Abstract
The ability to reprogram in vivo a somatic cell after differentiation is quite limited. One of the most impressive examples of such a process is transdifferentiation of pigmented epithelial cells (PECs) to lens cells during lens regeneration in newts. However, very little is known of the molecular events that allow newt cells to transdifferentiate. Histone B4 is an oocyte-type linker histone that replaces the somatic-type linker histone H1 during reprogramming mediated by somatic cell nuclear transfer (SCNT). We found that B4 is expressed and required during transdifferentiation of PECs. Knocking down of B4 decreased proliferation and increased apoptosis, which resulted in considerable smaller lens. Furthermore, B4 knockdown altered gene expression of key genes of lens differentiation and nearly abolished expression of γ-crystallin. These data are the first to show expression of oocyte-type linker histone in somatic cells and its requirement in newt lens transdifferentiation and suggest that transdifferentiation in newts might share common strategies with reprogramming after SCNT.—Maki, N., Suetsugu-Maki, R., Sano, S., Nakamura, K., Nishimura, O., Tarui, H., Del Rio-Tsonis, K., Ohsumi, K., Agata, K., Tsonis, P. A. Oocyte-type linker histone B4 is required for transdifferentiation of somatic cells in vivo.
Keywords: newt, lens regeneration, histone H1, γ-crystallin
As stem or progenitor cells are diverted to differentiate toward a particular lineage, they lose their multipotentiality. At the endpoint, when they are terminally differentiated, cells are thought to be unable to revert to multipotent state. Contrary to this finding, the case of newt lens regeneration is a prime paradigm of transdifferentiation at work. After lens removal, the iris pigmented epithelial cells (PECs) dedifferentiate and then change their fate to become lens cells. Four to 5 d after lentectomy, the pigmented cells at the tip of the dorsal iris shed their pigments and start proliferating, thus losing their original tissue characteristics. Depigmented cells are observed initially around d 8. At 13 or 14 d after lentectomy, these depigmented PECs have formed a vesicle, which is still undifferentiated and expresses no lens-specific markers. The last step marks the onset of transdifferentiation and the formation of the lost lens. After d 14, the posterior cells of the vesicle elongate and start expressing lens markers. The vesicle grows and differentiates to lens, which by d 20 has a considerable size and normal morphology. It is important to state here that while the ventral iris PECs undergo some of the initial events, they fail to transdifferentiate to lens. Transdifferentiation of PECs has been directly demonstrated by clonal culture experiments (1, 2).
Since transdifferentiation should involve large-scale reprogramming, we hypothesized that histones, which are known to regulate gene expression and reprogramming (3, 4), would be good candidates to explore such possibility. In this study, we focused on linker histones. Four types of linker histone, i.e., somatic-, oocyte-, testis-, and erythrocyte-type linker histones, have been identified (5, 6). Oocyte-type linker histone, which is known to be expressed during oogenesis and early embryogenesis (7,8,9,10,11,12), is believed to play an important role in the reprogramming mediated by SCNT into oocyte. After SCNT somatic-type linker histone H1 is replaced by oocyte-type linker histone (13,14,15), which allows chromatin to be remodeled by an ATP-dependent chromatin remodeling factor (16) and causes genome-wide chromatin decondensation (17). Here we show that oocyte-type linker histone B4 is recruited specifically into nucleus and is necessary for lens transdifferentiation.
MATERIALS AND METHODS
Cloning of newt B4 cDNA
Total RNA was extracted from Cynops pyrrhogaster ovary using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and poly(A) RNA was purified using oligotex-dT30 (Takara, Ohtsu, Japan). Ovary cDNA library was constructed using Universal RiboClone cDNA synthesis system (Promega, Madison, WI, USA). In this procedure, RT reaction was performed using random hexameric primers and cDNA was inserted into the EcoRV site in pBluescript SK(+). Using the ovary cDNA library, 4582 expressed sequence tags were obtained, and 2 clones showed a similarity with frog oocyte-type linker histone. 5′- and 3′-rapid amplification of cDNA ends RACE) was performed, and finally full-length C. pyrrhogaster B4 clone was obtained by end-to-end (from the initial methionine to stop codon) PCR. A partial sequence of Notophthalmus viridescens B4 was obtained from ovary by RT-PCR using primers based on the C. pyrrhogaster B4 sequence. 5′-and 3′-RACE was performed, and full-length N. viridescens B4 was cloned by end-to-end PCR (Supplemental Fig. 1).
Accession number of cloned cDNAs
Full or partial cDNAs were cloned by degenerate PCR and rapid amplification of cDNA ends. Accession numbers of cloned cDNAs are GQ890215 (C. pyrrhogaster B4), GQ890216 (N. viridescens B4), GQ844313 (N. viridescens nucleostemin), GQ844312 (N. viridescens MafB), GQ844314 (N. viridescens γ-crystallin), and GQ844315 (N. viridescens β-actin).
RT-PCR
Dorsal irises during lens regeneration were collected. RNAs were extracted using Trizol reagent (Invitrogen) for C. pyrrhogaster or Nucleospin (Macherey-Nagel, Düren, Germany) for N. viridescens. RT reaction was performed with a first-strand cDNA synthesis kit (GE Healthcare, Piscataway, NJ, USA) using an oligo(dT) primer. PCR for B4 was performed using the Ex Taq kit (Takara) and the following primers, Cynops-B4-RT-PCR-F, 5′-ATGGTACTGGAAGCCCTGAGG-3′; Cynops-B4-RT-PCR-R, 5′-ACGGAGTCCACAGACGGGTA-3′; Notophthalmus-B4-RT-PCR-F, 5′-GGCCACAGGAAGGTTTAAGCT-3′; Notophthalmus- B4-RT-PCR-R, 5′-CTCGACACCACTTTCTTGGCT-3′; Cynops-actin-RT-PCR-F, 5′-GAAGGTTATGCCCTGCCTCAT-3′; Cynops-actin-RT-PCR-R, 5′-TGAAGCTGTAGCCCCTCTCAGT-3′; Notophthalmus-actin-RT-PCR-F, 5′- TTATGCTCTGCCTCATGCCATCCT-3′; Notophthalmus-actin-RT-PCR-R, 5′- TTCGGCCGTAGTTGTGAAGCTGTA-3′.
Quantitative PCR
Regenerated lenses were collected using a mouth pipette. RNA was purified using Nucleospin (Macherey-Nagel). RT reaction was performed with a first-strand cDNA synthesis kit (GE Healthcare) using an oligo(dT) primer. qPCR was performed using a iQ SYBR Green supermix (Bio-Rad, Hercules, CA, USA), and the following primers: nucleostemin-qPCR-F, 5′-ATCTGCGGCCAAGAAAGAAACTGC-3′; nucleostemin-qPCR-R 5′-ATGTTTATCAGGAGCCTGGTCGGT-3′; MafB-qPCR-F, 5′-TGGACGCTCTAATCAACTCTGCCA-3′; MafB-qPCR-R; 5′-TTAGCAAGTAGCCTGGGTGCGTAT-3′; Pax6-qPCR-F, 5′-ATCGGAGGCAGCAAGCCCA-3′; Pax6-qPCR-R, 5′-AGATGGACGGACACTCGCGCTT-3′; γ-crystallin-qPCR-F, 5′-AATGATTCCATCAGGTCATGCCGC-3′; γ-crystallin-qPCR-R, 5′-TGTGGACAGTCTTCAGAGAACTCC-3′; RPL27-qPCR-F, 5′-ATTTATGAAACCCGGGAAGG-3′; RPL27-qPCR-R, 5′-CCAGGGCATGACTGTAAGGT-3′. To quantitate the expression of each gene, Ct values were compared to a standard curve generated using a series of dilutions of cloned cDNAs. The amount of mRNA was normalized to that of ribosomal protein L27. Specific PCR amplifications were confirmed by melting curve analysis.
Vivo-morpholino
Vivo-morpholinos specialized to enter cells in living animals were purchased from Gene Tools (Philomath, OR, USA). After lentectomy, B4 vivo-morpholino (5′-AGCAGTCTTCTTAGAAGCCATTTG-3′) or the control vivo-morpholino recommended by Gene Tools (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was intraperitoneally injected at 12.5 μg/g body weight every day until different samples were collected.
Antibodies
Rabbit polyclonal antibody against newt B4 was raised against a mixture of two peptides, ALRKNTDRKGAT and TDKDSAKPTAKRGKK (see Supplemental Fig. 1A) and affinity-purified using the peptides (Scrum Inc., Tokyo, Japan). The other primary antibodies used were histone H1 (V7013; Biomeda, South San Francisco, CA, USA), histone H3 (ab1791; Abcam, Cambridge, UK), and BrdU (MAB3510; Millipore, Billerica, MA, USA).
Immunohistochemistry
Eyeballs and ovaries were fixed with methanol-acetic acid solution (75% methanol and 25% acetic acid, v/v) at 4°C overnight, embedded in paraffin, and sectioned at 16 μm. After deparaffinization, sections were treated with permeable solution (0.05% Triton X-100, 0.05% saponin, and 2× SSC) for 1 h, rinsed with 2× SSC, and blocked in TNB buffer (0.1 M Tris-HCl, pH7.5; 0.15 M NaCl; and 0.5% blocking reagent) supplied with the TSA kit (Perkin Elmer, Waltham, MA, USA) for 1 h.
For detection of B4 and histone H1, the samples were incubated at 4°C overnight with a mixture of the primary antibodies, B4 antibody diluted 1:10 and histone H1 antibody diluted 1:100 with TNB buffer, and then incubated with the following secondary antibodies at room temperature for 90 min: Alexa Fluor 488-conjugated goat anti-rabbit IgG for B4 and Cy3-conjugated sheep anti-mouse IgG for histone H1. Nuclei were counterstained with Hoechst 33258. Images of stained tissue were taken using the BX-51 fluorescence microscope system (Olympus, Tokyo, Japan) equipped with a Cool SNAP cf2 camera (Photometrics, Tucson, AZ, USA). All images taken were saved in TIFF format. For measurement of signal intensity, the average signal intensity per pixel of B4, histone H1, and Hoechst33258 in each nucleus was measured using MetaMorph 7.1 software (Molecular Devices, Sunnyvale, CA, USA) without any image processing.
For BrdU staining, the samples were incubated at 4°C overnight with BrdU antibody diluted 1:100 with TNB buffer and then incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG at room temperature for 90 min. To detect apoptotic cells, ApopTag Plus In Situ Apoptosis Fluorescein Detection kit (Millipore) was used according to the manufacturer’s instruction.
Western blot analysis
Tissues were homogenized in 10 vol of 70% PBS. Two volumes of 0.6N HCl (final 0.2 N) was added to the homogenate and kept on ice 30 min to extract histones. The supernatant was recovered after centrifugation at 10,000 g for 10 min at 4°C and dialyzed twice with 0.1 N acetic acid for 30 min and twice with milli Q water for 30 min, followed by overnight incubation at 4°C. The extracted proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane (0.2 μm pore size, Invitrogen). The B4 band was probed with anti-B4 antibody followed by alkaline phosphatase-conjugated anti-rabbit IgG (Abcam). The histone H3 band was probed with anti-histone H3 antibody followed by alkaline phosphatase-conjugated anti-rabbit IgG (Abcam). The bands were visualized by incubation with NBT/BCIP solution (Roche, Basel, Switzerland). Intensity of the detected bands was measured using ImageJ 1.40g software (U.S. National Institutes of Health, Bethesda, MD, USA). The amount of each histone in B4 morpholino-treated iris sample was calibrated using a standard curve generated by dilutions of control morpholino-treated iris sample. The amount of B4 protein was normalized to that of histone H3.
Animal study compliance
All animal care and use protocols were in compliance with the Animal Experiment Handbook at the Kobe Center for Developmental Biology (RIKEN Kobe) and the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals.
RESULTS
Expression of B4 during Newt lens regeneration
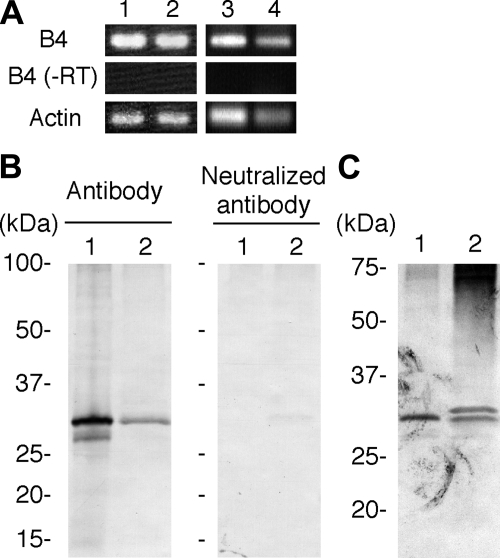
Initial immunohistochemical analysis using a Xenopus oocyte-type linker histone B4 antibody showed that antigens reacting with the antibody accumulate in nuclei of PECs during newt lens regeneration. This finding prompted us to clone a full-length B4 cDNA from two newt species, C. pyrrhogaster and N. viridescens (Supplemental Fig. 1). RT-PCR experiments clearly indicated that B4 is expressed during lens regeneration in both species (Fig. 1A). Antibody specific for newt B4 was raised using a mixture of newt B4 peptide sequences as antigens. Western blot analysis using this antibody also indicated that B4 is expressed during newt lens regeneration (Fig. 1B, C).
Figure 1.
Expression of B4 during lens regeneration. A) Detection of B4 expression by RT-PCR. Lane 1, C. pyrrhogaster ovary; lane 2, C. pyrrhogaster dorsal iris 10 d after lentectomy; lane 3, N. viridescens ovary; lane 4, N. viridescens dorsal iris 10 d after lentectomy. B, C) Detection of B4 protein by Western blot analysis. Ovary (lane 1) and dorsal iris 10 d after lentectomy (lane 2) were collected from C. pyrrhogaster (B) or N. viridescens (C), and histone proteins were extracted. Extracted proteins were analyzed by Western blotting using B4 peptide antibody (B, left panel). To confirm specific detection of B4 protein, the antibody was neutralized by incubation with antigen peptides prior to the primary antibody reaction (B, right panel). Note that the neutralizing antibody abolished bands indicating specificity of the antibody. Note that two bands, one band showing same mobility with the band detected in ovary and another band showing slightly less mobility, were detected in N. viridescens lens regenerating iris (C), which suggests alternative splicing or post-translational modifications of B4 in the iris.
Nuclear recruitment of B4 during lens regeneration
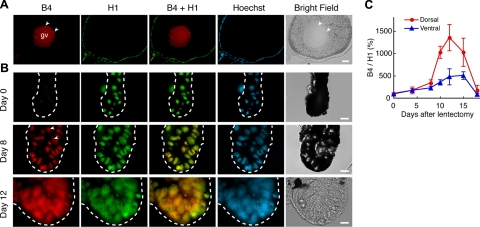
Having established the presence of B4 during regeneration, we decided to follow expression of B4 and H1 throughout a period of 20 d after lentectomy. The results are shown in Fig. 2. In ovaries, B4 localizes in the germinal vesicle, while H1 localizes in nucleus of follicle cells (Fig. 2A). In intact iris (d 0), B4 is virtually absent, while H1 is present in PECs. On d 8, when dedifferentiation is ongoing, PECs are positive for both B4 and H1. Similar patterns were observed at d 12 (Fig. 2B). A clear pattern emerged when we quantitated the expression and plotted the ratio of B4 to H1 (see Materials and Methods). Starting at d 8, the ratio of B4 to H1 clearly increases in the dorsal iris vesicle. Then, this ratio reaches a peak at d 12 before lens differentiation begins. Finally, it declines by d 18, when the transdifferentiation process is completed and what continues is the growth of the lens (Fig. 2C). Such a peak of the ratio is not seen in the ventral iris.
Figure 2.
Nuclear recruitment of B4 during lens regeneration. A) Immunostaining of ovary using B4 and H1 antibody. Arrowheads indicate nucleoli. Note detection of B4 in the germinal vesicle (gv) and H1 in nucleus of follicle cells. Localization of B4 in nucleoli is consistent with previous observations in mice (7, 18). B) Immunostaining during lens regeneration (0, 8, and 12 d after lentectomy) using B4 and H1 antibodies. Note that B4 is scarcely detected in intact iris. Note that the staining intensities of each panel are not comparative, because images were processed independently in order to show nuclear distribution of each antigen. C) Changes in the ratio of B4 to histone H1 during lens regeneration. After immunostaining, the intensity of B4 and H1 signals in each nucleus were measured, and the ratio of B4 to histone H1 was calibrated. Error bars = sd. Scale bars = 200 μm (A), 20 μm (B).
B4 knockdown affects proliferation and apoptosis
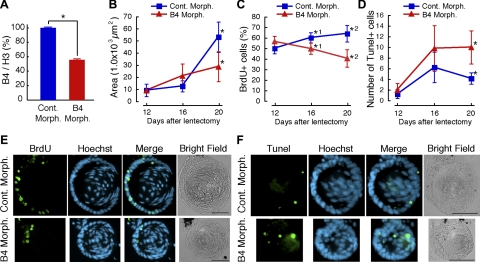
These striking patterns of B4 recruitment prompted us to examine its role in more details. For this, we proceeded by knocking down expression of B4, employing vivo-morpholino technology (19). To assess whether B4 morpholino reduces expression of B4, we injected morpholino every day intraperitonealy for 10 d after lentectomy. Then, dorsal irises were collected, and linker and core histones were extracted with hydrochloric acid. The extracted histones were analyzed by Western blotting. The amount of B4 was calculated based on the intensity of the detected band and normalized with that of histone H3. In irises from newts treated with the B4 morpholino, the amount of B4 protein was decreased by nearly 50% when compared to the levels in irises from newts injected with control morpholino (Fig. 3A).
Figure 3.
B4 knockdown affects proliferation and apoptosis. A) Decrease of B4 protein after B4 morpholino injection. After lentectomy, B4 vivo-morpholino was injected intraperitoneally every day in N. viridescens. On d 10, dorsal irises were collected, and the amount of B4 protein in the irises was measured by Western blotting. Amount of B4 was normalized with that of histone H3. *P = 0.000310 (control, n=2; B4, n=2). B–F) Effect of B4 knockdown in lens regeneration. B) Size of regenerated lens. *P = 0.00262 (control, n=7; B4, n=9). C) Number of BrdU-positive cells. *1: P = 0.0176 (control, n=4; B4, n=7); *2: P = 0.0000972 (control, n=7; B4, n=10). D) Number of Tunel-positive cells. *P = 0.00738 (control, n=6; B4, n=6). Error bars = sd. P values are from Student’s t test (2-tailed). E) BrdU immunostaining of lens from morpholino-injected newts 20 d after lentectomy. F) Tunel staining of lens from morpholino-injected newts 20 d after lentectomy. Scale bars = 100 μm. Cont. Morph., control vivo-morpholino; B4 Morph., B4 vivo-morpholino.
Following such a positive outcome of the morpholino treatment, we conducted a large-scale experiment by injecting morpholino and examining the regenerating lens for a period up to 20 d. We started seeing effects on lens differentiation and morphology after d 12, which correlated well with the expression and recruitment of B4. By d 20, we could conclude that the regenerated lens was considerably smaller in B4 morpholino-treated newts (Fig. 3B, E, F). On d 20, the lens size of B4 morpholino-injected newts was nearly half that of the control morpholino-injected newts (Fig. 3B). To investigate how B4 mediates such an effect on lens differentiation, we studied levels of cell proliferation and apoptosis in the regenerating lenses. BrdU was administered 24 h before fixation at different times, and its incorporation was analyzed by immunostaining. As expected, in control morpholino-injected newts the percentage of BrdU-positive cells was increased after 12 d. However, this percentage was significantly decreased in B4 morpholino-injected newts on d 16 and 20 (Fig. 3C, E). Next, Tunel staining was performed to examine whether lack of B4 leads to cell death. Indeed, it was shown that on d 20 significantly higher numbers of cells were undergoing apoptosis due to B4 morpholino treatment (Fig. 3D, F). Thus, so far our results clearly indicate an association between expression of B4 and lens transdifferentiation, which leads to a structurally normal lens. The reader should bear in mind that complete loss of B4 cannot result from the morpholino treatment; thus, even 50% decline in expression could elicit these results.
B4 is required for lens-specific gene expression during transdifferentiation
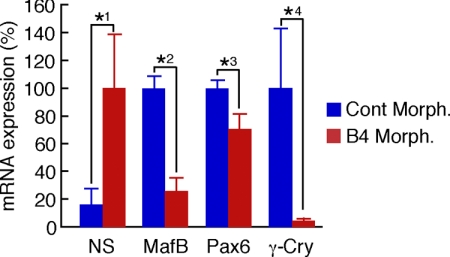
Despite the fact that the smaller lenses in B4 morpholino-injected newts can be attributed to an effect on proliferation and apoptosis, it is imperative to show whether expression of key genes known to regulate lens regeneration is affected by the B4 morpholino treatment. To examine this, we again injected newts for 20 d with B4 morpholino, collected only the regenerated lenses, isolated RNA, and examined the levels of several genes that are known to be structural and regulatory markers in lens differentiation and regeneration by qPCR. The expression of each gene was normalized with that encoding for ribosomal protein L27 so that we could account for the effect on lens size. Interestingly, we observed a dramatic down-regulation of γ-crystallin to 4% in regenerated lens from B4 morpholino-treated newts when compared to control. Likewise, we observed down-regulation of MafB and Pax-6, both known transcriptional factors that bind crystallin gene promoters (20, 21). On the contrary, we show up-regulation of nucleostemin (Fig. 4). Nucleostemin, a nucleolar protein that has been found in stem cells and cancer cells (22), is expressed in PECs during dedifferentiation and disappears after lens differentiation (23). Thus, it seems that the lens in B4 morpholino-treated animals retains the status of an earlier stage. From the effect of B4 knockdown on gene expression, it is clear that B4 is required for lens-specific gene expression during transdifferentiation.
Figure 4.
B4 is required for lens-specific gene expression during transdifferentiaion. Effects of B4 knockdown on gene expression were analyzed by qPCR. Clear indications show that B4 knockdown significantly down-regulates lens differentiation-specific markers, such as MafB, Prox1, and γ-crystallin (γ-Cry). However, nucleostemin (NS), a nucleolar protein related to stem cell-like state, is up-regulated. Error bars = sd. P values are from Student’s t test (2-tailed). *1: P = 0.0350 (n=4); *2: P = 0.0000249 (n=4); *3: P = 0.00342 (n=4); *4: P = 0.0298 (n=4). Cont. Morph., control vivo-morpholino; B4 Morph., B4 vivo-morpholino.
DISCUSSION
Transdifferentiation of PECs in newt lens regeneration is one of most obvious examples of in vivo reprogramming of somatic cells. It is also known that nuclear reprogramming is induced artificially by somatic cell nuclear transfer (SCNT) into oocytes, where genome-wide chromatin decondensation is mediated by replacement of linker histone H1 with histone B4. We hypothesized that similar events to SCNT might occur during reprogramming of PECs to lens cells. To test this hypothesis, we first cloned and examined expression of B4 during the process of lens regeneration. Indeed, we show that B4 is expressed in newt somatic cells. This is the first time that B4 was found in somatic cells. All previous studies in many animals, including mice, zebrafish, frogs, and sea urchins, have shown that B4 is specific to oocyte and early embryo before the onset of zygotic gene expression (7,8,9,10,11,12).
In addition to showing expression of B4 in PECs, we have also demonstrated that B4 has a function during lens transdifferentiation. Knocking down B4 induces apoptosis and negatively affects cell proliferation and lens differentiation, as shown by morphological as well as molecular criteria. Notably, we found that B4 regulates expression of key transcriptional factors, such as pax-6 and MafB, and of lens differentiation specific markers, such as γ-crystallin. However, knockdown of B4 up-regulates nucleostemin, a stem cell-specific marker, which is expressed during PEC dedifferentiation. However, the exact mechanisms whereby B4 acts on lens regeneration remain unclear. Nevertheless, we can offer two possible models. One is nonselective replacement. According to this, B4 would replace H1 nonselectively. This replacement would cause genome-wide chromatin decondensation similar to the reprogramming mediated by SCNT (16, 17). Such chromatin decondensation might allow transcriptional factors to interact with the promoter region of lens differentiation genes. The other model is selective replacement. In this case, B4 would affect expression of specific factors that need to interact with H1 for regulation. An example of such regulation has been shown in the case where Msx1 cooperates with H1b for repression of MyoD to inhibit muscle differentiation (24). To address those hypotheses, we need genome-wide ChIP-on-chip analysis. Such an experiment is not possible at present because the newt genome has not been sequenced.
Collectively, our expression and functional experiments identified a novel role of the linker histone B4. This is the first time that this oocyte-specific linker histone, which has been associated with reprogramming, was found to be expressed in adult somatic cells and, moreover, controls the process of transdifferentiation and lens regeneration. This finding suggests that reprogramming in germ cells and regenerating newt cells share similar strategies, thus providing a novel paradigm about cellular plasticity. In this case our results open new avenues for experimentation in regenerative biology.
Supplementary Material
Acknowledgments
The authors thank Y. Tanabe, and J. Hayashi for EST analysis. The authors are grateful to K. Ura for a helpful discussion. This work was supported by a KAKENHI grant (17657068) to N.M.; by the Naito Foundation and the Project for Realization of Regenerative Medicine as well as a Grant-in-Aid for Creative Research from the Japan Ministry of Education, Culture, Sports, Science, and Technology (17GS0318) to K.A., and by a U.S. National Institutes of Health grant (EY10540) to P.A.T.
References
- Abe S, Eguchi G. An analysis of differentiative capacity of pigmented epithelial cells of adult newt iris in clonal cell culture. Dev Growth Differ. 1977;19:309–317. doi: 10.1111/j.1440-169X.1977.00309.x. [DOI] [PubMed] [Google Scholar]
- Eguchi G, Okada T S. Differentiation of lens tissue from the progeny of chick retinal pigment cells cultured in vitro: a demonstration of a switch of cell types in clonal cell culture. Proc Natl Acad Sci U S A. 1973;70:1495–1499. doi: 10.1073/pnas.70.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Neelin J M, Callahan P X, Lamb D C, Murray K. The histones of chicken erythrocyte nuclei. Can J Biochem Physiol. 1964;42:1743–1752. doi: 10.1139/o64-185. [DOI] [PubMed] [Google Scholar]
- Godde J S, Ura K. Dynamic alterations of linker histone variants during development. Int J Dev Biol. 2009;53:215–224. doi: 10.1387/ijdb.082644jg. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hennebold J D, Macfarlane J, Adashi E Y. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Cho H, Wolffe A P. Xenopus laevis B4, an intron-containing oocyte-specific linker histone-encoding gene. Gene. 1994;143:233–238. doi: 10.1016/0378-1119(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Mandl B, Brandt W F, Superti-Furga G, Graninger P G, Birnstiel M L, Busslinger M. The five cleavage-stage (CS) histones of the sea urchin are encoded by a maternally expressed family of replacement histone genes: functional equivalence of the CS H1 and frog H1M (B4) proteins. Mol Cell Biol. 1997;17:1189–1200. doi: 10.1128/mcb.17.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw S, Vigneault C, Tremblay K, Sirard M A. Characterization of linker histone H1FOO during bovine in vitro embryo development. Mol Reprod Dev. 2006;73:692–699. doi: 10.1002/mrd.20448. [DOI] [PubMed] [Google Scholar]
- Wibrand K, Olsen L C. Linker histone H1M transcripts mark the developing germ line in zebrafish. Mech Dev. 2002;117:249–252. doi: 10.1016/s0925-4773(02)00173-9. [DOI] [PubMed] [Google Scholar]
- Muller K, Thisse C, Thisse B, Raz E. Expression of a linker histone-like gene in the primordial germ cells in zebrafish. Mech Dev. 2002;117:253–257. doi: 10.1016/s0925-4773(02)00174-0. [DOI] [PubMed] [Google Scholar]
- Teranishi T, Tanaka M, Kimoto S, Ono Y, Miyakoshi K, Kono T, Yoshimura Y. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Gao S, Chung Y G, Parseghian M H, King G J, Adashi E Y, Latham K E. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Becker M, Becker A, Miyara F, Han Z, Kihara M, Brown D T, Hager G L, Latham K, Adashi E Y, Misteli T. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol Biol Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci U S A. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P A, Kikyo N. Chromatin remodeling in nuclear cloning. Eur J Biochem. 2002;269:2284–2287. doi: 10.1046/j.1432-1033.2002.02887.x. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kihara M, Hennebold J D, Eppig J J, Viveiros M M, Emery B R, Carrell D T, Kirkman N J, Meczekalski B, Zhou J, Bondy C A, Becker M, Schultz R M, Misteli T, De La Fuente R, King G J, Adashi E Y. H1FOO is coupled to the initiation of oocytic growth. Biol Reprod. 2005;72:135–142. doi: 10.1095/biolreprod.104.032474. [DOI] [PubMed] [Google Scholar]
- Morcos P A, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. BioTechniques. 2008;45:613–614. doi: 10.2144/000113005. 616, 618. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Kashanchi F, Sax C M, Brady J N, Piatigorsky J. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol Cell Biol. 1995;15:653–660. doi: 10.1128/mcb.15.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Yasuda K. Characterization of the chicken L-Maf, MafB and c-Maf in crystallin gene regulation and lens differentiation. Genes Cells. 2002;7:693–706. doi: 10.1046/j.1365-2443.2002.00548.x. [DOI] [PubMed] [Google Scholar]
- Tsai R Y, McKay R D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, Takechi K, Sano S, Tarui H, Sasai Y, Agata K. Rapid accumulation of nucleostemin in nucleolus during newt regeneration. Dev Dyn. 2007;236:941–950. doi: 10.1002/dvdy.21027. [DOI] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.