Abstract
Cornelia de Lange Syndrome (CdLS) and manifests facial dysmorphic features, growth and cognitive impairment, and limb malformations. Mutations in three genes (NIPBL, SMC1A and SMC3) of the Cohesin complex and its regulators have been found in affected patients. Here, we present clinical and molecular characterization of 30 unrelated patients with CdLS. Eleven patients had mutations NIPBL (37%) and three patients had mutations in SMC1A (10%), giving an overall rate of mutations of 47%. Several patients shared the same mutation in NIPBL (p.R827GfsX2) but had variable phenotypes, indicating the influence of modifiers in CdLS. Patients with NIPBL mutations had a more severe phenotype than those with mutations in SMC1A or those without identified mutations. However, a high incidence of palate defects was noted in patients with SMC1A mutations. In addition, we observed a similar phenotype in both male and female patients with SMC1A mutations. Finally, we report the first patient with an SMC1A mutation and the Sandifer complex.
Keywords: Cornelia de Lange Syndrome (CdLS), NIPBL, SMC1A, SMC3, genes, mutations
Introduction
Mutations in NIPBL, a regulator of the Cohesin complex, were the first demonstrated cause of Cornelia de Lange Syndrome (CDLS1, MIM 122470) [Krantz et al., 2004; Tonkin et al., 2004]. This disorder is characterized by distinctive dysmorphic facial features, impairment of growth and cognitive development, limb malformations, and additional organ system involvement with variable expressivity [Ireland et al., 1993]. Most patients with CdLS have unaffected parents but familial cases have been reported [Russell et al., 2001]. Recently, mutations in the Cohesin structural components SMC1A and SMC3 were found in patients with atypical CdLS (CDLS2, MIM 300590 and CDLS3, MIM 610759) [Borck et al., 2007; Deardorff et al., 2007; Musio et al., 2006]. Eleven different SMC1A mutations in 14 unrelated patients have been reported. All patients had a mild to moderate CdLS phenotype [Borck et al., 2007; Deardorff et al., 2007; Musio et al., 2006]. Only a single mildly affected patient has been identified with a mutation in SMC3 [Deardorff et al., 2007].
Here, we report the identification of 14 additional mutations of the Cohesin complex genes NIPBL (11) and SMC1A (3) and discuss genotype-phenotype correlations in a cohort of 30 unrelated patients with CdLS.
Materials and Methods
Clinical Evaluation
Patients were enrolled in this study under an IRB-approved protocol. The evaluation included clinical and family history, physical examination and psychological evaluation. Probands were classified following the criteria proposed by Gillis et al. [2004]. Genotype-phenotype correlations were established using SPSS v14.0 program for Windows. We used the χ2 test of independence for the comparison between clinical findings and the presence of mutations in NIPBL and SMC1A. Post hoc analyses of the contingency table cells were based on adjusted residuals of Haberman. This is equivalent to a situation of multiple testing; therefore, Bonferroni corrections were calculated for the p-values of the cells.
Variant allele analysis
Genomic DNA was isolated from peripheral blood lymphocytes or cultured chorionic villi by standard protocols. The coding region and flanking intronic sequences of NIPBL (exons 2-47), SMC1A (exons 1-25) and SMC3 (exons 1-29) were screened for mutations by PCR amplification and bidirectional direct sequencing. Parental genotypes and 50 ethnically matched population controls were screened to assess whether each variant was de novo. Reference sequences used for SMC1A, SMC3 and NIPBL were RefSeq NM_006306, NM_005445 and NM_133433, respectively. When mRNA analysis was feasible, the Invitrogen RT-PCR kit was used. RT-PCR products were confirmed by sequencing. Details of the primers used and PCR conditions for NIPBL, SMC1A and SMC3 amplification are available upon request. Protein subsequence motifs for human NIPBL were analyzed using data from Uniprot [Wu et al., 2006] and Pfam [Finn et al., 2006] databases.
Results
Clinical Phenotypes
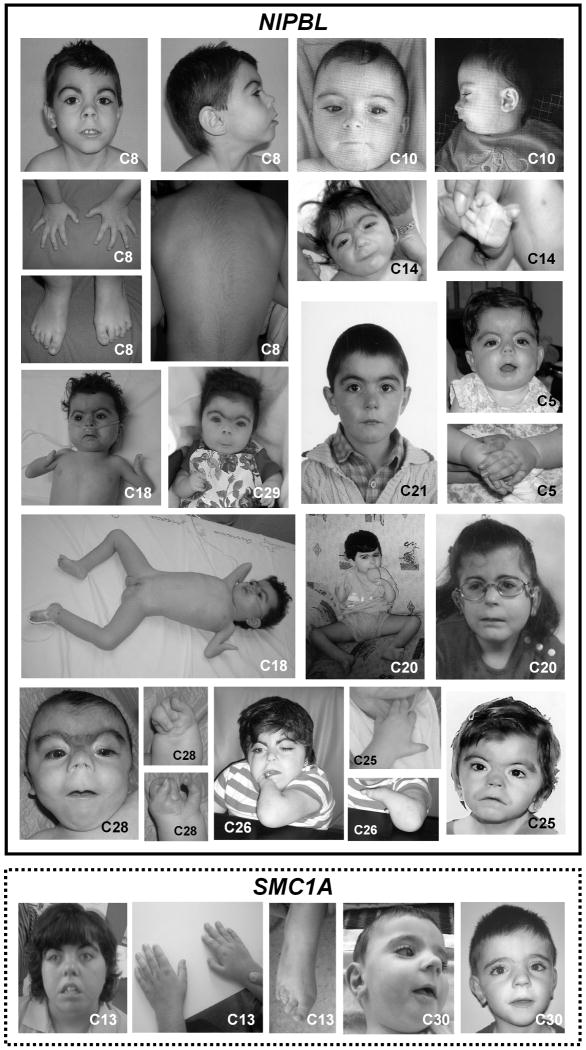
Thirty unrelated probands were studied. Twenty-seven were from Spain, two from Romania (patients C2 and C27), and one from Morocco (patient C18) (Table I). Additional molecular and clinical data of the 14 patients with identified mutations are included in Supplementary Table I (online material) and photographs are shown in Fig. 1.
Table I. Genotypes and Phenotypes of 30 patients with Cornelia de Lange syndrome.
| Clinical feature | Mutation negative n (Adjusted residuals) | Mutation positive | Total patients | |
|---|---|---|---|---|
| NIPBL n (Adjusted residuals) | SMC1A n (Adjusted residuals) | |||
| Sex | 11M / 19F | 2M / 1F | 4M / 7F | 30 |
| IUGR (Adjusted residuals) | 6 (-2.5) | 10 (2.5)** | 2 (0.2) | 18 |
| Growth Retardation | ||||
| Mild (>75%) | 13 (2.8)** | 2 (-3.8) | 3 (1.4) | 18 |
| Moderate (25-75%) | 2 (-2.8) | 9 (3.8)*** | 0 (-1.4) | 11 |
| Limb Abnormalities | ||||
| Mild (no reduction) | 15 (1.6) | 7 (-2.2) | 3 (0.8) | 25 |
| Moderate (>2 fingers) | 1 (-0.1) | 1 (0.4) | 0 (-0.5) | 2 |
| Severe (≤2 fingers) | 0 (-2.0) | 3 (2.4)* | 0 (-0.6) | 3 |
| Developmental Delay | ||||
| Mild (<2 years delayed) | 7 (1.3) | 3 (-0.2) | 0 (-1.7) | 10 |
| Moderate (>2 years delayed) | 5 (-0.9) | 3 (-0.5) | 3 (1.9) | 11 |
| Severe (profound delayed) | 0 (-1.1) | 1 (1.5) | 0 (-0.4) | 1 |
| Other clinical findings | ||||
| Microcephaly | 8 (-1.6) | 9 (1.6) | 2 (0.1) | 19 |
| Hirsutism | 7 (-0.7) | 7 (1.1) | 1 (-0.6) | 15 |
| Cardiac defect | 8 (-0.2) | 5 (-0.1) | 2 (0.5) | 15 |
| Gastroesophageal reflux | 5 (-1.0) | 5 (0.5) | 2 (1.0) | 12 |
| Deafness | 3 (-2.1) | 7 (2.2)* | 1 (-0.2) | 11 |
| Genitourinary anomalies | 4 (-1.4) | 6 (1.5) | 1 (-0.1) | 11 |
| Ophthalmological anomalies | 7 (0.9) | 3 (-0.8) | 1 (-0.1) | 11 |
| Palate anomalies | 5 (-1.4) | 5 (0.2) | 3 (2.1)* | 13 |
Evaluation of clinical findings in patients with CdLS with and without mutation in NIPBL or SMC1A. Growth curves are per CdLS curves.
Significant between 0.1<p<0.05,
Significant at p<0.05,
Significant at p<0.01, Bonferroni corrected p
FIG. 1.
Phenotypes of patients with CdLS and mutations in NIPBL and SMC1A. Individuals C5, C8, C10, C14, C18, C20, C21, C25, C26, C28 and C29 have mutations in NIPBL. Individuals C13 and C30 have mutations in SMC1A. Phenotype of individual C2 has been published elsewhere [Deardorff et al., 2007]. Patients with NIPBL mutations are surrounded by a solid line and patients with SMC1A mutations are surrounded by a dashed line.
Variant alleles
NIPBL
Thirty-eight variant alleles of NIPBL were identified in 30 unrelated probands (Supplementary Tables I, II and III, online material). Four prenatal diagnoses were normal. Eleven of the variants were apparently de novo mutations, nine of which had not been previously reported, giving a prevalence of 37% in this cohort (Supplementary Table I and Supplementary Fig. 1A, online material). Of these new mutations, two were nonsense: c.2146C→T (p.Q716X) and c.6880C→T (p.Q2294X), three were splice site: c.230+1G→A (p.L22QfsX3), c.4320+5G→C (p.V1414_A1440del) and c.4321G→T (p.F1442KfsX3/ p.V1441L), three were missense: c.6242G→C (p.G2081A), c.6269G→T (p.S2090I) and c.6449T→C (p.L2150P), and one was an in-frame deletion: c.5689_5691delAAT (p.N1897del).We also found two frameshift deletions that had been previously reported: c.2479_2480delAG (p.R827GfsX2) in seven unrelated patients [Bhuiyan et al., 2006; Gillis et al., 2004; Kaur et al., 2005; Selicorni et al., 2007] and c.7438_7439delAG (p.R2480KfsX5), which had been reported in one patient [Yan et al., 2006] (Supplementary Table I, online material). The mutation c.230+1G→A generated an alternative transcript with skipping of exon 3 to predict a truncated protein. The other intronic mutation (c.4320+5G→C) produced a variant without exon 19 to predict an in-frame deletion of 27 amino acid residues. However, the mutation c.4321G→T affected the first base of exon 20 and generated an alternative splicing with two transcripts, one bearing an exon 20 deletion to predict a truncated protein, and another normal-sized transcript predicted to yield p.V1441L protein (Supplementary Fig. 1A and 2, online material). Multiple sequence analysis in the neighborhood of mutated residues V1441, N1897, G2081, S2090I and L2150 wereconserved across species (rat, chicken, zebrafish) (Supplementary Fig. 1, online material). These changes were not detected in their respective parents or in 100 control alleles.
SMC1A
Among the 19 NIPBL-negative patients, we identified three mutations in SMC1A, giving a prevalence of 10% among the 30 patients reported here. We found an in-frame deletion c.802_804delAAG (p.K268del) in one woman, and a missense mutation c.2132G→A (p.R711Q) in one male (Supplementary Table I, Fig. 1 and Supplementary Fig. 3A). These residues were conserved (Supplementary Fig. 3B and 3C) and were not detected in parents or in 100 control alleles, suggesting de novo events. The other mutation, a c.587G→A (p.R196H) was previously reported [Deardorff et al., 2007], but the patient developed new clinical symptoms, which we describe in this report (Supplementary Table I and Supplementary Fig. 3A). Two novel and two previously reported SMC1A polymorphisms or variants of unknown significance were also observed [Deardorff et al., 2007] (Supplementary Tables II and III, online material).
SMC3
Twenty-nine sequence variants in SMC3 that may represent neutral polymorphisms were observed in 16 NIPBL-negative probands (Supplementary Tables II and III, online material).
Discussion
In this study of 30 patients with CdLS, we identified mutations in NIPBL (37%), or SMC1A (10%) (47% overall). Our NIPBL and SMC1A mutation rates were similar to prior reports [Bhuiyan et al., 2006; Borck et al., 2007; Deardorff et al., 2007; Gillis et al., 2004; Musio et al., 2006; Selicorni et al., 2007; Yan et al., 2006]. To carry out accurate genotype-phenotype correlations in this cohort, the NIPBL-positive probands were classified into three groups: truncating mutations, splice-site mutations, and missense or in-frame deletion mutations. Truncating mutations presumably result in haploinsuffiency and, as expected, led to more severe phenotypes. Interestingly, patient 20, who had severe bilateral limb defects, carried the mutation p.R827GfsX2, which had been previously identified in seven patients of distinct ethnic origins. This is currently the most frequent CdLS-causing mutation worldwide [Bhuiyan et al., 2006; Gillis et al., 2004; Kaur et al., 2005; Selicorni et al., 2007]. In addition, phenotypes associated with this mutation are quite variable, with limb defects ranging from mild to severe. This indicates the influence of modifiers in the expressivity of CdLS [Gillis et al., 2004].
As in a previous report we noted a splice-site mutation with a variable phenotype [Selicorni et al., 2007]. Patient C10, who had a mild phenotype, had the mutation p.L22QfsX3, which generates both a normal transcript and a predominant aberrant alternatively spliced transcript with an exon 3 deletion. The pathogenic effect of this mutation, in addition to the early truncation of the protein, may be explained by disruption of NIPBL interaction with mau-2 in the Cohesin complex [Seitan et al., 2006]. Another interesting splice-site mutation is p.V1414_A1440del (c.4320+5G→C), which generated an aberrant spliced transcript with deletion of exon 19. This small exon codifies 27 amino acids located within the more preserved carboxy terminal half of the protein, although it has not been associated with any functional domain. Loss of exon 19 in patient C28 led to severe limb defects. To date, only one other single mutation, p.V1414_A1440del (c.4320+2T→A), which deleted exon 19 and was associated with severe limb malformations has been reported [Schoumans et al., 2007]. The finding of another splice-site mutation, p.F1442KfsX3/ p.V1441L (c.4321G→T), which caused the deletion of exon 20 and mild symptoms, suggests this domain of the protein is important in limb formation.
As expected, patients with missense mutations or in-frame deletions (when compared to patients with nonsense or frameshift mutations) had milder clinical features, with one exception; patient C18, who had bilateral single-digit hypomelia and carried the mutation p.L2150P. The presence of severe phenotypes in patients carrying missense mutations suggests the involvement of functional domains of the protein. Among the 147 mutations previously reported in NIPBL, only two missense mutations have been associated with severe limb defects [Schoumans et al., 2007; Tonkin et al., 2004]. Surprisingly, one of them, p.T2146P, is located four amino acids away (in the 5′ side) from the mutation p.L2150P mutation and lies in the protein-interaction region HEAT repeat [Schoumans et al., 2007] (Supplementary Fig. 1A and 1E, online material). The Drosophila (Nipped-BNC39) mutation Nipped-BI1510 (equivalent to V2147 en NIPBL) produces an increase in the number of “nicks” in the wings. These alterations are currently considered equivalent to those seen in the human hands [Gause et al., 2008]. The mutated residue I1510 in Nipped-B is also conseved in NIPBL (V2147) and it is close (in the 3′direction) to mutation T2146 and to three amino acids (in the 5′direction) of the new mutation L2150 (Supplementary Fig. 1A and 1E, online material). The similarities in the effects of the change in these residues indicate that they are part of an amino acid-sequence involved in limb development.
More recently, it has been reported that in NIPBL the sequence between residues 1838 and 2000, included in the predicted region HEAT repeat interacts with histone deacetylases 1 and 3 [Jahnke et al., 2008]. The novel mutation p.N1897del is located in the middle of this sequence, two amino acids away from mutation p.R1895T, which affects that interaction [Jahnke et al., 2008]. The clinical findings associated with this mutation and with the four reported that affect that domain [Borck et al., 2007; Gillis et al., 2004; Jahnke et al., 2008; Selicorni et al., 2007], include mild involvement of the limbs and suggest that the NIPBL-deacetylase interaction has a minor role in limb development.
The two new mutations identified in the SMC1A gene, one missense and one in-frame deletion, suggest that more severe mutations of the protein are incompatible with life [Liu and Krantz, 2008]. Both mutations are located in the predicted coiled-coil structure. The deletion of residue K268 (p.K268del) would cause a shift in the relative position of the accompanying residues, leading to displacement of corresponding residues of the two anti-parallel helices that form the coiled coil. The missense mutation p.R711Q affects arginine R711, which was mutated in other patient previously reported (p.R711W) [Deardorff et al., 2007]. It is interesting that five out of eleven mutations in the SMC1A gene affect positively-charged arginine residues, and it has been proposed that these residues may be involved in an interaction with DNA [White and Erickson, 2009].
In this series, we found no mutations in the SMC3 gene. Instead, we found a large number of polymorphisms or variants of unknown significance.
The study of the clinical features of this cohort showed that the growth retardation, limb malformations, and even hearing loss, are more likely to be present in patients carrying NIPBL mutations, compared to patients with SMC1A mutations. In contrast, patients with mutations in SMC1A had a higher incidence of high palate anomalies, a finding not previously reported. The remaining clinical manifestations in patients with mutations in SMC1A were usually moderate to mild, and similar to those of patients in whom no mutation was found (Table I). Of the three major clinical features of CdLS, mental retardation is the only one that is consistently present in the three groups of patients analyzed. This indicates that additional genes implicated in CdLS may produce mild or moderate phenotypes with mental retardation as the most relevant feature [Deardorff et al., 2007].
Another interesting finding is the presence of the Sandifer complex in a 3-year-old boy (C30) carrying a mutation in the SMC1A gene. Although the Sandifer complex has been previously reported in patients with CdLS [Sommer, 1993] we report the first patient with this manifestation and with an identified genetic mutation. Besides, this patient had recently developed epilepsy, for which he was being treated with an anticonvulsivant. Finally, the female patient with SMC1A mutation did not appear to have milder clinical manifestations than the males carrying a mutation in the same gene, indicating that in our cohort of SMC1A mutated cases, gender is irrelevant to expressivity.
Supplementary Material
Acknowledgments
We thank the families who participated in this study and the Spanish Cornelia de Lange Syndrome Foundation. We thank Drs. Angel Alonso and Alberto Valiente from the Service of Genetics, Hospital Virgen del Camino, Pamplona, Spain for providing samples. This study was supported by grants from: The Spanish Ministry of Health-Fondo de Investigación Sanitaria (FIS) (Ref.# PI061343); the Diputación General de Aragón (DGA) (Grupo Consolidado B20); Spanish Ministry of Education and Science (Ref.# SAF2004-06843); the Comunidad de Madrid's Activities Program R&D Groups in Biosciences (Ref.# S-BIO-0260/2006-COMBACT); the Ajut de Suport als Grups de Recerca de Catalunya (Ref.# 2005SGR-00733); CIBER Fisiopatología de la Obesidad y Nutrición is an initiative of Instituto de Salud Carlos III, Spain and the NICHD (5PO1HD052860) (IDK). We also thank Biomol-Informatics S.L. (www.biomol-informatics.com) for bioinformatic counselling; and the Fundación Ramón Areces for financial support to CBMSO. MC and JCK are the recipients of fellowships from Banco Santander Central Hispano, and MCGR from the Diputación General de Aragón (Ref.# B120/2006).
References
- Bhuiyan ZA, Klein M, Hammond P, van Haeringen A, Mannens MM, Van Berckelaer-Onnes I, Hennekam RC. Genotype-phenotype correlations of 39 patients with Cornelia de Lange syndrome: the Dutch experience. J Med Genet. 2006;43:568–575. doi: 10.1136/jmg.2005.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Zarhrate M, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Incidence and Clinical Features of X-linked Cornelia de Lange Syndrome Due to SMC1L1 Mutations. Hum Mutat. 2007;28:205–206. doi: 10.1002/humu.9478. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pié J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in Cohesin Complex Members SMC3 and SMC1A Cause a Mild Variant of Cornelia de Lange Syndrome with Predominant Mental Retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID. NIPBL Mutational Analysis in 120 Individuals with Cornelia de Lange Syndrome and Evaluation of Genotype-Phenotype Correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland M, Donnai D, Burn J. Brachmann-de Lange syndrome. Delineation of the clinical phenotype. Am J Med Genet. 1993;47:959–964. doi: 10.1002/ajmg.1320470705. [DOI] [PubMed] [Google Scholar]
- Jahnke P, Xu W, Wulling M, Albrecht M, Gabriel H, Gillessen-Kaesbach G, Kaiser FJ. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008;36:6450–6458. doi: 10.1093/nar/gkn688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, DeScipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID. Precocious Sister Chromatid Separation (PSCS) in Cornelia de Lange Syndrome. Am J Med Genet Part A. 2005;138A:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Russell KL, Ming JE, Patel K, Jukofsky L, Magnusson M, Krantz ID. Dominant Paternal Transmission of Cornelia de Lange Syndrome: A New Case and Review of 25 Previously Reported Familial Recurrences. Am J Med Genet. 2001;104:267–276. doi: 10.1002/ajmg.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J, Wincent J, Barbaro M, Djureinovic T, Maguire P, Forsberg L, Staaf J, Thuresson AC, Borg A, Nordgren A, Malm G, Anderlid BM. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet. 2007;15:143–149. doi: 10.1038/sj.ejhg.5201737. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 Homologs Link Sister Chromatid Cohesion to Cell and Axon Migration Guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, Sforzini C, Tarantino E, Memo L, Scarano G, Larizza L. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- Sommer A. Occurrence of the Sandifer complex in the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1026–1028. doi: 10.1002/ajmg.1320470719. [DOI] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- White GE, Erickson HP. The Coiled Coils of Cohesin Are Conserved in Animals, but Not In Yeast. PLoS One. 2009;4:e4674. doi: 10.1371/journal.pone.0004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Saifi GM, Wierzba TH, Withers M, Bien-Willner GA, Limon J, Stankiewicz P, Lupski JR, Wierzba J. Mutational and Genotype-Phenotype Correlation Analyses in 28 Polish Patients with Cornelia de Lange Syndrome. Am J Med Genet Part A. 2006;140A:1531–1541. doi: 10.1002/ajmg.a.31305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.