Abstract
MYC genes are deregulated in a plurality of human cancers. Through direct and indirect mechanisms the MYC network regulates the expression of >15% of the human genome, including both protein-coding and non-coding RNAs. This complexity has complicated efforts to define the principal pathways mediating MYC’s oncogenic activity. MYC plays a central role providing for the bioenergetic and biomass needs of proliferating cells, and polyamines are essential cell constituents supporting many of these functions. The rate-limiting enzyme in polyamine biosynthesis, ODC, is a bona fide MYC target, as are other regulatory enzymes in this pathway. A wealth of data link enhanced polyamine biosynthesis to cancer progression, and polyamine-depletion may limit malignant transformation of pre-neoplastic lesions. Studies using transgenic cancer models also supports that the effect of MYC on tumor initiation and progression can be attenuated through repression of polyamine production. High-risk neuroblastomas (an often lethal embryonal tumor in which MYC activation is paramount) deregulate numerous polyamine enzymes to promote expansion of intracellular polyamine pools. Selective inhibition of key enzymes in this pathway, e.g., using DFMO and/or SAM486, reduces tumorigenesis and synergizes with chemotherapy to regress tumors in pre-clinical models. Here we review the potential clinical application of these and additional polyamine-depletion agents to neuroblastoma and other advanced cancers in which MYC is operative.
Key terms: Putrescine, Spermidine, Spermine, experimental therapeutics, MYC, MYCN, embryonal tumors
BACKGROUND
The MYC proto-oncogenes, which include MYC, MYCN and MYCL, are among the most frequently deregulated genes in cancer
They encode highly homologous basic-helix-loop-helix leucine zipper transcription factors that are biologically redundant but differentially expressed spatiotemporally. MYC genes function through heterodimerization with Max and operate within a network of related proteins to regulate transcription through interactions at E-box sequences within promoters of diverse target genes (1). Many estimates have the MYC network governing expression of >15% of all human genes (2, 3) and a growing roster of non-coding RNAs (4). A simplified gene-specific model of transcriptional regulation has been expanded with the appreciation that MYC genes also contribute to global chromatin regulation. Loss of MYCN in neural stem cells, for example, leads to aberrant nuclear structure mimicking a heterochromatin state accompanied by widespread histone modifications (5). Such higher-order regulatory activities may explain MYC’s profound influence on transcription and the diversity in putative targets genes identified across different model systems.
MYC activity is tightly regulated through transcriptional and post-translational mechanisms, with rapid degradation of Myc protein in concert with cell cycle exit. In many cancers MYC genes are deregulated through genomic translocation or amplification events that lead to supraphysiologic Myc expression. Although mutations in Myc have been identified in Burkitt’s lymphoma cells (accompanying rather than replacing activating translocations (6)), Myc oncogenesis typically results from deregulated overexpression of wild-type protein. Such large-scale biological reprogramming of cells through enforced expression of this promiscuous transactivator and chromatin regulator is highly oncogenic and the ubiquity of MYC activation across tumor types makes it an attractive cancer target.
Inhibiting grossly deregulated transcription factors remains a daunting therapeutic challenge, yet pharmaceutical successes continue to whittle away at the list of domains considered “undruggable”, so direct Myc antagonism may be an achievable goal. However, a secondary concern for such an approach is that systemic interference with Myc might be quite toxic as it is indispensable for cell cycle entry in response to mitogenic signals. This fear has been partially allayed by evidence that a profound dominant negative Myc contruct can be activated globally in mice without undue toxicity (Gerard Evan, AACR 2009). An alternative approach to interfering directly with Myc is to define the critical downstream pathways necessary for its oncogenic activity. Among these may be more immediately tractable drug targets that exploit cancer-specific aspects of Myc activity with a greater therapeutic index.
An improved understanding of Myc biology has emerged from high-dimensional assays that provide global transcriptome and/or Myc-chromatin binding site data. These platforms have generated daunting lists of genes and chromatin binding sites that underscore the widespread involvement of Myc in diverse biological processes. Still, patterns are discernible within this complexity. The most conserved set of MYC target genes function in ribosomal biogenesis and protein metabolism and processing, and this is true for both MYC (3) and MYCN (7). Additionally, programs that direct carbon assimilation, anabolic pathways, and bioenergetics are all targeted by Myc (3). Thus, Myc orchestrates a program redirecting metabolism to provide for the energetic needs of the cell through augmented aerobic glycolysis (8) and glutaminolysis (9); and the biomass needs through enhanced synthesis and processing of RNA, DNA, protein, lipid and polyamine precursors.
Polyamines are multifunctional polycations found in nearly all living organisms
They support biological processes through stabilization of anionic macromolecules and modulate DNA:protein and protein:protein interactions. A detailed understanding of polyamine activities is hampered by the fact that they participate in mainly transient ionic interactions that are difficult to study. Still, polyamine homeostasis is essential for cell survival and depletion activates cellular checkpoints that constrain proliferation or induce apoptosis (10). Reduced polyamines are seen in post-mitotic and senescent cells, while enhanced polyamine biosynthesis accompanies normal as well as oncogenic proliferation (11, 12). That polyamine biosynthesis may be instructive in the cancer process, rather than simply a consequence of increased proliferation, emerged as molecular studies linked numerous cancer genes directly to polyamine metabolism (13, 14). For example, ornithine decarboxylase (ODC1), the rate-limiting enzyme for polyamine biosynthesis, is a MYC target gene (15) and bona fide oncogene. Odc can substitute for Myc and cooperate with Ras to transform cells in vitro (16) and in vivo (17). Thus, enhanced polyamine synthesis is essential to oncogenic signaling and may be specifically required to support Myc-governed functions.
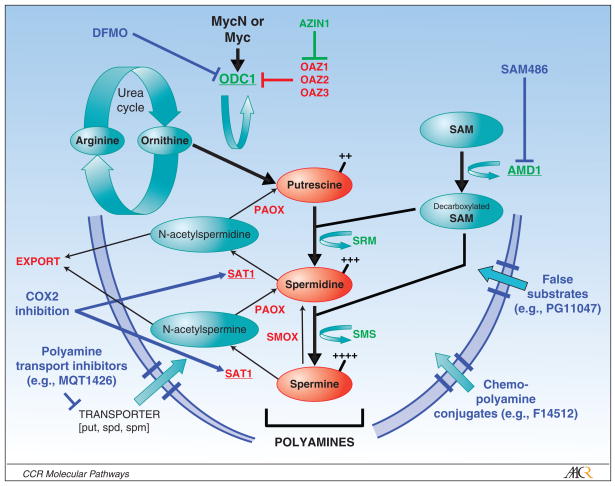
Intracellular polyamine levels are modulated through tightly regulated synthetic, catabolic, uptake and export pathways [Figure 1, and reviewed in (13, 14, 18)]. The rate-limiting biosynthetic enzymes are ornithine decarboxylase (ODC1) and S-adenosylmethionine decarboxylase (AMD1). Odc homodimers decarboxylate ornithine, a urea cycle product, to the diamine putrescine; while Amd decarboxylates S-adenosylmethionine to provide the aminopropyl donor for conversion of putrescine to spermidine and spermine. These latter conversions are mediated by the aminopropyltransferases spermidine synthase (SRM) and spermine synthase (SMS), respectively. Multiple levels of control are exercised over Odc and Amd activities. They are highly transcriptionally regulated, with ODC1 being a direct MYC target (15), and are post-translationally regulated with the shortest half-lives (10–30 minutes) of any mammalian enzyme. Odc is rapidly degraded through a process initiated by Odc antizymes that bind monomeric Odc and present the protein to the 26S proteasome for degradation independent of ubiquitylation, while also inhibiting polyamine uptake. These antizymes (OAZ1, OAZ2, OAZ3) are themselves responsive to intracellular polyamines to provide a negative feedback loop (19). Further, two mammalian antizyme inhibitors have been identified [AZIN1 (20) and, more recently, ADC (21)] that encode enzymatically inactive Odc homologues that compete to neutralize the antizymes and therefore constitute a positive regulator of Odc activity (22). This level of control underscores the importance of modulating Odc activity to ensure cellular fitness.
Figure 1. Schematic of polyamine metabolism required to support cell proliferation and therapeutic opportunities in this pathway.
Putrescine (diamine), spermidine (triamine) and spermine (tetramine) are the major polyamines. Ornithine derived from the urea cycle provides the initial substrate for Odc mediated decarboxylation to putrescine. Amd1 provides the aminopropyl donor to support SRM and SMS mediated conversion to higher order polyamines, respectively. Pro-synthetic polyamine enzymes are shown in GREEN; catabolic enzymes are shown in RED (underlined enzymes are highly regulated with among shortest half-life of any mammalian enzyme. Polyamine therapeutics and their sites of action are in BLUE (described in the text). Those shown are in pre-clinical or early phase clinical development as cancer therapeutics. ODC1, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermidine synthase; AMD1, S-adenosylmethionine decarboxylase; AZIN1, Odc Antizyme inhibitor; SMOX, spermidine oxidase; PAOX, polyamine oxidase; SAT1, spermine/spermidine N-acetyltransferase; OAZ1,2,3, Odc Antizymes.
Polyamine catabolism occurs via acetylation of spermidine or spermine by the readily inducible spermidine/spermine-N-acetyltransferase, SAT1 (23). Acetylated polyamines may be exported from the cell through specific transmembrane solute carriers to reduce intracellular levels (24). Alternatively, acetylated spermine and spermidine may be converted through PAOX oxidase activity to spermidine and putrescine, respectively; while SMOX oxidase activity can convert spermine to spermidine directly. These conversions allow for homeostatic control over the repertoire of natural polyamines, which may be important to maintain functions unique to select polyamines (such protein translation, in which spermidine acts as a gate-keeper by regulating the activity of eIF5A (25); Figure 2). Finally, an as yet uncharacterized energy-dependent polyamine transporter functions to selectively import polyamines present in the microenvironment through dietary intake, export from neighboring cells, or synthesis by intestinal flora. This pathway can restore polyamine levels under conditions of biosynthetic blockade and may potentially undermine therapeutic efforts to diminish intracellular polyamines.
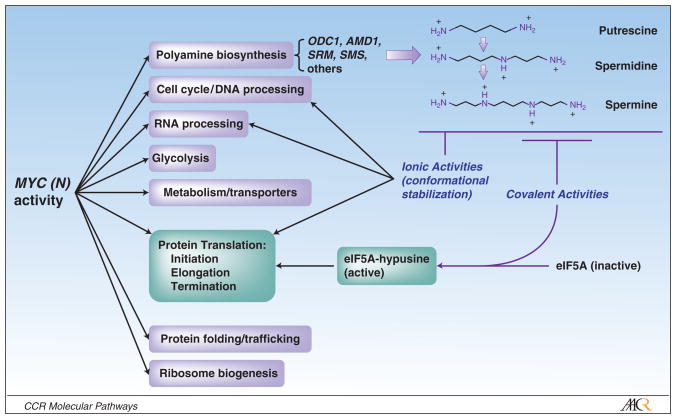
Figure 2. Myc-polyamine axis and functional synergies.
Myc genes govern diverse programs to support proliferation. Polyamines, mediated by Myc activity, support many Myc functions through ionic stabilization of key macromolecules in these processes. Spermidine has an additional vital function: eIF5A is a universal elongation factor that uniquely supports all three steps of protein translation (56), and is active only following hypusination at lysine 51 (25). This post-translational modification (in which spermidine alone acts as an aminobutyl donor) is not found in any other eukaryotic protein, and is essential for eIF5A activity. This pathway links Myc-driven polyamine homeostasis to an essential role supporting protein translation. Additional roles for polyamines in free-radical scavenging, formation of cytotoxic products, and additional covalent bond-mediated activities have been experimentally defined but are not shown (57).
This multitude of regulatory controls highlights the critical need for polyamine homeostasis, and as polyamines operate downstream of Myc to support proliferation they provide an intriguing cancer target. Indeed, substantial effort has been spent over 30 years to leverage polyamine disruption as an anti-cancer strategy with limited success initially. Inhibitors acting at nearly every step in this pathway have been developed and investigated and despite encouraging pre-clinical results with diverse agents and cancer models, translation to clinical utility has been slow [comprehensively reviewed in (13)]. Initial studies testing single polyamine-targeting drugs were disappointing, although not completely without successes as activity has been seen in hematolymphoid (26, 27) and CNS neoplasms (28). To date, however, little attention has been paid to the potential for cancer genotype-specific responses, analogous to synthetic-lethal interactions in yeast. Studies using transgenic cancer models suggest that Myc deregulation may provide an Achille’s heel for cancer cells through a requirement for polyamine sufficiency that can be targeted therapeutically and embryonal cancers may be particularly vulnerable.
CLINICAL-TRANSLATIONAL ADVANCES
Neuroblastoma is a childhood embryonal tumor that frequently presents with high-risk clinical and genetic features
Despite maximally intensive therapy survival remains dismal and innovative treatment approaches are needed [reviewed in (29)]. Several recurrent genomic alterations correlate with outcome, and of these, MYCN amplification is most strongly correlated with advanced disease and treatment failure (30, 31). In high-risk neuroblastomas that lack MYCN amplification, MYC is frequently deregulated instead (32–34) suggesting MYC signaling may be essential for the high-risk phenotype. Still, despite over 20 years of recognition that MYC deregulation is a seminal event in neuroblastoma no molecularly targeted therapy has emerged to leverage this discovery. Polyamine homeostasis, deregulated downstream of MYC genes, may provide such a target.
First, polyamine regulators are aberrantly expressed in high-risk neuroblastomas to coordinately augment biosynthesis and reduce catabolism. ODC1 mRNA is significantly higher in high-risk tumors while the antizyme OAZ2 is reduced, consistent with polyamine enhancement [note: unlike OAZ1, OAZ2 may not deliver Odc to the proteasome for degradation, yet it is equipotent at inhibiting Odc activity and polyamine uptake (35)]. Moreover, every prosynthetic enzyme (including AMD1, SRM, and SMS) is markedly upregulated in the highest-risk subset with MYCN amplification, while, conversely, there is reduced SMOX (36). A similar pattern is seen when evaluating neuroblastoma cell lines in comparison with fetal adrenal or neural tissues (36, 37). Second, ODC1 expression correlates with outcome in neuroblastoma, independent of MYCN amplification, supporting that this upregulation has functional consequences (36). In addition to direct transactivation by MYC genes, ODC1 expression is influenced by a functional promoter polymorphism at the A317G SNP (38, 39). In neuroblastoma the higher expressing genotypes have an inferior survival, particularly when analysis is restricted to the MYCN non-amplified tumors, again supporting functional validity for this pathway (Michelle Haber, AACR 2008 and personal communication).
Studies using a transgenic model of neuroblastoma supports a requirement for polyamines in tumor initiation, progression and therapy response
Mice carrying a neural-crest targeted MYCN transgene (TH-MYCN model) develop lethal neuroblastoma with complete penetrance in the homozygous state, and ~40% penetrance in the hemizygous state (40, 41). Tumors arise stochastically within peripheral sympathetic ganglia and recapitulate human neuroblastoma features, with cooperative genetic alterations at chromosome regions orthologous to those in the human disease (40, 41). Importantly, tumors arise in an appropriate microenvironment to recapitulate the heterotypic cell interactions important to cancer propagation and provides a relevant therapeutics testing platform (42). As with human neuroblastomas, TH-MYCN tumors demonstrate altered polyamine regulator expression compared with sympathetic ganglia, with upregulated ODC1, AZIN, AMD1, SRM, SMS and downregulated OAZ2, SMOX and SAT1 (manuscript in preparation). Thus, the model likely reflects the polyamine pools, pathway flux, and compensatory mechanisms present in human neuroblastoma.
Treating TH-MYCN mice with α-difluoromethylornithine (DFMO), a suicide inhibitor of Odc, increases tumor free survival. Moreover, tumor penetrance is reduced in hemizygous mice pre-emptively treated, supporting a requirement for Odc in tumor initiation downstream of MycN (36, 37). Of note, no tumors arose following DFMO withdrawal consistent with a finite vulnerable period for embryonal oncogenesis. This differs from the Eμ-Myc lymphoma model where protection from lymphomagenesis required persistent Odc inactivation (43). Neuroblastomas that arise under DFMO exposure may activate compensatory mechanisms to circumvent polyamine depletion. Since upregulated Amd1 accompanies Odc inhibition in mammalian cells, as confirmed in neuroblastoma (44), we tested the ability of DFMO and SAM486 (4-amidinoindan-1-one-2′-amidinhydrazone; a competitive Amd1 inhibitor from Novartis) to synergize in this model. Neuroblastoma penetrance was further reduced, including in homozygous mice, so optimized polyamine depletion contributes to markedly improved efficacy (AACR 2009, manuscript in preparation). A more practical test, however, requires treatment of established tumors. DFMO treatment of TH-MYCN mice harbouring clinically detected neuroblastomas extends time to tumor progression and augments efficacy of numerous chemotherapeutics supporting that this strategy may have clinical relevance (36). Select DFMO and chemotherapy combinations improved survival as well, implying that these synergistic effects went beyond cytostasis.
Initial data using human neuroblastoma cell lines grown as xenografts in immunodeficient mice are similarly supportive that polyamine disruption interferes with tumor progression (Michelle Haber, personal communication) although these studies will need to be extended to assess the role MYCN amplification plays in modulating sensitivity to these inhibitors. It is encouraging that both DFMO and SAM486 inhibit neuroblastoma cell line growth in vitro independent of MYCN amplification ((36) and data not shown), likely reflecting a commonality of Myc deregulation in the high-risk phenotype (33). Mechanistically, Odc inhibition reduces Rb phosphorylation at Ser795 and Ser807/811 through loss of MycN-mediated repression of p27Kip1, leading to G1 growth arrest. Coincident with this, Akt phosphorylation at Ser473 and GSK3B at Ser9 are induced promoting survival (44, 45). Although DFMO similarly abolished p27Kip1 induction in the Eμ-Myc lymphoma model in vivo, the effects in the TH-MYCN neuroblastoma model may instead be mediated through transcriptional upregulation of p21Cip1 (37).
Strategies to integrate polyamine depletion therapeutics into neuroblastoma treatment are warranted
It is unlikely that traditional phase 1 and 2 studies for patients with relapsed or refractory high-risk neuroblastoma using only polyamine-targeted agents would generate enthusiasm. These pathway targeted compounds might best be tested on a backbone of cytotoxic drugs to improve or restore chemoresponsiveness, as shown with the mouse model, and such clinical trials are in development. However, maximizing the impact of such strategies requires a more complete understanding of tumor-specific polyamine flux and the compensatory mechanisms used to escape blockade in these pathways. For example, many tumor cells respond to Odc inhibition with enhanced polyamine uptake from the microenvironment (46). Radiolabeled spermidine uptake from neuroblastoma cell lines is not induced during DFMO or SAM486 exposure although it remains possible that this represents accommodation to prolonged tissue culture as polyamine content is nominal in culture media. Neuroblastoma cell lines established from the TH-MYCN model, however, do induce uptake from 2- to 6-fold under such conditions. Despite this, they remain sensitive to polyamine depletion in vivo.
There are potential opportunities for improving polyamine depletion responses for tumors with inducible polyamine uptake. First, compounds that antagonize polyamine uptake, such as D-lysine spermine (MQT-1426) and N1-spermyl-L-lysinamide (ORI202), are under pre-clinical development (47) and may cooperate with DFMO and other biosynthesis inhibitors to more profoundly reduce polyamine levels and improve therapeutic responses in vivo (48). Alternatively, “Trojan horse” approaches with polyamine-chemotherapy conjugates have been used and could cooperate with polyamine biosynthesis inhibitors. In this approach, DFMO or similar agents induce polyamine depletion in cancer cells that subsequently upregulate polyamine transport. This targets delivery of polyamine-chemotherapy conjugates preferentially to tumor cells. Numerous such conjugates have been developed, but a subsidiary benefit of delivering a DNA-interacting cytotoxic in this manner is that the polyamine moiety not only enhances tumor-specific uptake but also DNA binding through cationic interactions. The spermine-podophyllotoxin conjugate F14512 (Pierre Fabre) has superior cytotoxicity in cells with enhanced polyamine uptake in vitro (IC50 in the nM range) and regressed breast carcinoma xenografts in vivo (49), providing strong proof of principal for this approach.
Beyond inhibiting biosynthesis and impeding import, another therapeutic option is to enhance catabolism and/or export of polyamines. For example, NSAIDs influence polyamine acetylation and export through upregulation of SAT1 via PPARγ (50). Although anti-cancer effects attributable to this drug class are pleiotropic, cox-inhibitor mediated apoptosis in cancer cells can be rescued by polyamine supplementation, implicating this pathway (50). This may have therapeutic relevance as sulindac and DFMO together have documented efficacy in reducing adenoma recurrence in an at-risk population as demonstrated in a large chemoprevention trial (51). SAT1 induction also occurs downstream of platinator and other chemotherapeutics, in association with additional polyamine regulator changes to repress polyamine content, and this may provide synergy for select polyamine depletion-chemotherapeutic combinations (52). SAT1 acetylates polyamines and co-localizes with the putative export solute transporter SLC3A2 to facilitate export (24), leading to increased polyamine metabolic flux ~5-fold through upregulated biosynthetic enzyme activity in efforts to restore homeostasis (53). This provides another opportunity for synergy between SAT1 inducers (e.g., NSAIDs, cisplatinum) and biosynthesis inhibitors (e.g., DFMO, SAM486) that prevent this response.
Finally, there is great interest in analogues that mimic native polyamines in homeostatic regulation. Such compounds can utilize the polyamine transporter to concentrate in cancer cells leading to compensatory downregulation of polyamine biosynthesis and upregulation of catabolism, while not substituting functionally for natural polyamines. Agents of this class include assorted symmetrically and non-symmetrically substituted analogues, conformationally restricted analogues, oligoamines and macrocyclic polyamine analogues. These agents are reviewed in (13) and are in pre-clinical and clinical studies. Of these, PG-11047 (a second-generation conformationally restricted analogue) has been shown to have activity in vivo in pre-clinical non-small cell lung carcinoma models (54), although recent testing against pediatric tumors through the Pediatric Preclinical Testing Program (pptp.stjude.org; (55)) showed minimal activity as a single agent against neuroblastoma (J. Maris, AACR 2009).
Optimism remains that polyamine depletion can be exploited therapeutically and the agents discussed herein represent only a partial list of those under active drug development. DFMO (Eflornithine) has gained FDA approval for trypanosomiasis and has been the most rigorously tested agent in this class. Although it has demonstrated potent activity in colorectal adenoma chemoprevention it has yet to demonstrate potency against more advanced cancers. Whether combination therapies that deprive cancer cells of major compensatory pathways will synergize is not yet known, but many rational combinations to do just that exist. Transitioning this class of agents from scientific discovery to preclinical anti-cancer activity appears to have been achieved, but bridging the chasm to demonstrate clinical utility remains a significant challenge.
Acknowledgments
The author’s laboratory is supported by NIH CA97323 and the Richard and Sheila Sanford Chair in Pediatric Oncology (to M.D.H.) and the Children’s Neuroblastoma Cancer Foundation (N.F.E.). The authors thank Susan Gilmour, Michelle Haber, Murray Norris, Glenn Marshall and Andre Bachmann for helpful discussions.
References
- 1.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nature Reviews Molecular Cell Biology. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell BC, Cheung AF, Simkevich CP, et al. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278:12563–73. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- 4.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genetics. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genetics. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 7.Boon K, Caron HN, van_Asperen R, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–93. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osthus RC, Shim H, Kim S, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 9.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences USA. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas T, Thomas TJ. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell and Molecular Life Sciences. 2001;58:244–58. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson G, Heby O. Polyamine and nucleic acid concentrations in Ehrlich ascites carcinoma cells and liver of tumor-bearing mice at various stages of tumor growth. J Natl Cancer Inst. 1972;48:165–72. [PubMed] [Google Scholar]
- 12.Russell D, Snyder SH. Amine synthesis in rapidly growing tissues: ornithine decarboxylase activity in regenerating rat liver, chick embryo, and various tumors. Proceedings of the National Academy of Sciences U S A. 1968;60:1420–7. doi: 10.1073/pnas.60.4.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature Reviews Drug Discovery. 2007;6:373–90. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 14.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nature Reviews Cancer. 2004;4:781–92. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 15.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–8. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moshier JA, Dosescu J, Skunca M, Luk GD. Transformation of NIH/3T3 cells by ornithine decarboxylase overexpression. Cancer Res. 1993;53:2618–22. [PubMed] [Google Scholar]
- 17.Smith MK, Trempus CS, Gilmour SK. Co-operation between follicular ornithine decarboxylase and v-Ha-ras induces spontaneous papillomas and malignant conversion in transgenic skin. Carcinogenesis. 1998;19:1409–15. doi: 10.1093/carcin/19.8.1409. [DOI] [PubMed] [Google Scholar]
- 18.Basuroy UK, Gerner EW. Emerging concepts in targeting the polyamine metabolic pathway in epithelial cancer chemoprevention and chemotherapy. Journal of Biochemistry (Tokyo) 2006;139:27–33. doi: 10.1093/jb/mvj022. [DOI] [PubMed] [Google Scholar]
- 19.Matsufuji S, Matsufuji T, Miyazaki Y, et al. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koguchi K, Kobayashi S, Hayashi T, Matsufuji S, Murakami Y, Hayashi S. Cloning and sequencing of a human cDNA encoding ornithine decarboxylase antizyme inhibitor. Biochim Biophys Acta. 1997;1353:209–16. doi: 10.1016/s0167-4781(97)00106-1. [DOI] [PubMed] [Google Scholar]
- 21.Kanerva K, Makitie LT, Pelander A, Heiskala M, Andersson LC. Human ornithine decarboxylase paralogue (ODCp) is an antizyme inhibitor but not an arginine decarboxylase. Biochem J. 2008;409:187–92. doi: 10.1042/BJ20071004. [DOI] [PubMed] [Google Scholar]
- 22.Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008;17:793–802. doi: 10.1110/ps.073427208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Celano P, Mank AR, Pegg AE, Casero RA., Jr Characterization of a full-length cDNA which codes for the human spermidine/spermine N1-acetyltransferase. Biochem Biophys Res Commun. 1991;179:407–15. doi: 10.1016/0006-291x(91)91385-p. [DOI] [PubMed] [Google Scholar]
- 24.Uemura T, Yerushalmi HF, Tsaprailis G, et al. Identification and characterization of a diamine exporter in colon epithelial cells. J Biol Chem. 2008;283:26428–35. doi: 10.1074/jbc.M804714200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proceedings of the National Academy of Sciences U S A. 1983;80:1854–7. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pless M, Belhadj K, Menssen HD, et al. Clinical efficacy, tolerability, and safety of SAM486A, a novel polyamine biosynthesis inhibitor, in patients with relapsed or refractory non-Hodgkin’s lymphoma: results from a phase II multicenter study. Clin Cancer Res. 2004;10:1299–305. doi: 10.1158/1078-0432.ccr-0977-03. [DOI] [PubMed] [Google Scholar]
- 27.Siimes M, Seppanen P, Alhonen-Hongisto L, Janne J. Synergistic action of two polyamine antimetabolites leads to a rapid therapeutic response in childhood leukemia. Int J Cancer. 1981;28:567–70. doi: 10.1002/ijc.2910280506. [DOI] [PubMed] [Google Scholar]
- 28.Levin VA, Hess KR, Choucair A, et al. Phase III randomized study of postradiotherapy chemotherapy with combination alpha-difluoromethylornithine-PCV versus PCV for anaplastic gliomas. Clin Cancer Res. 2003;9:981–90. [PubMed] [Google Scholar]
- 29.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 30.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 31.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–6. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 32.Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proceedings of the National Academy of Sciences U S A. 2008;105:14094–9. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Mazanek P, Dam V, et al. Deregulated Wnt/B-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene. 2008;27:1478–88. doi: 10.1038/sj.onc.1210769. [DOI] [PubMed] [Google Scholar]
- 34.Westermann F, Muth D, Benner A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biology. 2008;9:R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu C, Lang DW, Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem. 1999;274:26425–30. doi: 10.1074/jbc.274.37.26425. [DOI] [PubMed] [Google Scholar]
- 36.Hogarty MD, Norris MD, Davis K, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–45. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rounbehler RJ, Li W, Hall MA, Yang C, Fallahi M, Cleveland JL. Targeting ornithine decarboxylase impairs development of MYCN-amplified neuroblastoma. Cancer Res. 2009;69:547–53. doi: 10.1158/0008-5472.CAN-08-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Harris RB, Rosson D, Boorman D, O’Brien TG. Functional analysis of human ornithine decarboxylase alleles. Cancer Res. 2000;60:6314–7. [PubMed] [Google Scholar]
- 39.Martinez ME, O’Brien TG, Fultz KE, et al. Pronounced reduction in adenoma recurrence associated with aspirin use and a polymorphism in the ornithine decarboxylase gene. Proceedings of the National Academy of Science U S A. 2003;100:7859–64. doi: 10.1073/pnas.1332465100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansford LM, Thomas WD, Keating JM, et al. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proceedings of the National Academy of Science U S A. 2004;101:12664–9. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss WA, Aldape K, Bishop JM. Targeted expression of NMYC causes neuroblastoma in transgenic mice. The EMBO Journal. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chesler L, Goldenberg DD, Seales IT, et al. Malignant progression and blockade of angiogenesis in a murine transgenic model of neuroblastoma. Cancer Res. 2007;67:9435–42. doi: 10.1158/0008-5472.CAN-07-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson JA, Keller UB, Baudino TA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–44. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 44.Wallick CJ, Gamper I, Thorne M, et al. Key role for p27Kip1, retinoblastoma protein Rb, and MYCN in polyamine inhibitor-induced G1 cell cycle arrest in MYCN-amplified human neuroblastoma cells. Oncogene. 2005;24:5606–18. doi: 10.1038/sj.onc.1208808. [DOI] [PubMed] [Google Scholar]
- 45.Koomoa DL, Yco LP, Borsics T, Wallick CJ, Bachmann AS. Ornithine decarboxylase inhibition by alpha-difluoromethylornithine activates opposing signaling pathways via phosphorylation of both Akt/protein kinase B and p27Kip1 in neuroblastoma. Cancer Res. 2008;68:9825–31. doi: 10.1158/0008-5472.CAN-08-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiler N, Delcros JG, Moulinoux JP. Polyamine transport in mammalian cells. An update Int J Biochem Cell Biol. 1996;28:843–61. doi: 10.1016/1357-2725(96)00021-0. [DOI] [PubMed] [Google Scholar]
- 47.Weeks RS, Vanderwerf SM, Carlson CL, et al. Novel lysine-spermine conjugate inhibits polyamine transport and inhibits cell growth when given with DFMO. Exp Cell Res. 2000;261:293–302. doi: 10.1006/excr.2000.5033. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Weeks RS, Burns MR, Boorman DW, Klein-Szanto A, O’Brien TG. Combination therapy with 2-difluoromethylornithine and a polyamine transport inhibitor against murine squamous cell carcinoma. Int J Cancer. 2006;118:2344–9. doi: 10.1002/ijc.21621. [DOI] [PubMed] [Google Scholar]
- 49.Barret JM, Kruczynski A, Vispe S, et al. F14512, a potent antitumor agent targeting topoisomerase II vectored into cancer cells via the polyamine transport system. Cancer Res. 2008;68:9845–53. doi: 10.1158/0008-5472.CAN-08-2748. [DOI] [PubMed] [Google Scholar]
- 50.Babbar N, Ignatenko NA, Casero RA, Jr, Gerner EW. Cyclooxygenase-independent induction of apoptosis by sulindac sulfone is mediated by polyamines in colon cancer. J Biol Chem. 2003;278:47762–75. doi: 10.1074/jbc.M307265200. [DOI] [PubMed] [Google Scholar]
- 51.Meyskens FL, McLaren CE, Pelot D, et al. Difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas: a randomized placebo-controlled, double-blind trial. Cancer Prevention Research. 2008 doi: 10.1158/1940-6207.CAPR-08-0042. (Online first 2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varma R, Hector S, Greco WR, et al. Platinum drug effects on the expression of genes in the polyamine pathway: time-course and concentration-effect analysis based on Affymetrix gene expression profiling of A2780 ovarian carcinoma cells. Cancer Chemother Pharmacol. 2007;59:711–23. doi: 10.1007/s00280-006-0325-3. [DOI] [PubMed] [Google Scholar]
- 53.Kramer DL, Diegelman P, Jell J, Vujcic S, Merali S, Porter CW. Polyamine acetylation modulates polyamine metabolic flux, a prelude to broader metabolic consequences. J Biol Chem. 2008;283:4241–51. doi: 10.1074/jbc.M706806200. [DOI] [PubMed] [Google Scholar]
- 54.Hacker A, Marton LJ, Sobolewski M, Casero RA., Jr In vitro and in vivo effects of the conformationally restricted polyamine analogue CGC-11047 on small cell and non-small cell lung cancer cells. Cancer Chemother Pharmacol. 2008;63:45–53. doi: 10.1007/s00280-008-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houghton PJ, Morton CL, Tucker C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatric Blood and Cancer. 2007;49:928–40. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 56.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–21. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seiler N, Raul F. Polyamines and apoptosis. Journal of Cellular and Molecular Medicine. 2005;9:623–42. doi: 10.1111/j.1582-4934.2005.tb00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]