This work was supported in part by grants from the National Human Genome Research Institute.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Gumucio DL, et al. Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human gamma and epsilon globin genes. Mol. Cell. Biol. 1992;12:4919–4929. doi: 10.1128/mcb.12.11.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 3.Dubchak I, et al. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennacchio LA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 5.Boffelli D, et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 6.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha S, Blanchette M, Tompa M. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004;5:170. doi: 10.1186/1471-2105-5-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dermitzakis ET, et al. Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment. Genome Res. 2004;14:852–859. doi: 10.1101/gr.1934904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 10.Eddy SR. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 12.Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 13.InternationalHuman Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.International Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 15.Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Maglott DR, Katz KS, Sicotte H, Pruitt KD. NCBI’s LocusLink and RefSeq. Nucleic Acids Res. 2000;28:126–128. doi: 10.1093/nar/28.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha VK, et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl Acad. Sci. USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston SD, et al. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- 23.Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nature Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 26.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 27.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontana W, et al. RNA folding and combinatory landscapes. Phys. Rev. E. 1993;47:2083–2099. doi: 10.1103/physreve.47.2083. [DOI] [PubMed] [Google Scholar]
- 30.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambros V, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;30(January) doi: 10.1038/nature03315. advance online publication, (doi:10.1038/nature03315). [DOI] [PubMed] [Google Scholar]
- 34.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 36.Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 37.Margulies EH, et al. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc. Natl Acad. Sci. USA. doi: 10.1073/pnas.0409882102. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.