Abstract
Affinity Grids are electron microscopy (EM) grids with a pre-deposited lipid monolayer containing functionalized Nickel-nitrilotriacetic acid (Ni-NTA) lipids. Affinity Grids can be used to prepare His-tagged proteins for single-particle EM from impure solutions or even directly from cell extracts. Here we introduce the concept of His-tagged adaptor molecules, which eliminate the need for the target protein or complex to be His-tagged. The use of His-tagged protein A as adaptor molecule allows Affinity Grids to be used for the preparation of virtually any protein or complex provided that a specific antibody is available or can be raised against the target protein. The principle is that the Affinity Grid is coated with a specific antibody that is recruited to the grid by His-tagged protein A. The antibody-decorated Affinity Grid can then be used to isolate the target protein directly from a cell extract. We first established this approach by preparing negatively stained specimens of both native ribosomal complexes and ribosomal complexes carrying different purification tags directly from HEK-293T cell extract. We then used the His-tagged protein A/antibody strategy to isolate RNA polymerase II (RNAP II), still bound to native DNA, from HEK-293T cell extract, allowing us to calculate a 25-Å resolution density map by single-particle cryo-EM.
Keywords: Monolayer purification, Affinity Grid, single-particle electron microscopy, ribosome, RNA polymerase II
Strategy for the use of Affinity Grids for non-His-tagged proteins and complexes
Single-particle electron microscopy (EM) is a versatile technique that can be used to determine the structure of macromolecular complexes (e.g., 1). Much effort has been directed in the last decade towards automating data collection (e.g., 2, 3) and advancing image processing techniques to achieve near-atomic resolution (reviewed in 4, 5). As a result of the numerous technical advances, the biochemical purification of macromolecular complexes suitable for structural analysis has now become a bottleneck in single-particle EM. Monolayer purification was introduced as a new method to prepare single-particle EM specimens directly from cell extracts without the need for biochemical purification of the target protein or complex 6. The underlying principle is that His-tagged target proteins contained in a cell extract are specifically recruited to functionalized Nickel-nitrilotriacetic acid (Ni-NTA) lipids that are part of a lipid monolayer formed over the cell extract. The lipid monolayer and the attached His-tagged proteins can then be transferred to an EM grid and prepared by negative staining or vitrification for EM imaging. The Affinity Grid was subsequently developed to simplify the use of monolayer purification by having the Ni-NTA lipid-containing monolayer pre-deposited on the EM grid 7. In addition, the Affinity Grid proved to be an even milder preparation method than monolayer purification, to avoid particle clustering often seen in specimens prepared by monolayer purification, and to make monolayer purification more suitable for the preparation of membrane proteins.
The application of both monolayer purification and Affinity Grid depends on the target protein carrying a His tag, which mediates the interaction with the Ni-NTA lipids in the monolayer. To extend its use to proteins and complexes that are not His-tagged, we exploited the fact that complexes can be assembled on the Affinity Grid in a step-wise fashion. This feature makes it possible to use His-tagged adaptor molecules to specifically recruit non-His-tagged target proteins to the Affinity Grid. Any molecule can serve as adaptor as long as it interacts specifically with the target protein and can be His-tagged. To avoid having to generate a specific His-tagged adaptor for each target protein, we produced His-tagged protein A, a surface protein expressed by S. aureus that interacts strongly with the constant domain of mammalian immunoglobulin G (IgG) antibodies 8. Hence, provided that a specific antibody is available or can be raised against the target protein, His-tagged protein A makes the Affinity Grid a viable option to prepare any protein or complex for single particle EM directly from cell extracts without prior biochemical purification (Fig. 1a).
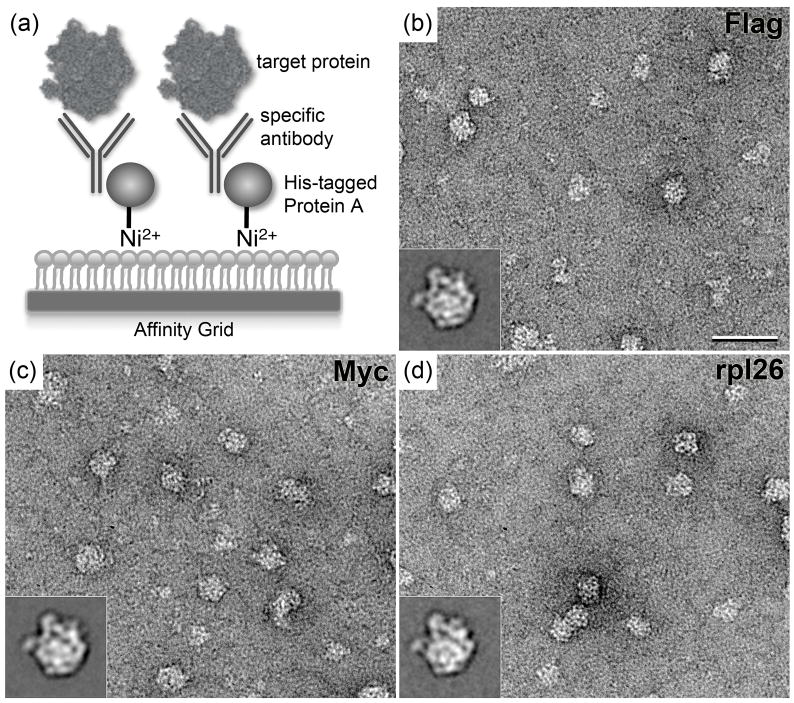
Figure 1. Principle of the recruitment of untagged complexes to the Affinity Grid using the His-tagged protein A/antibody strategy, and application to 60S ribosomal subunits.
(a) Schematic drawing of the recruitment of target complexes to the Affinity Grid by the His-tagged protein A/antibody adaptor system. (b–d) Representative images and class averages (insets) of negatively stained 60S ribosomal subunits recruited to the Affinity Grid by antibodies against Flag tag (b), Myc tag (c) and rpl26 (d). Scale bar is 60 nm and the side length of the insets is 43 nm. A construct of rpl3 containing an N-terminal tandem Flag-Myc tag was transfected into HEK-293T cells as described in 6. Affinity Grids were prepared according to 7. 3-μl aliquots first of His-tagged protein A (0.1 mg/ml) and then the respective IgG antibody (0.1 mg/ml) were applied to an Affinity Grid for 1 minute each. The excess solution was removed and a 3-μl aliquot of the HEK-293T cell extract was applied. After blotting, the sample was negatively stained according to 6. Specimens were examined using an FEI Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates under low-dose conditions at a nominal magnification of 67,000× and a defocus value of about −1.5 μm. Imaging plates were read out with a DITABIS Micron scanner (DITABIS Digital Biomedical Imaging System AG, Pforzheim, Germany) according to 23. The digitized images were binned over 2 × 2 pixels for a final sampling of 4.5 Å/pixel at the specimen level.
Testing the His-tagged protein A/antibody strategy with ribosomal complexes
Ribosomal complexes were chosen to test the His-tagged protein A/antibody strategy, because they were already used to establish the monolayer purification and Affinity Grid techniques 6, 7. Antibodies against the Flag and Myc tags were purchased from Sigma-Aldrich (St. Louis, MO) and Covance (Emeryville, CA), respectively. The antibody against subunit 26 (residues 129–145) of the large ribosomal subunit 26 (rpl26) was purchased from Abcam, Inc. (Cambridge, MA). To express His-tagged protein A, primers were designed for the protein A gene, which was then amplified from S. aureus genomic DNA purchased from ATCC (accession number 700699 D-5). The amplified gene was subcloned into the pET-15b vector, which includes an N-terminal His tag. The construct was expressed in E. coli (BL21 strain) and the recombinant protein was purified by conventional Nickel affinity and gel filtration chromatography as previously described for His-tagged rpl3 6. Mammalian HEK-293T cells were transiently transfected with a construct of the rpl3 subunit containing an N-terminal tandem Flag-Myc tag. Tandem-tagged rpl3 was produced by introducing a Myc-tagged rpl3 construct into the pCMV-Tag 2A vector (Agilent Technologies, Santa Clara, CA), which adds an N-terminal Flag tag 9. After 48 hours a cell extract was prepared as described previously 6.
To prepare ribosomal complexes from the HEK-293T cell extract, Affinity Grids with a monolayer containing 2% Ni-NTA lipids were first incubated for one minute with 3 μl of a 0.1 mg/ml solution of His-tagged protein A and then for another minute with 3 μl of a 0.1 mg/ml solution of one of the three antibodies, anti-Flag, anti-Myc and anti-rpl26 IgGs. The solution containing unbound protein A and antibodies was gently removed from the grid with a Hamilton syringe, and 3 l aliquots of the HEK-293T cell extract (containing ribosomal complexes with incorporated Flag-Myc tagged rpl3) were added to the grids. After a 2-minute incubation, the grids were washed and prepared for EM imaging by negative staining as described 6.
In establishing the method we adjusted the incubation steps of the protocol to optimize particle binding while minimizing background. We found that optimizing conditions for the incubations with His-tagged protein A and antibody were most critical for minimizing the background. Although some background of unoccupied protein A and/or IgG antibody always remained, 1-minute incubations with 0.1 mg/ml solutions of His-tagged protein A and antibody resulted in the lowest background. Since the interactions between the His tag of protein A and the Ni-NTA lipids and between protein A and the constant part of IgG antibodies may be affected to some degree by the buffer conditions, different antibody concentrations can be tested (typically 0.01–0.2 mg/ml). The affinities of antibodies for their antigens may be most variable, and it is therefore important to test for how long the antibody-decorated Affinity Grid is best incubated with the cell extract containing the target protein (typically 2–5 minutes) and whether the concentration of the cell extract needs to be adjusted. We found that for all three antibodies we tested to recruit ribosomal complexes to Affinity Grids, anti-Flag, anti-Myc and anti-rpl26 IgGs, a 2-minute incubation with HEK-293T cell extract yielded a particle density suitable for single-particle analysis (Figs. 1b–d). According to the 3D model of the mammalian ribosome 10, the epitopes for the antibodies should be equally accessible and so the three antibodies seem to have similar affinities for their antigens.
Images of negatively stained specimens showed that all preparations contained complexes ~30 nm in size and displaying features consistent with the mammalian 60S ribosomal subunit (Figs. 1b–d). For each antibody-specific sample, 56 images were recorded, and ~8000 particles were selected (8112 particles for the specimen prepared with anti-Flag antibody, 7825 particles for that prepared with anti-Myc antibody, and 7947 particles for that prepared with anti-rpl26 antibody). After windowing the particles into individual 96 × 96 pixel images, the particles were classified into 20 classes using the SPIDER software package 11. Each data set was subjected to 10 rounds of multi-reference alignment followed by K-means classification. The references used in the first alignment were randomly selected from the raw images.
Each set of classes produced averages that displayed features consistent with the 60S ribosomal subunit (insets in Figs. 1b-d and Supplementary Fig. 1). Although there is some variation, the class averages show that the 60S ribosomal subunits adsorbed to the EM grid with a strongly preferred orientation that is the same in all three sets of averages. According to the placement of X-ray crystal structures into the 8.9-Å cryo-EM map of the 60S ribosomal subunit 10, subunits rpl3 and rpl26 are located in orthogonal positions in the 60S subunit. It is therefore somewhat unexpected that EM specimens prepared with anti-Myc and anti-Flag antibodies (both on rpl3) and that prepared with anti-rpl26 antibody show predominantly the same view of the 60S ribosomal subunit. Preferred orientations are often introduced by the negative staining procedure, in particular during the drying of the specimen 12, 13. It thus appears that the protein A/antibody linkage of the 60S ribosomal subunits to the Affinity Grid provides sufficient flexibility for the complex to adopt its preferred orientation on the grid. The notion of high linker flexibility is consistent with our previous finding that 50S ribosomal subunits, recruited to an Affinity Grid through His-tagged rpl3, adopted almost randomly distributed orientations in vitrified specimens 7.
Application of the His-tagged protein A/antibody strategy to RNA polymerase II
After establishing the His-tagged protein A/antibody strategy with the 60S ribosomal subunit, we wanted to use the method to isolate an unrelated biological complex from the same cell extract for analysis by cryo-EM. We chose to prepare native human RNA polymerase II (RNAP II), a 12-subunit complex with a molecular weight of ~550 kDa. The overall composition and structure of RNAP II are conserved from yeast to human (e.g., 14, 15, 16, 17).
We incubated Affinity Grids with a monolayer containing 20% Ni-NTA lipids with His-tagged protein A and an antibody against the flexible C-terminal domain linker region of Rpb1 (yeast subunit nomenclature). After incubation with HEK-293T cell extract, the grid was blotted and vitrified by quick-freezing into liquid ethane. Images of the vitrified specimen showed particles with dimensions and features consistent with those previously seen in specimens of purified yeast RNAP II 18. Many of the particles were, however, attached to long strands, ~16 Å in diameter, presumably representing nucleic acid polymers (Fig. 2a).
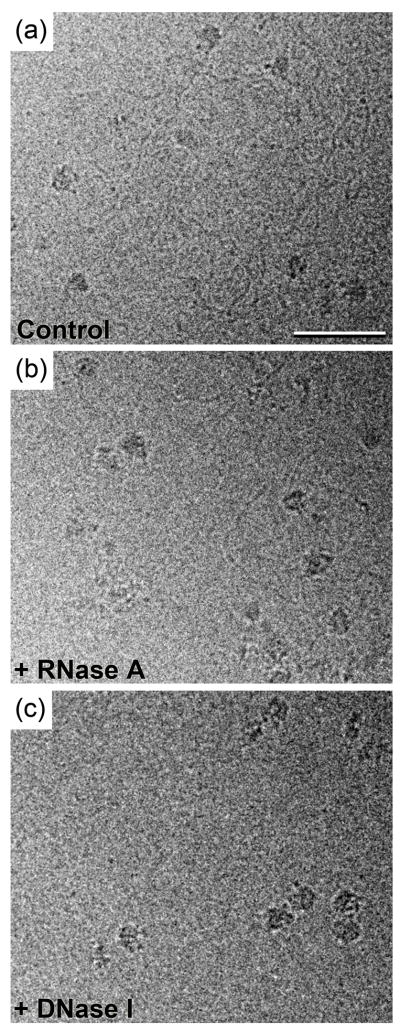
Figure 2. Images of vitrified RNAP II purified onto Affinity Grids using His-tagged protein A and an antibody against Rpb1.
(a) Image of vitrified RNAP II prepared onto an Affinity Grid from HEK-293T cell extract, showing that many of the complexes are associated with nucleic acid strands. (b) Treatment of the cell extract (0.1 ml of ~3 mg/ml protein) with 10 units of RNase A for 30 minutes prior to application to the Affinity Grid did not eliminate the strands. (c) Treatment of the cell extract with 10 units of DNase I for 30 minutes prior to application to the Affinity Grid eliminated the majority of the strands, identifying the strands to be DNA. Vitrified specimens of RNAP II were prepared according to 7 and transferred into an FEI F20 electron microscope equipped with a field emission gun using an Oxford cryo-specimen holder, maintaining a temperature of −180°C. Samples were examined at an acceleration voltage of 200 kV and images were recorded on Kodak SO-163 film at a nominal magnification of 50,000× using low-dose procedures and a defocus ranging from −2 to −4 μm. The film was developed and digitized as described 6 for a final sampling of 4.2 Å/pixel at the specimen level. Scale bar is 50 nm.
To identify the strands associated with the RNAP II particles, we prepared vitrified Affinity Grid samples with cell extracts that were treated with DNase I (Sigma-Aldrich, St. Louis, MO), RNase A (Roche Applied Science, Indianapolis, IN) or lysis buffer (as a control). Cryo-EM images of all samples showed RNAP II particles. In the control sample prepared with cell extract treated with lysis buffer, most RNAP II particles were still associated with nucleic acid strands (not shown, but similar to Fig. 2a), as were the RNAP II particles in the sample prepared with RNase A-treated cell extract (Fig. 2b). In contrast, the sample prepared with DNase I-treated cell extract lacked all but a few thin fibers (~10 Å in diameter, potentially mRNA) (Fig. 2c). We therefore conclude that the majority of strands associated with the RNAP II complexes were DNA molecules.
Cryo-EM density map of RNAP II
We selected 28,380 particles of RNAP II still attached to DNA strands from 216 images of vitrified samples and classified them into 100 classes. The resulting class averages indicated that RNAP II adopted many different orientations in the vitrified ice layer (Supplementary Fig. 2a). To produce a reference model, we used the EMAN software package 3 to generate a 30-Å reference map based on the crystal structure of the yeast RNAP II elongation complex, which contains 19 bases of template DNA, 7 bases of non-template DNA and 10 residues of single-stranded RNA (PDB entry 1Y77) 19. We then used FREALIGN version 7.05 20 to align the particle images to the reference model and to calculate a CTF-corrected density map that served as new reference model. The orientation parameters were further refined over 10 alignment cycles in FREALIGN. The Fourier shell correlation (FSC) curve with the final data set containing 22,704 particles indicated a resolution of 25 Å (Supplementary Fig. 2b), according to the FSC = 0.5 criterion 21, and the final density map was low-passed filtered to this resolution (Fig. 3). A plot of the Euler angles shows that the ice-embedded particles adopted randomly distributed orientations (Supplementary Fig. 2c), demonstrating that the protein A/antibody linker provides sufficient flexibility to obtain the different views needed to calculate a fully defined 3D reconstruction. Comparisons between raw images, class averages and re-projections from the density map suggest that the 3D reconstruction is consistent with the projection data (Supplementary Fig. 2d).
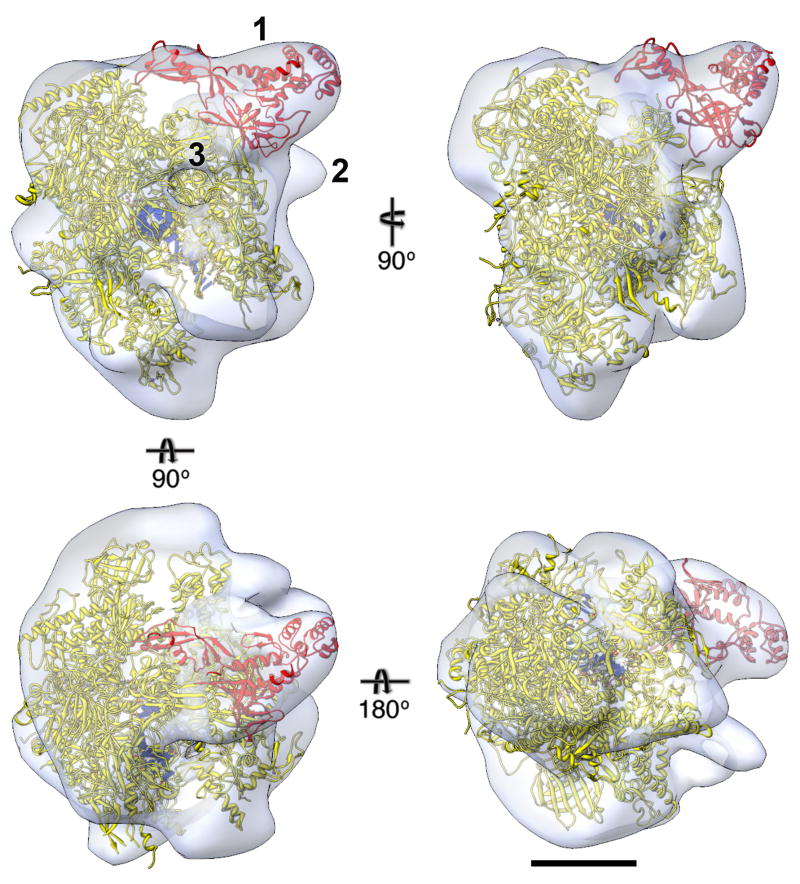
Figure 3. EM density map of human RNAP II with docked crystal structure.
Different views of the final density map (white translucent surface) of human RNAP II filtered to 25 Å resolution calculated from 22,704 particles, into which the crystal structure of yeast RNAP II was placed (Rpb4/7 in red, other subunits in yellow, nucleic acid in blue; PDB entry 1Y77 19). Label “1” indicates the region of the density map into which the atomic model of Rpb4/7 was moved. Label “2” indicates density not accounted for by the crystal structure, which may represent the C-terminal domain of Rpb1 and/or part of the antibody against Rpb1 used to recruit RNAP II to the Affinity Grid. Label “3” indicates the exit channel for the newly synthesized RNA. Scale bar is 5 nm.
Our cryo-EM density map of the human RNAP II accommodated the full yeast elongation complex, but required some adaptations of the crystal structure. The model for the Rpb4/7 heterodimer was outside the density map, but was located directly adjacent to an unoccupied region of the map. We therefore manually placed the Rpb4/7 structure into the unoccupied density by a rigid body movement (labeled “1” in Fig. 3, Supplementary Fig. 2e). The fact that the Rpb4/7 heterodimer was not seen in several crystal structures of the yeast complex (e.g., 14, 15, 22) suggests that it is flexibly attached to RNAP II, providing justification for its movement into our density map. Indeed, the Rpb4/7 also had to be moved to fit into the density in a previous 3D reconstruction of human RNAP II obtained with cryo-negatively stained specimens 17.
The second region of our density map that differs from the crystal structure of the yeast complex is an area of extra density adjacent to the C-terminal domain of Rpb1 (labeled “2” in Fig. 3). This extra density was also seen in the previous EM map of human RNAP II 17, which was attributed to the flexible C-terminal domain of Rpb1. In our map, this unoccupied density may also represent some portion of the antibody used to recruit the complex to the Affinity Grid, as it binds to this region of Rpb1. Additional small differences between our density map and the manually fit crystal structure may be accounted for by missing loops in the crystal structures and small differences between the yeast and human complexes.
The DNA binding groove was unoccupied in the previous EM reconstruction obtained with biochemically purified RNAP II. As we selected RNAP II particles that were still associated with DNA, our density map includes density for the DNA. Furthermore, the density map also visualizes the exit channel for the RNA (labeled “3” in Fig. 3), which is seen at the expected position, i.e., over the single-stranded RNA and upstream of the DNA fragment seen in the crystal structure of yeast RNAP II.
Conclusions and outlook
We have established a method based on a His-tagged adaptor molecule that allows the use of Affinity Grids to prepare non-His-tagged proteins and complexes from cell extracts. We chose His-tagged protein A as adaptor because it can be universally used for any protein or complex provided that there is a specific antibody for the target, but many other adaptor molecules can be considered. For example, His-tagged ligands can be used to recruit the corresponding receptors or His-tagged avidin could be used to recruit biotinylated targets. Any such adaptor system is useful for the preparation of single-particle specimens, but if the target protein is of pharmacological interest, structural knowledge of antibody-protein or ligand-receptor interactions can also provide valuable information to guide efforts in drug development.
The averages of the negatively stained 60S ribosomal subunits (Figs. 1b-d and Supplementary Fig. 1) and the angular distribution plot of the vitrified RNAP II particles (Supplementary Fig. 2c) suggest that the linker system provides substantial flexibility. Hence, the target complex can adopt preferred orientations in negatively stained specimens and randomly distributed orientations in a vitrified ice layer. Specimens prepared with the Affinity Grid using an adaptor system thus have similar characteristics as conventionally prepared specimens.
Because of its ease and versatility, the protein A/antibody adaptor system for the isolation of native complexes from cell extracts using the Affinity Grid could be a very helpful tool for high-throughput structure determination of macromolecular complexes by single-particle EM.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant PO1 GM62580 (to Stephen C. Harrison). T.W. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy 30 years. Q Rev Biophys. 2009;42:139–158. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stagg SM, Lander GC, Pulokas J, Fellmann D, Cheng A, Quispe JD, Mallick SP, Avila RM, Carragher B, Potter CS. Automated cryoEM data acquisition and analysis of 284742 particles of GroEL. J Struct Biol. 2006;155:470–481. doi: 10.1016/j.jsb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Walz T. The advent of near-atomic resolution in single-particle electron microscopy. Annu Rev Biochem. 2009;78:723–742. doi: 10.1146/annurev.biochem.78.070507.140543. [DOI] [PubMed] [Google Scholar]
- 5.Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. Proc Natl Acad Sci U S A. 2008;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: a pre-fabricated EM grid for monolayer purification. J Mol Biol. 2008;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsgren A, Sjoquist J. “Protein A” from S. aureus”. I Pseudo-immune reaction with human gamma-globulin. J Immunol. 1966;97:822–827. [PubMed] [Google Scholar]
- 9.Dukovski D, Li Z, Kelly DF, Mack E, Walz T. Structural and functional studies on the stalk of the transferrin receptor. Biochem Biophys Res Commun. 2009;381:712–716. doi: 10.1016/j.bbrc.2009.02.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 12.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 15.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 16.Chung WH, Craighead JL, Chang WH, Ezeokonkwo C, Bareket-Samish A, Kornberg RD, Asturias FJ. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- 17.Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Craighead JL, Chang WH, Asturias FJ. Structure of yeast RNA polymerase II in solution: implications for enzyme regulation and interaction with promoter DNA. Structure. 2002;10:1117–1125. doi: 10.1016/s0969-2126(02)00813-4. [DOI] [PubMed] [Google Scholar]
- 19.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 20.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 22.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Hite RK, Cheng Y, Walz T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J Electron Microsc. 2009;59:53–63. doi: 10.1093/jmicro/dfp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.