Abstract
Epithelial stem cells, such as those present in mammalian skin, intestine, or mammary gland, are tissue stem cells capable of both long-term self-renewal and multi-lineage differentiation. Here we review studies implicating epigenetic control mechanisms in mammalian epithelial stem cell development and homeostasis. We also provide an update of recent progresses in the involvement of canonical Wnt signaling and note an interesting link between the Wnt pathway and chromatin regulation in epithelial stem cells. We anticipate that epigenetic and epigenomic studies of these cells will increase exponentially in the near future.
Keywords: EPITHELIAL STEM CELL, EPIGENETICS, WNT SIGNALING, CHROMATIN
Epigenetic regulation without alteration of DNA sequence underlies the progression of stem cells through different stages of development and differentiation. The control of epigenetic status of a cell involves, among other mechanisms, covalent modification of histones (e.g., acetylation, methylation, phosphorylation, ubiquitination, sumoylation), and DNA methylation. Chromatin regulation can occur at two non-mutually exclusive levels: (1) individual genomic loci, as exemplified by epigenetic events leading to gene transcription or silencing, (2) global chromatin configuration across the genome. The recent breakthroughs of reprogramming differentiated somatic cells such as skin fibroblasts into induced pluripotent stem cells using a small number of transcription factors, and the elucidation of DNA methylation and histone modification landscapes of embryonic stem (ES) cells have shed light onto a unique epigenetic strategy used to support pluripotency [Gan et al., 2007; Meissner et al., 2008]. Interestingly, the so-called “bivalent” chromatin domains, characterized by coexistence of both repressive (e.g., histone H3 trimethyl lysine 27 or H3K27me3) and active (e.g., histone H3 trimethyl lysine 4 or H3K4me3) histone marks, have been found in regulatory genes of not only pluripotent ES cells, but also lineage-restricted cells such as resting neurons, CD4+ T cells, fibroblasts and hematopoietic stem/progenitor cells [Shen and Orkin, 2009; and references therein]. Such domains are hypothesized to indicate a poised chromatin state that prepares pluripotent or multipotent stem cells for subsequent differential gene expression upon choosing among multiple downstream cell fates.
Despite rapid progress in epigenetic studies of embryonic and other somatic stem cells, the epigenomes of epithelial stem cells remains to be elucidated. Considering that epithelium is one of the major tissue targets of tumorigenesis, such studies are fundamentally important for human health. In this article, we review the scant literature implicating epigenetic control modules in mammalian epithelial stem cell development and homeostasis, noting particularly a link between Wnt signaling and chromatin regulation, with the anticipation that epigenetic and epigenomic studies of these cells will increase exponentially in the near future. We focus our discussion on leading stem cell models, namely skin, intestine, and mammary gland.
EPITHELIAL STEM CELLS
Epithelial cells line surfaces and cavities of organs throughout the body, providing functions of protection, sensation, secretion, and absorption that are indispensable for living organisms. Unlike nervous and muscle tissues that have little turnover, epithelial tissues experience extensive life-long homeostasis and regeneration. This remarkable repopulation capacity is attributed to long-lived stem cells residing in adult epithelial tissues. Characteristically, these stem cells are capable of generating both daughter stem cells for self-renewal and committed progenitors that can differentiate into lineage-specific progenies. The speed and pattern of epithelial turnover vary from tissue to tissue [Blanpain et al., 2007], implicating tissue-specific epigenetic regulation of epithelial stem cell homeostasis.
Although the existence of epithelial stem cells has been long recognized, bona fide molecular markers for different types of adult stem cells remain elusive until recently. To circumvent the obstacle, scientists have adopted two main methods for the initial identification of candidate stem cells. The well-known DNA label-retention assay is based on the assumption that stem cells are generally quiescent and retain the BrdU label of genomic DNA much longer than their rapidly cycling progenies. An elegant variant of this strategy is histone H2B-GFP labeling, as used in tracking the hair follicle stem cells (HFSCs) [Tumbar et al., 2004]. The other approach is to “borrow” surrogate markers from already characterized tissues. For example, Sca1 (stem cell antigen 1) and Prominin 1 (CD133), markers of hematopoietic stem cells, have been adopted to experimentally enrich for mammary progenitor cells and intestinal stem/progenitor cells, respectively [Welm et al., 2002; Zhu et al., 2009]. The definitive identification of adult stem cells entails two gold-standard strategies: transplantation and genetic lineage tracking, which are designed to assess self-renewal and multi-potency, two important features of adult stem cells. In a transplantation assay, candidate stem cells selected by molecular markers are transplanted into recipient animals, and the repopulation of a functional tissue from a single donor cell would demonstrate the identity of a bona fide stem cell. An example of this powerful approach is the discovery that a single isolated mammary stem cell is able to generate an entire mammary gland upon transplantation [Shackleton et al., 2006]. In lineage tracing experiments, stem cells are genetically labeled in situ, after which the fate of the labeled stem cells and their clonal progeny are monitored over space and time.
While cell-intrinsic factors are clearly important for stem cell characteristics, the activities of stem cells are largely instructed by the in vivo microenvironment called the stem cell niche that influences stem cell survival, self-renewal, and differentiation. The stem cell niche consists of various components and interactions including growth factors, cytokines, cell–cell contacts, and cell–matrix adhesion components. In adult tissues, this niche is often necessary for maintaining a pool of quiescent epithelial stem cells [Fuchs, 2009], thus preventing their exhaustion or deregulated expansion that may result in tumorigenesis. The canonical Wnt signaling pathway (see below) has emerged as an important stem cell pathway that acts both directly on stem cells and indirectly on the niche to modulate stem cell fate [Rattis et al., 2004].
EPIGENETIC REGULATION OF EPITHELIAL STEM CELLS
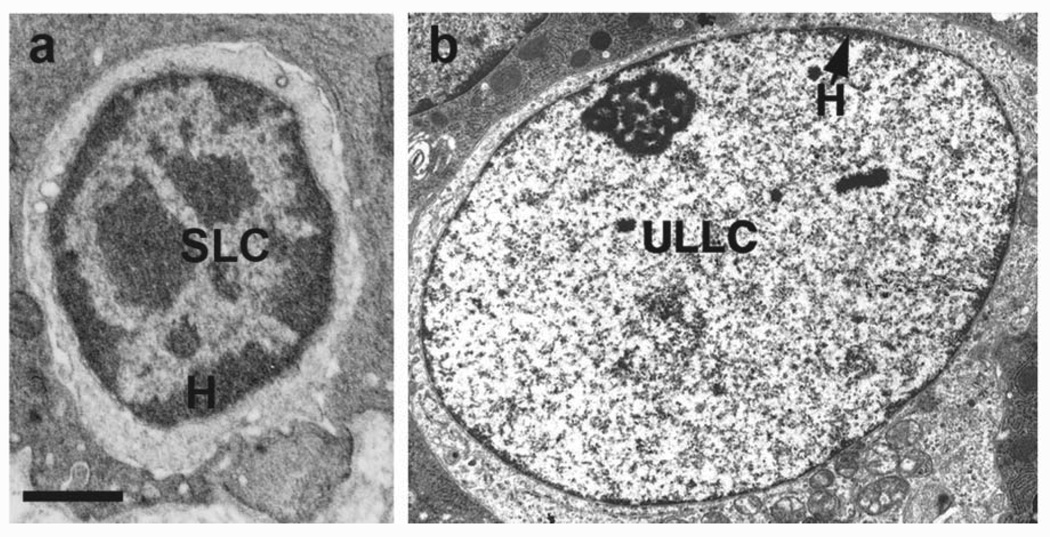
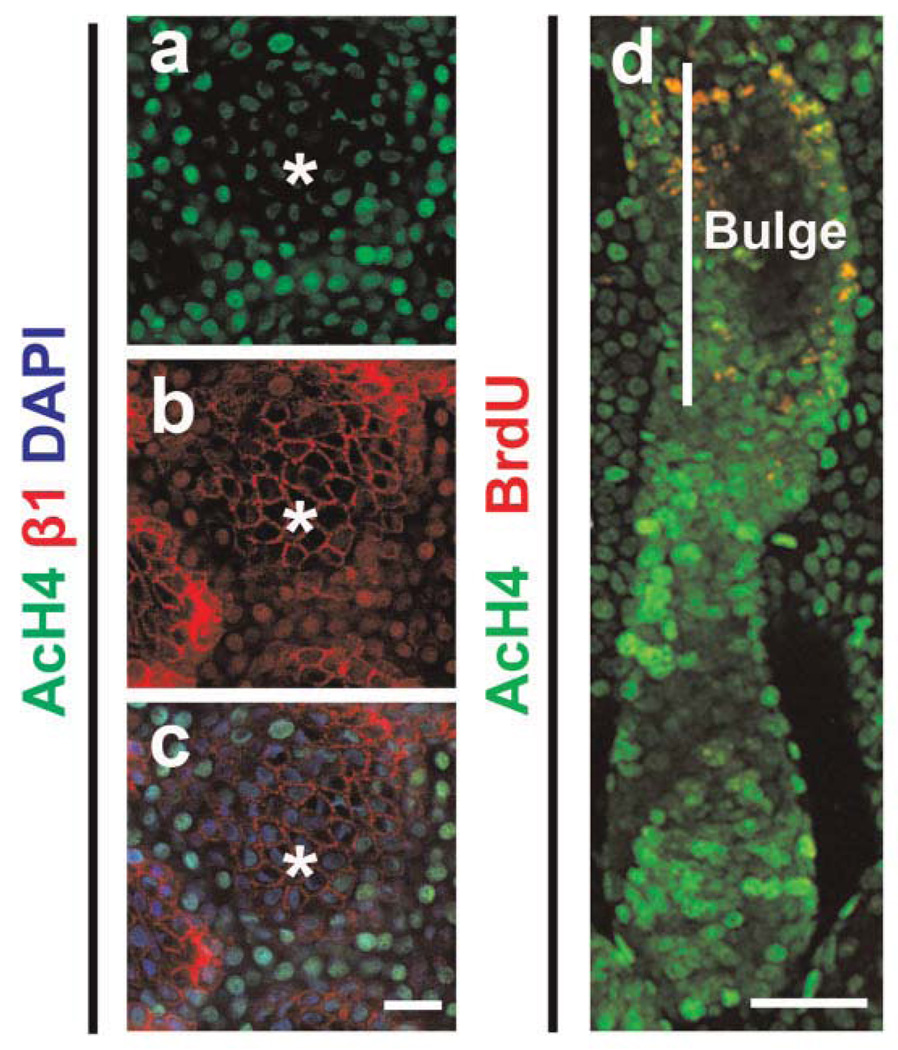
Although scarce, there have been studies, with the earliest dating back to more than 10 years ago, that highlight the unique feature of global chromatin configuration in epithelial stem cells. For example, Chepko and Smith reported electron microscopic studies noting that mammary epithelial cells undergo chromatin organizational changes as they evolve through the different stages of differentiation [Chepko and Smith, 1997; Chepko and Dickson, 2003]. The “small light cells” (SLC, putative multipotent mammary stem/early progenitor cells) contain dense clumps of heterochromatin, whereas heterochromatin disappears in the “undifferentiated large light cells” (ULLC, putative committed progenitor cells) but forms again upon differentiation towards a luminal fate (Fig. 1). Recently, Fiona Watt’s group examined the global patterns of histone modifications in mammalian skin using immunostaining, providing a first glimpse at the “histone code” that associates with quiescent stem cells present in human interfollicular epidermis as well as mouse hair follicle bulge [Frye et al., 2007, Fig. 2]. This “histone code” appears to be characterized by high levels of histone H3 lysine 9 and histone H4 lysine 20 (H4K20) trimethylation and low levels of histone H4 acetylation and H4K20 mono-methylation. Interestingly, tampering with the “code” by application of inhibitors of histone deacetylases (HDAC) or overexpression of c-Myc results in altered proliferation/differentiation characteristics of epidermal stem cells. Despite these intriguing morphological and biochemical observations, study of the molecular regulation of epithelial stem cell epigenetics is still in its infancy.
Fig. 1.
Chromatin changes in rat mammary epithelium. a: SLC, small light cell; b: ULLC, undifferentiated large light cell; H, heterochromatin. (Reprinted with permission from Elsevier Publishers, Limited, p 244 of Chepko and Smith, 1997, and p 87 of Chepko and Dickson, 2003.)
Fig. 2.
Quiescent stem cells in human interfollicular epidermis (a–c) and mouse hair follicle bulge (d) contain lower levels of histone H4 acetylation (AcH4) than their transit amplifying progeny. β1, β1 integrin, which is highly expressed in epidermal stem cells; DAPI, nuclei staining; BrdU, label retaining cells in the bulge. Asterisks, clusters of epidermal stem cells. (Reprinted with permission from Public Library of Science (PLoS), p e763 of Frye et al., 2007.)
Reverse genetic approaches have allowed scientists to probe into the functional involvement of important chromatin regulators in epithelial stem cells. Polycomb group (PcG) proteins are evolutionally conserved epigenetic gene silencers, and B mi 1 is a component of the polycomb repression complex 1 (PRC1) that recognizes H3K27me3 and ubiquitinates histone H2A, resulting in gene repression [Wang et al., 2004]. There is growing evidence that B mi 1 is a key regulator of the self-renewal of adult stem cells, including hematopoietic, neural, and mammary stem cells [Sparmann and van Lohuizen, 2006]. As a downstream target of sonic hedgehog signaling, B mi 1 mediates its effect on the self-renewal of normal and malignant human mammary stem cells [Liu et al., 2006]. Ablation of Bmi1 in mice leads to reduced activity of mammary stem cells in transplantation assays, a defect that can be partially rescued by additional inactivation of the downstream target locus Ink4a/Arf, which encodes cell cycle inhibitors p16Ink4a and p19Arf [Pietersen et al., 2008]. Moreover, loss of Bmi1 causes premature differentiation of lobuloalveoli in virgin mice independently of p16Ink4a and p19Arf, while overexpression of Bmi1 suppresses pregnancy-induced lobuloalveolar differentiation. Together, these results demonstrate a dual function for Bmi1 in promoting mammary stem/progenitor cell proliferation and preventing premature differentiation, and illustrate the flexibility of epigenetic silencing in mammary stem cell fate choices. Recently, Bmi1 expression has been shown to mark a presumably slow cycling population of intestinal stem cells (ISCs) [Sangiorgi and Capecchi, 2008], presenting an intriguing but speculative connection between epigenetic silencing and ISC quiescence.
Enhancer of zeste homolog 2 (Ezh2) is another PcG member, and a methyltransferase component of the polycomb repressive complex 2 (PRC2) composed of itself, Eed, and Suz12 [Sparmann and van Lohuizen, 2006]. Functioning within the complex, Ezh2 trimethylates primarily H3K27 to initiate gene repression. In a recent study, Ezh2 has been found to mark embryonic progenitor cells of the epidermis, and its genetic ablation leads to reduced progenitor cell proliferation accompanied by aberrantly elevated expression of cell cycle inhibitors Ink4A and Ink4B, as well as premature induction of late-differentiation genes [Ezhkova et al., 2009]. Conversely, H3K27me3 demethylase JMJD3 (jumonji domain containing 3) occupies epidermal differentiation genes as PcG binding disappears, and is both necessary and sufficient for terminal differentiation [Sen et al., 2008]. These studies collectively underscore the importance of epigenetic repression and derepression in controlling the balance between epidermal stem/progenitor cell proliferation and differentiation.
EZH2 as part of the PRC2 complex has also been shown to recruit DNA methyltransferases (DNMTs) to cognate target genes, providing a direct link between the two modes of epigenetic silencing, namely histone modification (H3K27 methylation) and DNA methylation [Vire et al., 2006]. The importance of DNA methylation in helping epithelial progenitor cells preserve cellular memory through repeated cell divisions has begun to be addressed. Enriched in epidermal progenitor cells but not in differentiating cells, DNMT1 is required cell-intrinsically for maintaining epidermal progenitor cell proliferation and for preventing premature terminal differentiation [Sen et al., 2010]. Conversely, the depletion of Gadd45, which interacts with DNA repair-associated DNA demethylase complexes, inhibits the induction of select differentiation genes upon differentiation cues, whereas Gadd45 overexpression compromises progenitor cell proliferation and results in premature differentiation. Correlating with such functional outputs, many epidermal differentiation genes have been found to be methylated at their promoters in proliferating progenitor cells, but demethylated upon differentiation [Sen et al., 2010]. Thus, epigenetic silencing of select differentiation genes is used as a prominent mechanism to suppress terminal differentiation in proliferating progenitor cells. To date, the involvement of epigenetic activating mechanisms in controlling epidermal proliferation and differentiation has yet to be directly elucidated. Moreover, it remains to be determined whether similar epigenetic mechanisms govern the maintenance of other epithelial stem/progenitor cell populations. Along this line, interesting expression patterns have been described for epigenetic silencers including HDAC1, HDAC2, HDAC3, and DNMT1 during intestinal stem/progenitor cell differentiation [Vincent and Van Seuningen, 2009].
Wnt/β-CATENIN SIGNALING: AN EPIGENETIC UPDATE
In canonical Wnt signaling, binding of Wnt ligands to Frizzled receptors/low-density lipoprotein related protein (LRP) 5 or LRP6 co-receptors initiates a cascade of intracellular events leading to stabilization of the central molecule β-catenin and its subsequent nuclear translocation. Within the nucleus, β-catenin forms complexes with DNA sequence-specific transcription factors of lymphocyte enhancer factor/T cell factor (LEF/TCF) family to regulate the expression of the Wnt target genes. Many of such target genes are silenced in the Wnt-inactive state by LEF/TCF-associated transcriptional co-repressor complexes including Groucho and CtBP, which are then replaced by β-catenin upon Wnt activation. The activated β-catenin also recruits myriad nuclear co-factors to the LEF/TCF-binding loci. Many of these factors are important regulators of histone modification, including histone acetyltransferase (HAT) complexes such as CBP/p300 and TRRAP/TIP60, SETdomain histone methyltransferase (HMT) of H3K4 such as MLL1 and MLL2 (mixed-lineage leukemia 1 and 2) complexes, the SWI/SNF (switch/sucrose non-fermentable) family of ATPases (e.g., BRG1 or brahma-related gene 1) involved in nucleosome repositioning, Hyrax/parafibromin—component of the PAF1 (RNA polymerase II associated factor 1) complex that interacts with RNA polymerase II and facilitates histone modification, and finally the Pygopus (Pygo) family of chromatin effector proteins via B-cell CLL/lymphoma 9 (BCL9) protein [Mosimann et al., 2009].
Pygo is an evolutionally conserved family of plant homeodomain (PHD)-containing proteins with important roles in animal development [Jessen et al., 2008]. Initially identified in genetic screens for Wg (Wnt counterpart) signaling components in Drosophila, dPygo is essential for transcriptional activation of Wg targets during Drosophila development. This activity is downstream of Arm/β-catenin and mediated by the assembly of a chain of activation complex, and/or by dPygo-dependent nuclear retention of Arm/β-catenin [Jessen et al., 2008]. Mammals have two Pygo homologs, Pygo1 and Pygo2, both of which have been shown to bind directly to K4 di- or trimethylated histone H3 (H3K4me2/3) via the conserved C-terminal PHD domain [Fiedler et al., 2008; Gu et al., 2009]. Furthermore, Pygo2 interacts with WDR5 (WD repeat-containing protein 5), a core subunit of H3K4 HMT complexes including MLL1 and MLL2, facilitating its chromatin association [Gu et al., 2009]. Consistently, Pygo2 is required for optimal trimethylation of H3K4 in MCF10A cells, both globally and at specific Wnt/β-catenin target loci. Pygo2 is also reported to associate with HAT activity and facilitates histone acetylation [Nair et al., 2008; Andrews et al., 2009]. In vivo, genetic ablation of Pygo2 results in dramatic reduction in Wnt signaling output [Li et al., 2007; Gu et al., 2009], yet in vitro, whether Pygo2 activates reporter gene expression remains uncertain. This may not be surprising given that the establishment of histone modification and actual transcriptional activation or silencing can be uncoupled.
The critical involvement of chromatin events in Wnt target gene transcription [Mosimann et al., 2009] now illuminates a previously underappreciated link between Wnt signaling and the epigenetic control of epithelial stem cell homeostasis (see below). Further strengthening this link is the recent finding that β-catenin converges with telomerase, another central regulator of stem cell maintenance and activation, on interaction with BRG1 and activation of downstream target genes [Park et al., 2009].
Wnt SIGNALING IN MODEL EPITHELIAL STEM CELLS
In this section, we first present a brief overview of the function of Wnt signaling in two leading epithelial stem cell models: those of the intestine and hair follicle (readers are referred to more comprehensive reviews on the topic [Blanpain et al., 2007; Barker et al., 2008]). We then focus on discussing recent advances regarding the involvement of Wnt signaling in mammary epithelial stem cells.
WNT SIGNALING IN EPITHELIAL STEM CELLS OF THE INTESTINE
The intestinal tract is lined with rapidly self-renewing epithelia, composed of invaginating crypts and protruding villi that contain ISCs and terminally differentiated cells, respectively. Previous studies in mice have provided strong evidence that Wnt signaling is required for the normal homeostasis of ISCs (Table I) [Barker et al., 2008; and references therein]. Specifically, abrogation of Wnt pathway by deletion of TCF4, or by transgenic overexpression of Wnt inhibitor Dickkopf 1 (DKK1) results in a dramatic reduction in proliferation of crypt cells. Conversely, constitutive activation of Wnt pathway results in massive proliferation of intestinal stem/progenitor cells and the onset of intestinal tumorigenesis.
TABLE I.
Summary of Selected Publications on the Involvement of Wnt Signaling in the Regulation of Epithelial Stem/Progenitor Cells
| Mutation | Tissue type | Phenotype | Refs. |
|---|---|---|---|
| Wnt1 transgenic | Mammary gland | Increased stem/progenitor cells, tumorigenesis |
Li et al. (2003), Liu et al. (2004), Stingl et al. (2006), Vaillant et al. (2008) |
| Hair follicle | Increased proliferation followed by exhaustion of stem cells |
Castilho et al. (2009) | |
| LRP5/6 knockout | Mammary gland | Reduced stem/progenitor cells | Lindvall et al. (2006), Lindvall et al. (2009) |
| Dkk1 transgenic | Mammary gland | Block in development | Andl et al. (2002) |
| Hair follicle | Failure of hair follicle formation | Andl et al. (2002) | |
| Intestine | Reduced epithelial proliferation, loss of crypts | Kuhnert et al. (2004), Pinto et al. (2003) | |
| Conditional β-catenin knockout | Hair follicle | Loss of hair follicle formation | Huelsken et al. (2001) |
| Activated β-catenin transgenic | Hair follicle | New hair follicle formation, tumor | Gat et al. (1998) |
| Mammary gland | Increased proliferation, tumor | Li et al. (2003), Teuliere et al. (2005) | |
| Intestine | Intestine tumor formation | Harada et al. (1999) | |
| Conditional Pygo2 knockout | Mammary gland | Decreased stem/progenitor cells | Gu et al. (2009) |
| Tcf4 knockout | Intestine | Depletion of stem cells | Korinek et al. (1998) |
| Lef1 knockout | Hair follicle | Loss of hair follicle formation | van Genderen et al. (1994) |
| Mammary gland | Loss of mammary gland | van Genderen et al. (1994) | |
| Lef1 transgenic | Hair follicle | Increased hair follicle formation | Zhou et al. (1995) |
Given the intimate link between Wnt signaling and stem cell maintenance, Clevers and coworkers screened Wnt target genes and identified Lgr5 or GPR49, an orphan G protein-coupled receptor, as a marker of ISCs [Barker et al., 2007]. Lgr5 is restrictively expressed in the columnar base cells located on the crypt bottom of murine small intestine, where the niche for gut stem cells is thought to be located. Lineage tracking experiment using inducible Lgr5-Cre knocking and Rosa26-LacZ reporter mice demonstrated that cells of all epithelial lineages of the small intestine and colon were LacZ-labeled over long periods of time, providing strong evidence that Lgr5 marks long-lived and multipotent ISCs. Recently, by using Lgr5 as an ISC marker, the same group reported that a single Lgr5-labeled ISC, when cultured in vitro in the presence of Wnt agonist R-Spondin and other factors, gives rise to a crypt-villus structure without a mesenchymal niche [Sato et al., 2009], further underscoring the critical importance of Wnt signaling in maintaining ISC identity.
WNT SIGNALING IN HAIR FOLLICLE EPITHELIAL STEM CELLS
Wnt signaling plays an important role in the maintenance and activation of HFSCs, which support the extensive regeneration of hair during postnatal life (Table I) [Blanpain et al., 2007; and references therein]. Specifically, inhibition of the Wnt pathway by conditional ablation of β-catenin or by ectopic expression of DKK1 specifically in epithelia blocks hair follicle formation during embryogenesis, with the former also causing a loss of the postnatal HFSC niche. Conversely, cytokeratin 14 (K14) promoter-driven expression of a truncated, non-degradable form of β-catenin (ΔN-β-catenin) in transgenic mice results in de novo hair follicle morphogenesis and tumors. Elegant follow-up studies further suggest that transient, moderate expression of active (non-degradable) forms of β-catenin in quiescent HFSCs induces precocious entry into anagen and subsequent differentiation [Van Mater et al., 2003; Lowry et al., 2005]. Consistent with such a role in HFSC activation, the expression of the Wnt/β-catenin pathway target Lgr5 has been shown to mark a cycling population of HFSCs [Jaks et al., 2008]. In addition to its role in normal homeostasis of HFSCs, Wnt signaling has also been implicated in the regulation of skin stem cells during wound repair, where it facilitates de novo hair follicle formation from the committed interfollicular epidermal stem cells [Ito et al., 2007].
It is important to recognize that Wnt ligands do not always function through β-catenin and that downstream factors may act independently of Wnt signaling. TCF3 and TCF4 have emerged as factors that are necessary and/or sufficient to maintain primitive, undifferentiated skin progenitor cells, as enforced expression of TCF3 suppresses epidermal, hair follicle and sebaceous grand differentiation, whereas double deletion of TCF3 and TCF4 compromises the long-term maintenance and repair of epidermis and hair follicles [Nguyen et al., 2009; and references therein]. At least some of the functions mediated by TCF3 or TCF4 are Wnt-independent, and a detailed look at how β-catenin and TCF factors may differentially bind and affect the epigenetic status of target loci will be of interest. Finally, a recent study showed that persistent expression of Keratin 5 promoter-driven Wnt1 in skin-induced hair follicle hyperproliferation as well as stem cell exhaustion and aging in a manner that is dependent on the mTOR (mammalian target of rapamycin) pathway, pointing out the multivalent function of Wnt signaling in the homeostasis of skin stem cells [Castilho et al., 2009].
WNT SIGNALING IN MAMMARY EPITHELIAL STEM CELLS
Fueled first by embryonic mammary progenitor cells and subsequently by postnatal stem/progenitor cells residing in the terminal end bud (TEB) of elongating ducts, mammary development proceeds from mid-embryogenesis to 10–12 weeks after birth, culminating in the generation of a mature gland capable of additional morphogenesis during pregnancy/lactation. The presence of adult mammary stem cells (MaSCs), which are capable of both self-renewal and differentiation into at least three different cell lineages (basal/myoepithelial, ductal luminal, and alveolar epithelial cells), was first demonstrated half a century ago and their characteristics/distribution/activity have been subsequently studied using different methods [Stingl, 2009]. A breakthrough came when two research groups published the isolation of highly enriched mouse MaSCs using two sets of surface markers, namely Lin−/CD29high/CD24+ or Lin−/CD49fhigh/CD24+, by fluorescence activated cell sorting, and the demonstration of their activity in reconstituting a mammary gland upon transplantation [Shackleton et al., 2006; Stingl et al., 2006]. Although to date the exact niche for these self-renewing and multipotent stem cells remains to be defined, a great deal has been learned about their molecular characteristics and regulation [for comprehensive review, see Visvader, 2009].
As in intestine and hair follicle, Wnt signaling is one of the most critical signaling pathways that regulate MaSCs [Visvader, 2009; and references therein]. In fact, a recent gene expression profiling of highly pure MaSC population compared with their direct progenies implicated the high activity of Wnt-associated signaling network in MaSCs [Pece et al., 2010]. Moreover, mice with compromised Wnt/β-catenin signaling, including those with DKK1 over-expression or deletion of LRP5/6 or LEF1, fail to properly develop mammary glands. In the converse experiments, aberrant activation of Wnt pathway in mouse mammary tumor virus (MMTV)-Wnt1 or MMTV-ΔN-β-catenin transgenic mice results in mammary tumors and/or an expansion of the mammary stem/progenitor pool (Table I). Interestingly, a fine comparative analysis of MMTV-Wnt1 and MMTV-ΔN-β-catenin mammary tumors has suggested that while ΔN-β-catenin targets an alveolar progenitor population, Wnt1, by virtue of its secreted nature, targets the neighboring basal cells including those with stem/early progenitor cell characteristics [Teissedre et al., 2009]. Consistently, overexpression of ΔN-β-catenin in basal mammary epithelial cells leads to amplification of primitive basal-type cells that do not express lineage markers [Teuliere et al., 2005]. Collectively, these studies demonstrate the MaSC-containing basal population to be a primary target of active Wnt signaling. This said, there is also evidence suggesting that Wnt1 expressed under the control of MMTV might cause dedifferentiation of committed luminal progenitor cells to a basal, more stem-like state [Vaillant et al., 2008].
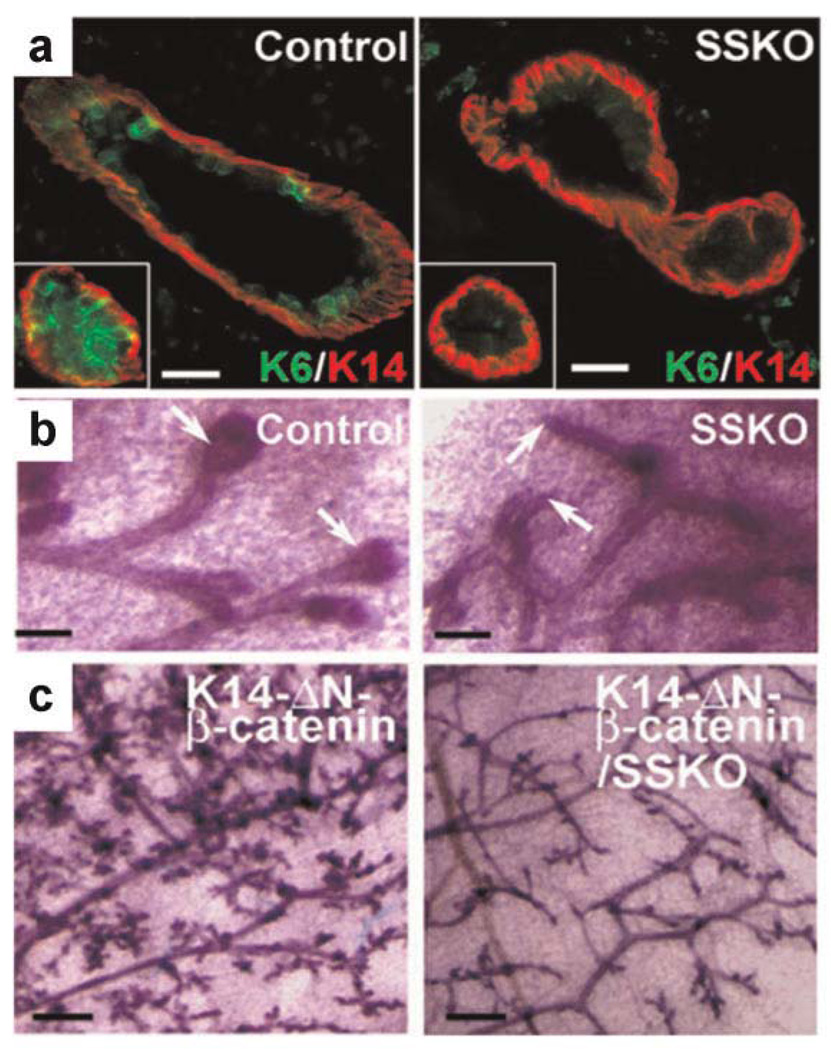
Recently, we have obtained genetic evidence that epigenetic activator Pygo2 is expressed in both embryonic and postnatal mammary progenitor cells and plays an important role in the expansion of mammary stem/progenitor cells [Gu et al., 2009]. Ablation of Pygo2 by complete or K14-Cre-specific gene knockout in mice impairs mammary morphogenesis and regeneration likely due to the impairment of self-renewing expansion of mammary stem/progenitor cells (Fig. 3). This role is linked to Wnt signaling because loss of Pygo2 results in reduced Wnt signaling output, as assessed by both artificial Wnt reporter and endogenous Wnt target gene expression. More importantly, loss of Pygo2 completely rescues the precocious mammary outgrowth induced by ΔN-β-catenin overexpression under a K14 promoter. Underpinning the epigenetic nature of Pygo2 function, a mutant Pygo2 protein containing a point mutation in its PHD domain that affects its ability to bind H3K4me3 but not BCL9/β-catenin is no longer able to promote colony formation by cultured mammary epithelial cells. Moreover, deletion of the PHD domain, which results in loss of both H3K4me3 and BCL9/β-catenin binding, yielded a dominant negative effect in this assay, suggesting that the regulation of mammary cell proliferation by Pygo2 requires proper interaction with both H3K4me3 and the BCL9/β-catenin complex. Our study has uncovered the first in vivo connection between Wnt signaling and the epigenetic regulation in epithelial stem cells, and has now paved the way for future work to examine how Wnt signaling interacts with the epigenetic machinery to control epithelial stem cell homeostasis.
Fig. 3.
Pygo2 is required for mammary epithelial stem/progenitor cell expansion and K14-ΔN-β-catenin-induced mammary hyperplasia. a: Reduced presence of K6+ progenitor cells in 8-week-old Pygo2-deficient mammary duct and ductal termini (inset). b: Carmine red-stained whole-mount mammary gland preparations from 3-week-old female mice, showing absence of TEB structures (arrows) in the mutant. c: Whole-mount carmine-stained mammary glands of 12-week-old K14-ΔN-β-catenin and K14-ΔN-β-catenin/K14-Cre/Pygo2floxed/− (SSKO) littermates (Gu et al., 2009; originally published in J Cell Biol 185:811– 826).
FUTURE PERSPECTIVES
During the past few years, there has been impressive progress in the identification, isolation, and molecular characterization of epithelial stem cells. Such advances in combination with elegant genetic tools have allowed us to begin to investigate the genetic control of the proliferation and lineage differentiation of these cells. In keeping with the blooming field of epigenetics, efforts have also been initiated to venture into the epigenetic control, both DNA methylation and histone modification, of epithelial stem cells. We predict that the next several years will present both challenging and rewarding opportunities for this scientific journey.
Genetic studies of epigenetic regulators, especially those with a known involvement in ES cells or other stem cell systems, and of epigenetic components of Wnt signaling, will continue to rise. For example, little is known about MLLs’ involvement in somatic epithelial stem cells. A recent report showed that MLL1 is important for neuronal differentiation of neural stem cell [Lim et al., 2009], raising the possibility of similar functions in other stem cell systems. Given the potentially wide-spread involvement of the epigenetic components in development, tissue-, stage-, and/or cell-type specific gene ablation will likely to be essential in uncovering their functions in epithelial stem cells.
Epithelial stem cells in different tissues react differently to canonical Wnt signaling activation. In addition, even cells within the same tissue respond differently to Wnt signaling depending on their differentiation status. There is also strong evidence that the level and duration of Wnt signaling can dictate the specific type of downstream cellular response of the same cells. For example, low levels of Wnt signaling facilitate HFSC activation, whereas medium levels promote HFSC terminal differentiation and high levels stimulate tumorigenesis [Blanpain et al., 2007]. While multiple mechanisms (e.g., different usage of Wnt ligand-receptor/co-receptor combinations or cross-talk with other signaling pathways) likely underlie this context dependence, reciprocal interaction between Wnt signaling and the chromatin status of the “receiving” cells is conceivably an important determinant. What are the specific epigenetic patterns that confer the desired cellular specificity to Wnt signaling? What epigenetic landscaping changes are elicited by Wnt signaling? What are the specific target genes that are differentially regulated and what epigenetic mechanisms are utilized to achieve such intricate regulation? These are all interesting questions worthy of future pursuit.
The “histone code” hypothesis states that combinations of histone modifications specify chromatin structure and gene activity to achieve/maintain specific cellular states [Jenuwein and Allis, 2001]. Literature on the use of ChIP-chip, ChIP-seq, or MeDIP (methylated DNA immunoprecipitation) where chromatin DNA that is immunoprecipitated with an antibody against a chromatin-bound protein, modified histones, or CpG-methylated DNAs are hybridized to an array or subjected to high-throughput sequencing [Park, 2009; Pomraning et al., 2009], to map genome-wide factor occupancy or histone marks is exploding. However, such techniques have not been widely applied to the study of epithelial stem cells. Genome-wide ChIP-chip or ChIP-seq studies using antibodies against modified histones or myriad transcription factors such as those involved in Wnt signaling will rise in number to better understand the regulatory gene network that governs the self-renewal and lineage differentiation of epithelial stem cells. The generation and comparison of chromatin landscapes of normal and experimentally or genetically manipulated cells will provide insights into the epigenetic control of epithelial stem cells and uncover potential mechanisms of stem cell plasticity. Although the field has made considerable progress in the prospective identification of various epithelial stem cells, the limitation in specificity of available markers and the remaining heterogeneity of the resulting populations will undoubtedly complicate such large-scale analysis. Therefore, the identification of novel and highly specific stem cell markers for different epithelial tissues remains a long-term task for stem cell biologists. Moreover, technological advances, such as improving the sensitivity and robustness and lessening the cost of chromatin-state map analysis, will be necessary for the routine analysis of a small population of highly pure stem cells and their differentiated progenies.
As epigenetic and epigenomic studies unfold, there will be increasing recognition of the importance of non-coding DNAs, such as intergenic, intron, and repetitive regions in epithelial stem cell regulation. Potential involvement of non-coding RNAs will likely emerge in such analysis, and genetic follow-up studies will surface. Whether and how developmental signaling pathways like Wnt signaling regulate the epigenetic states of such non-coding sequences, and whether and how they might in turn regulate signaling strength and specificity in epithelial stem cells might constitute a new research direction in the Wnt signaling field.
In summary, the stage is now set for exploring the relationship between epigenetic mechanisms, developmental signaling pathways, and stem-cell-associated processes such as quiescence, self-renewal, lineage selection, and terminal differentiation. Specifically, the analysis of the effect of Wnt signaling on the epigenomes of epithelial stem cells promises to be highly informative both in terms of basic stem cell biology and in terms of offering new therapeutic tools to manipulate the epigenetic state of these cells.
ACKNOWLEDGMENTS
We are indebted to Gil Smith, Gloria Chepko, Fiona Watt and coworkers for allowing us to use their published images, and to Gloria Chepko for discussions and critical reading of the manuscript. We apologize to those whose publications are not cited here due to space limitation imposed by the journal. Work in the Dai Laboratory has been supported by DOD grant W81XWH-04-1-0516, NIH Grants R01-AR47320, R01-GM083089, and K02-AR51482 to X.D. B.G. is a recipient of CBCRP postdoctoral fellowship (14FB-0129).
Abbreviations used
- H3K27me3
histone H3 trimethyl lysine 27
- H3K4me3
histone H3 trimethyl lysine 4
- ES
embryonic stem (cell)
- SLC
small light cell
- ULLC
undifferentiated large light cell
- H4K20
histone H4 lysine 20
- HDAC
histone deacetylases
- PcG
polycomb group
- PRC1
polycomb repression complex 1
- Ezh2
zeste homolog 2
- PRC2
polycomb repressive complex 2
- DNMT
DNA methyltransferase
- LRP
low-density lipoprotein related protein
- LEF
lymphocyte enhancer factor
- TCF
T cell factor
- HAT
histone acetyltransferase
- HMT
histone methyltransferase
- Pygo
Pygopus
- BCL9
B-cell CLL/lymphoma 9
- PHD
plant homeodomain
- HFSCs
hair follicle stem cells
- TEB
terminal end bud
- MaSC
mammary stem cells
- ISCs
intestinal stem cells
- DKK1
Dickkopf 1
- K14
cytokeratin 14
- MMTV
mouse mammary tumor virus
REFERENCES
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dcv Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Andrews PG, He Z, Popadiuk C, Kao KR. The transcriptional activity of Pygopus is enhanced by its interaction with cAMP-response-element-binding protein (CREB)-binding protein. Biochem J. 2009;422:493–501. doi: 10.1042/BJ20090134. [DOI] [PubMed] [Google Scholar]
- Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dcv. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: Turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepko G, Dickson RB. Ultrastructure of the putative stem cell niche in rat mammary epithelium. Tissue Cell. 2003;35:83–93. doi: 10.1016/s0040-8166(02)00107-6. [DOI] [PubMed] [Google Scholar]
- Chepko G, Smith GH. Three division-competent, structurally-distinct cell populations contribute to murine mammary epithelial renewal. Tissue Cell. 1997;29:239–253. doi: 10.1016/s0040-8166(97)80024-9. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One. 2007;2:e763. doi: 10.1371/journal.pone.0000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: Epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. Dc Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Gu B, Sun P, Yuan Y, Moraes RC, Teng A, Teng A, Agrawal A, Rheaume C, Bilanchone V, Veltmaat JM, Takemaru K, Millar S, Lee EY, Lewis MT, Li B, Dai X. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811–826. doi: 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Ito M, Yang Z, AndI T, Cui C, Kim N, Millar SE, Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jessen S, Gu B, Dai X. Pygopus and the Wnt signaling pathway: A diverse set of connections. Bioessays. 2008;30:448–456. doi: 10.1002/bies.20757. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf1. Proc Natl Acad Sci USA. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Rheaume C, Teng A, Bilanchone V, Munguia JE, Hu M, Jessen S, Piccolo S, Waterman ML, Dai X. Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis. 2007;45:318–325. doi: 10.1002/dvg.20299. [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, Tan LK, Rosen JM, Varmus HE. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc NatI Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mil1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindvall C, Evans NC, Zylstra CR, Li Y, Alexander CM, Williams BO. The Wnt signaling receptor Lrp5 is required for mammary ductal stem cell activity and Wnt1-induced tumorigenesis. J Biol Chem. 2006;281:35081–35087. doi: 10.1074/jbc.M607571200. [DOI] [PubMed] [Google Scholar]
- Lindvall C, Zylstra CR, Evans N, West RA, Dykema K, Furge KA, Williams BO. The Wnt coreceptor Lrp6 is required for normal mouse mammary gland development. PLoS One. 2009;4:e5813. doi: 10.1371/journal.pone.0005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc NatI Acad Sci USA. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of betacatenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: Regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, Sassone-Corsi P, Dai X. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol. 2008;320:446–455. doi: 10.1016/j.ydbio.2008.05.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Venteicher AS, Hong JY, Choi J, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, McLaughlin M, Veenstra TD, Nusse R, McCrea PD, Artandi SE. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Rattis FM, Voermans C, Reya T. Wnt signaling in the stem cell niche. Curr Opin Hematol. 2004;11:88–94. doi: 10.1097/01.moh.0000133649.61121.ec. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kuj ala, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shen X, Orkin SH. Glimpses of the epigenetic landscape. Cell Stem Cell. 2009;4:1–2. doi: 10.1016/j.stem.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217:229–241. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Teissedre B, Pinderhughes A, Incassati A, Hatsell SJ, Hiremath M, Cowin P. MMTV-Wntl and - DeltaN89beta-catenin induce canonical signaling in distinct progenitors and differentially activate Hedgehog signaling within mammary tumors. PLoS One. 2009;4:e4537. doi: 10.1371/journal.pone.0004537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuliere J, Faraldo MM, Deugnier MA, Shtutman M, Ben-Ze’ev A, Thiery JP, Glukhova MA. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132:267–277. doi: 10.1242/dev.01583. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Van Mater D, Kolligs FT, Dlugosz AA, Fearon ER. Transient activation of beta-catenin signaling in cutaneous keratinocytes is sufficient to trigger the active growth phase of the hair cycle in mice. Genes Dev. 2003;17:1219–1224. doi: 10.1101/gad.1076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Van Seuningen I. Epigenetics, stem cells and epithelial cell fate. Differentiation. 2009;78:99–107. doi: 10.1016/j.diff.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- WeIm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]