Abstract
Introduction
Tumor immune tolerance can derive from the recruitment of suppressor cell populations, including myeloid-derived suppressor cells (MDSC). In cancer patients, increased MDSC correlate with more aggressive disease and a poor prognosis.
Experimental Design
Expression of 15 immune factors (TGFβ, IL-1β, IL-4, IL-6, IL-10, GM-CSF, M-CSF, IDO, FLT3L, c-kit L, iNOS, ARG-1, TNFα, COX2, VEGF) by MDSC-inducing human solid tumor cell lines was evaluated by RT-PCR. Based upon these data, cytokine mixtures were then tested for their ability to generate suppressive CD33+ cells from healthy donor PBMC in vitro by measuring their ability to inhibit the proliferation of, and IFNγ production by, fresh autologous human T cells after CD3/CD28 stimulation. Induced MDSC were characterized with respect to their morphology, surface phenotype, and gene expression profile.
Results
MDSC-inducing cancer cell lines demonstrated multiple pathways for MDSC generation, including over-expression of IL-6, IL-1β, COX2, M-CSF, and IDO. CD33+ cells with potent suppressive capacity were best generated in vitro by GM-CSF and IL-6, and secondarily by GM-CSF + IL-1β, PGE2, TNFα, or VEGF. Characterization studies of cytokine-induced suppressive cells revealed CD33+CD11b+CD66b+HLA-DRlowIL-13Rα2int large mononuclear cells with abundant basophilic cytoplasm. Expression of iNOS, TGFβ, NOX2, VEGF, and/or ARG-1 was also up-regulated and transwell studies showed suppression of autologous T cells to be contact dependent.
Conclusion
Suppressive CD33+ cells generated from PBMC by GM-CSF and IL-6 were consistent with human MDSC. This study suggests that these cytokines are potential therapeutic targets for the inhibition of MDSC induction in cancer patients.
INTRODUCTION
Myeloid-derived suppressor cells (MDSC) have recently been recognized as a subset of innate immune cells whose function can alter adaptive immunity and produce immunosuppression (1). These cells are a heterogeneous population of immature cells derived from the lineage of dendritic cells, macrophages, and granulocytes (2). In mice, MDSC are identified by CD11b+, IL-4Rα+, and GR-1+ expression, with recognized granulocytic and monocytic subsets (2). Human MDSC, on the other hand, are less well-defined but are generally agreed to be suppressive, myeloid-derived (CD33+), CD11b+, and non-lineage determined (Lin−; CD3−CD19−CD56−CD14−) with poor antigen presentation (HLA-DR−) (3,4). Granulocyte marker CD66b has also been reported on human MDSC (5).
MDSC are absent or rare in healthy hosts but may naturally accumulate in situations of trauma and sepsis to temper immune responses (2,6). MDSC are also observed to accumulate in the setting of many tumors as key contributors to tumor immune tolerance (2). In murine tumor models and cancer patients, MDSC are found in increased numbers in the tumor microenvironment, peripheral blood, liver, and tumor-draining lymph nodes, and their concentrations correlate with increased stage and metastatic disease (7,8). The specific pathways by which tumors recruit, expand, and activate MDSC remain unknown, but increasing evidence exists for the involvement of interleukin (IL)-1β, IL-6, cyclooxygenase 2 (COX2)-generated PGE2, high concentrations of granulocyte-macrophage colony stimulating factor (GM-CSF), macrophage (M)-CSF, vascular endothelial growth factor (VEGF), IL-10, transforming growth factor beta (TGFβ), indoleamine 2,3-dioxygenase (IDO), FLT3 ligand (FLT3L), and stem cell factor (c-kit L) (2,9,10). Co-culture of immune competent cells with tumor cell lines has been shown to induce tolerogenic DC or MDSC in vitro (11–13 and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). Thus, further examination of the immune modulatory factors expressed by MDSC-inducing cancers and tumor cell lines could identify the cytokine(s) necessary for generation of this suppressor cell population and provide therapeutic targets for MDSC inhibition.
Because MDSC comprise a heterogeneous population of cells, identification of unique surface markers for MDSC, particularly in humans, has been elusive and definition of MDSC by suppressive function remains important (4). However, expression of CCAAT/enhancer-binding protein (C/EBP)-beta, a member of basic region-leucine zipper transcription factors that regulate immune and inflammatory response genes, has been proposed as a transcriptional marker of activated and functionally suppressive MDSC in mice, analogous to FoxP3 expression in regulatory T cells (14,15). MDSC mediate T cell suppression through a variety of mechanisms, including arginase 1 (ARG-1)-mediated local arginine depletion, inducible nitric oxide synthase (iNOS) and NADPH oxidase (NOX2) production of reactive oxygen and nitrogen species, VEGF expression, and cysteine depletion (16–19). In addition to direct T cell suppression, recent evidence suggests a role for MDSC in the expansion of CD4+CD25+FoxP3+ regulatory T cells (Tregs) in the tumor microenvironment through both TGFβ dependent and independent pathways (11,20). The expression of programmed death ligand 1 (PDL1) is increased on the surface of MDSC in some murine tumors models, though the role of this and related ligands in MDSC-mediated suppression remains unclear (21,22). Through these mechanisms, MDSC have been observed to mediate both antigen-specific and nonspecific immune tolerance in cancer (23).
Previously we demonstrated the in vitro generation of human MDSC from healthy donor peripheral blood mononuclear cells (PBMC) through direct co-culture with human solid tumor cell lines (13 and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). The tumor-educated CD33+ cells showed a phenotype typical of human MDSC (CD33+ HLA-DRlow CD66b+ IL13Rαint) and exhibited potent suppressive ability against autologous CD8+ T cells in an antigen non-specific manner. In this study, we examine the ways in which select cytokines induce MDSC from healthy donor PBMC. Using the data generated by tumor-induced MDSC co-cultures, specific cytokine mixtures were developed for the in vitro generation of human CD33+ MDSC from non-suppressive PBMC. Characterization of the morphology, phenotype, suppressive function, and gene expression of the resulting suppressor cells showed a population consistent with human MDSC. These studies shed light on potential therapeutic targets to abrogate MDSC accumulation in the setting of tumor immune tolerance.
MATERIALS AND METHODS
Cell Lines and Cell Culture
Tumor cell lines were obtained from the American Type Tissue Collection or gifted to the Epstein laboratory. Notable gifts include the SW cell lines from the Scott and White Clinic (Temple, TX) and the Panc 4.14 cell line from Dr. Elizabeth Jaffee (Johns Hopkins Medical Center, Baltimore, MD). Tumor cell line authenticity was performed by cytogenetic and surface marker analysis performed at ATCC or in our laboratory. All cell lines were maintained at 37°C in complete medium (RPMI-1640 with 10% FCS (Hyclone, Inc., Logan, UT), 2mM L-Glutamine 100U/mL Penicillin, and 100µg/mL Streptomycin) in T-25 flasks in humidified 5% CO2 incubators and passaged 2–3 times per week by light trypsinization.
In vitro generation of human MDSC
i. Cytokine Induction
Human PBMC were isolated from healthy volunteer donors by venipuncture, followed by differential density gradient separation (Ficoll Hypaque, Sigma, St. Louis, MO). PBMC were cultured in T-25 flasks at 5×105 cells/mL in complete medium for seven days, supplemented with cytokines as indicated. Recombinant human cytokines used for induction include: IL-1β (10ng/mL, Sigma), IL-6 (10ng/mL, Sigma), PGE2 (1µg/mL, Sigma), TGFβ1 (2ng/mL, R&D Systems, Minneapolis, MN), TNFα (10ng/mL, R&D), VEGF (10ng/mL, Sigma), and GM-CSF (10ng/mL, R&D). For combination-cytokine induction experiments, the cytokines used are indicated in the Results section. GM-CSF was added to the mixtures of cytokines to support cell viability, as described previously (24). PBMC cultured in medium alone were run in parallel as a control for each donor. Cultures were run in duplicate, and medium and cytokines were refreshed every two-three days. For all studies, USC Institutional Review Board approval was obtained and a total of 18 male and 7 female donors were used under protocol HS-06-00579.
ii. MDSC Isolation
After one week, all cells were collected from PBMC cultures. Adherent cells were removed using non-protease cell detachment solution Detachin (Genlantis, San Diego, CA). CD33+ cells were isolated from each culture using anti-CD33 magnetic microbeads and LS column separation (Miltenyi Biotec, Bergisch Gladbach, Germany) per manufacturer’s instructions. The purity of isolated cell populations was found to be greater than 90% by flow cytometry.
Quantitative RT-PCR for gene expression of tumor cell lines and MDSC
For gene expression studies, tumor cell lines were collected from culture flasks by trypsinization and cytokine-induced CD33+ cells isolated from whole PBMC cultures by fluorescence activated cell sorting after Cytokine Induction described above. RNA was isolated from cells using Qiagen RNeasy kits (RNeasy Mini to isolate tumor RNA, and RNeasy Micro to isolate MDSC RNA), followed by DNase (TurboDNase, Applied Biosciences) treatment. For quantitative RT-PCR, 100ng of DNase-treated RNA was amplified with gene specific primers using one-step Power SYBR green RNA-to-Ct kit (Applied Biosciences) and run in an Mx300P Strategene thermocycler. Data were acquired and analyzed using MxPro software (Stratagene). Gene expression was normalized to a housekeeping gene (GAPDH) or myeloid cell marker (CD33), and fold change determined relative to human standard RNA (Stratagene). Primer sequences were obtained from the NIH qRT-PCR database (http://primerdepot.nci.nih.gov) and were synthesized by the USC Core Facility (25, Table I).
Table I.
Gene specific primers for quantitative RT-PCR.
| Gene | Forward primer (5'-to-3') | Reverse primer (5'-to-3') |
|---|---|---|
| ARG-1 | GTTTCTCAAGCAGACCAGCC | GCTCAAGTGCAGCAAAGAGA |
| B7H4 | CCCTGAAATACCAAAGCCAA | AGCTCCACTCAGCCAGTACC |
| CD33 | TTCCTCCTGTGGGTCTTCAC | CTTTCCAGGAGATGGCTCAG |
| C/EBPβ | AGCTGCTCCACCTTCTTCTG | AGCGACGAGTACAAGATCCG |
| c-kit L | TCCTGCAGATCCCTTCAGTT | ACACCACTGTTTGTGCTGGA |
| COX2 | GTTTTGACATGGGTGGGAAC | CCCTCAGACAGCAAAGCCTA |
| FLT3L | GGCCTCTAGCCAACTTCCTC | CAGTCCCCAGGACCTGCT |
| GAPDH | TTAAAAGCAGCCCTGGTGAC | CTCTGCTCCTCCTGTTCGAC |
| GM-CSF | GTCTCACTCCTGGACTGGCT | ACTACAAGCAGCACTGCCCT |
| IDO | TCAGGCAGATGTTTAGCAATG | GGCACACGCTATGGAAAACT |
| IL-10 | TCAAACTCACTCATGGCTTTGT | GCTGTCATCGATTTCTTCCC |
| IL-1β | GGAGATTCGTAGCTGGATGC | GAGCTCGCCAGTGAAATGAT |
| IL-4 | AGCGAGTGTCCTTCTCATGG | CAGCCTCACAGAGCAGAAGA |
| IL-6 | CTGCAGCCACTGGTTCTGT | CCAGAGCTGTGCAGATGAGT |
| iNOS | ATTCTGCTGCTTGCTGAGGT | TTCAAGACCAAATTCCACCAG |
| MCSF | CCAGCAACTGGAGAGGTGTC | GCGCTTCAGAGATAACACCC |
| NOX2 (NCF1) | CCTGCCTCAATAGGGAACATT | TCGTACCCAGCCAGCACTAT |
| PDL1 | TATGGTGGTGCCGACTACAA | TGCTTGTCCAGATGACTTCG |
| PDL2 | TGACTTCAAATATGCCTTGTTAGTG | GAAGAGTTCTTAGTGTGGTTATATG |
| TGFβ | GCAGAAGTTGGCATGGTAGC | CCCTGGACACCAACTATTGC |
| TNFα | AGATGATCTGACTGCCTGGG | CAGCCTCTTCTCCTTCCTGA |
| VEGF | CACACAGGATGGCTTGAAGA | AGGGCAGAATCATCACGAAG |
Characterization of MDSC
Suppression Assay
The suppressive function of cytokine-induced CD33+ cells was measured by their ability to inhibit the proliferation of autologous T cells, as described previously (13 and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). Fresh T cells isolated from PBMC of autologous donors by anti-CD3 microbeads and magnetic LS column separation (Miltenyi Biotec) were CFSE-labeled (2.5µM, Sigma) and seeded in 96-well plates at 2×105 cells/well. CD33+ cells from the above Cytokine Induction cultures were added to these wells at a 1:4 or 1:2 ratio. T cell stimulation was provided by anti-CD3/CD28 stimulation beads (Invitrogen) and IL-2 (100U/mL, R&D Systems, Minneapolis, MN). Suppression Assay wells were analyzed by flow cytometry for proliferation of T cells after three days. For each donor and assay run, controls included T cells cultured alone with and without T cell stimulation, and T cells cultured with CD33+ cells from medium-only culture. Each CD33+ sample was run in duplicate and data were acquired as percent proliferation for 15,000 events of live, lymphoid-gated cells. Samples were run on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) and data acquisition and analysis were performed using CellQuestPro software (BD) at the USC Flow Cytometry core facility.
IFNγ cytokine bead array
Interferon gamma (IFNγ) production by T cells in the MDSC Suppression Assay was measured as a correlate of T cell activation. Supernatant was collected at the conclusion of the Suppression Assay from each sample and stored at −80°C until analysis. Protein levels of IFNγ were measured using BD Cytometric Bead Array human IFNγ flex set and human soluble protein master buffer kit (BD), per manufacturer’s instructions. Samples were run on a FACSCalibur flow cytometer (BD) and data acquisition and analysis were performed using CellQuestPro and FCAP Array software (BD).
Morphology of MDSC
Wright-Giemsa staining (Protocol Hema 3, Fisher, Kalamazoo, MI) of CD33+ cell cytospin preparations was performed to assess the morphology of cytokine-induced CD33+ cells. Freshly isolated PBMC and CD33+ cells from healthy donors were prepared in parallel for comparison. Observation, evaluation, and image acquisition were performed using Leica DM2500 microscope (Leica Microsystems, www.leica-microsystems.com) connected to an automated, digital SPOT RTke camera and SPOT Advanced Software (SPOT Diagnostic Instrument Inc., www.diaginc.com). Images were resized and brightened for publication using Adobe Photoshop software (Adobe, www.adobe.com).
Flow cytometry analyses of cell phenotypes
The phenotype of in vitro-generated MDSC was evaluated for expression of CD33, HLA-DR, CD11b, CD11c, CD66b, CD68, CD14, and IL-13Rα2 and compared to whole PBMC and non-induced CD33+ cells. Changes in PBMC subpopulations during cytokine induction and the expansion of Treg cells in CD33+-T cell co-cultures also were measured by flow cytometry. For staining, cells were collected from flasks using Detachin (Genlantis) to minimize cell surface protein digestion, washed twice with FACS buffer (2% FCS in PBS), and 106 cells were resuspended in 100ul FACS buffer. Cells were stained for 1hr on ice with cocktails of fluorescently-conjugated monoclonal antibodies or isotype-matched controls, then washed twice with FACS buffer, and resuspended in FACS buffer for analysis. For intracellular staining, cell surface staining was performed first, followed by buffer fixation/permeabilization (eBioscience, San Diego, CA) and intracellular staining. Antibodies used were from BD: CD4 (RPA-T4), CD8 (RPA-T8), CD11b (ICRF44), CD11c (B-ly6), CD14 (M5E2), CD25 (M-A251), CD33 (HIM3–4, WM53), CD66b (G10F5), FoxP3 (259D/C7), HLA-DR (G46-6); Santa Cruz Biotech: IL-13Rα2 (B-D13); Miltenyi Biotec: CD25 (4E3); and eBioscience: CD1a (HI149), CD3 (OKT3), CD20 (2H7), CD56 (MEM188), CD68 (Y1/82A), and isotype controls. Samples were run on a FACSCalibur flow cytometer (BD) and data acquisition and analysis were performed as above. Samples were run in duplicate and PBMC cultured in medium alone were run in parallel for comparison. Data were acquired as the fraction of labeled cells within a live-cell gate set for 15,000 events.
Transwell assays
The MDSC Suppression Assay was modified to test contact dependency of MDSC-mediated T cell suppression. For these studies, cytokine induced CD33+ cells isolated from Cytokine Induction cultures and fresh autologous CFSE-labeled T cells were co-cultured at a 1:4 ratio in the presence of T cell stimuli for 3 days, as described above. In deviation from the methods above, transwell plates (0.4µm pores, Griener Bio-One, Wemmel, Belgium) were used for these studies, with T cells cultured in plate wells and CD33+ cells cultured in transwell inserts to inhibit direct T cell – CD33+ cell contact.
Cytokine induction of CD33+ MDSC from T cell-depleted PBMC
To determine the role, if any, of T cells, the Cytokine Induction Assay was repeated for select cytokine mixtures using T cell-depleted PBMC from healthy volunteers, rather than whole PBMC. For these studies, PBMC obtained after density gradient separation were depleted of T cells using anti-CD3 magnetic microbeads and LD column separation (Miltenyi Biotec), per manufacturer’s instructions. Subsequent culture, cell isolation, and function evaluation of cytokine induced CD33+ cells from these cultures was performed as described in the MDSC Isolation and Suppression Assays above.
Statistical analysis
For MDSC Suppression Assays, CFSE fluorescence was measured by flow cytometry to determine the percent proliferation for each sample relative to T cell controls. Where possible, mean relative proliferation and SD were calculated and graphed using Microsoft Excel software (Microsoft, Redmond, WA). A change in mean T cell proliferation in the presence or absence of tumor-educated MDSC was tested for statistical significance using the student t test for independent samples. For all analyses in which multiple experimental samples were compared against one another (e.g. qRT-PCR), a one-way ANOVA was performed followed by Dunnett post-test analysis using GraphPad Prism (La Jolla, CA). For all statistical tests, significance level α = 0.05.
RESULTS
Human solid tumor cell lines over-express multiple putative MDSC-promoting factors
In this study, cancer cell line induction of human CD33+ MDSC was used as a model for tumor induction of human MDSC. Previously, we screened 100 human solid tumor cell lines for their ability to induce functionally suppressive CD33+ MDSC from healthy donor PBMC using a novel co-culture method (13 and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). In that study we reported 46 cancer cell lines capable of generating CD33+ human MDSC. Here, we selected 17 of the 46 MDSC-inducing cancer cell lines, as well as 6 non-MDSC inducing control cancer cell lines, to examine the mechanisms by which tumors generate MDSC. Gene expression levels of putative MDSC inducing immune-modulatory factors were compared for both groups of cancer cell lines using quantitative RT-PCR techniques (Figure 1). Gene expression by tumor cell lines was examined for 15 immune modulatory factors: ARG-1, IL-1β, IL-4, IL-6, IL-10, iNOS, c-kit L, COX2, FLT3L, GM-CSF, IDO, M-CSF, TGFβ, TNFα, and VEGF.
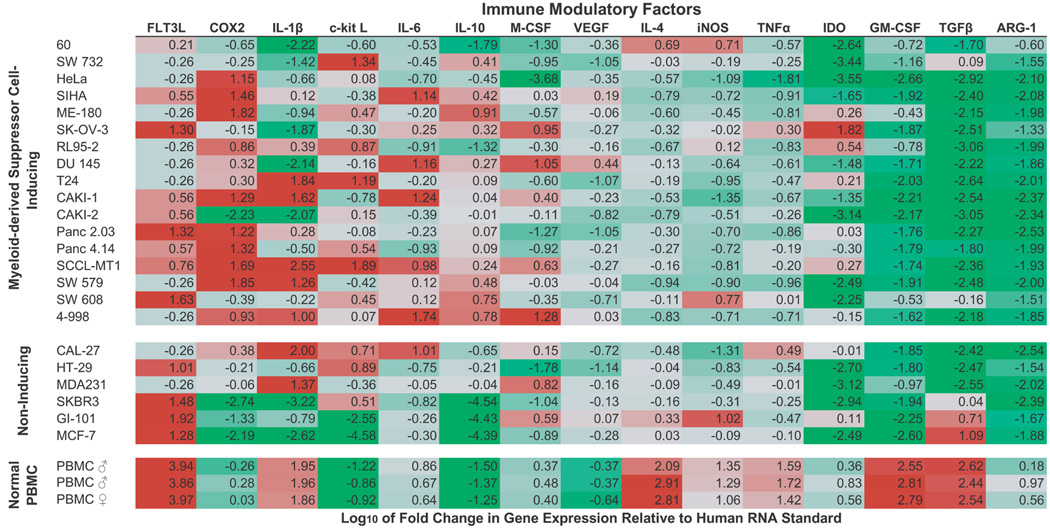
Figure 1. Expression of putative MDSC-inducing factors by human solid tumor cell lines and PBMC.
The fold change in gene expression of putative MDSC-inducing immune modulatory factors was determined by quantitative RT-PCR techniques for MDSC-inducing and non-inducing human solid tumor cell lines. In addition, the expression of these factors by freshly isolated PBMC from healthy volunteer donors was analyzed in parallel. Due to the large variation in gene expression levels amongst tumor cell lines, the log10 fold change is shown (e.g. fold increases of 10 and 100 are represented as a log10 changes of 2.0 and 3.0, respectively). Genes whose expression was higher in the tumor cell line than the reference sample are shown in red (90th percentile); those whose expression was lower than the reference sample are shown in green (10th percentile). Note, the differential expression of factors COX2, IL-1β, and IL-6 in MDSC-inducing and non-inducing cell lines and the high expression of GM-CSF in normal donor PBMC compared to tumor cell lines.
All tumor cell lines examined were found to have a statistically significant increase in expression of at least one of these factors relative to a reference human RNA standard; however no single factor was shown to be critical for MDSC induction (Figure 1). MDSC-inducing tumor cell lines showed increased expression of COX2, IL-1β, IL-6, M-CSF, and IDO (Figure 1). FLT3L and c-Kit L expression appeared to be increased in both groups of tumor cell lines, suggesting that these are not singular distinguishing factors for MDSC induction. TGFβ, GM-CSF, and ARG-1 showed consistent down-regulation amongst MDSC-inducing tumor cell lines. These results suggest multiple pathways of MDSC induction for the tested human cancer cell lines.
Human CD33+ MDSC are generated in vitro by soluble immune modulatory factors
Using MDSC-induction by cancer cell lines as a model, cytokine mixtures were designed for the in vitro generation of human MDSC from healthy donor PBMC. Briefly, PBMC were cultured for one week in the presence or absence of immune modulatory factors GM-CSF and IL-1β, IL-6, VEGF, TGFβ, TNFα, or PGE2 (as a proxy for COX2 over-expression (26)), and then CD33+ cells were isolated and tested for suppressive function by MDSC Suppression Assays. The ability of cytokine-induced CD33+ to inhibit autologous T cell proliferation and IFNγ production was evaluated at cell ratios of 1:2 and 1:4 (Figure 2A, B). Multi-cytokine combinations were also tested for the induction of CD33+ MDSC.
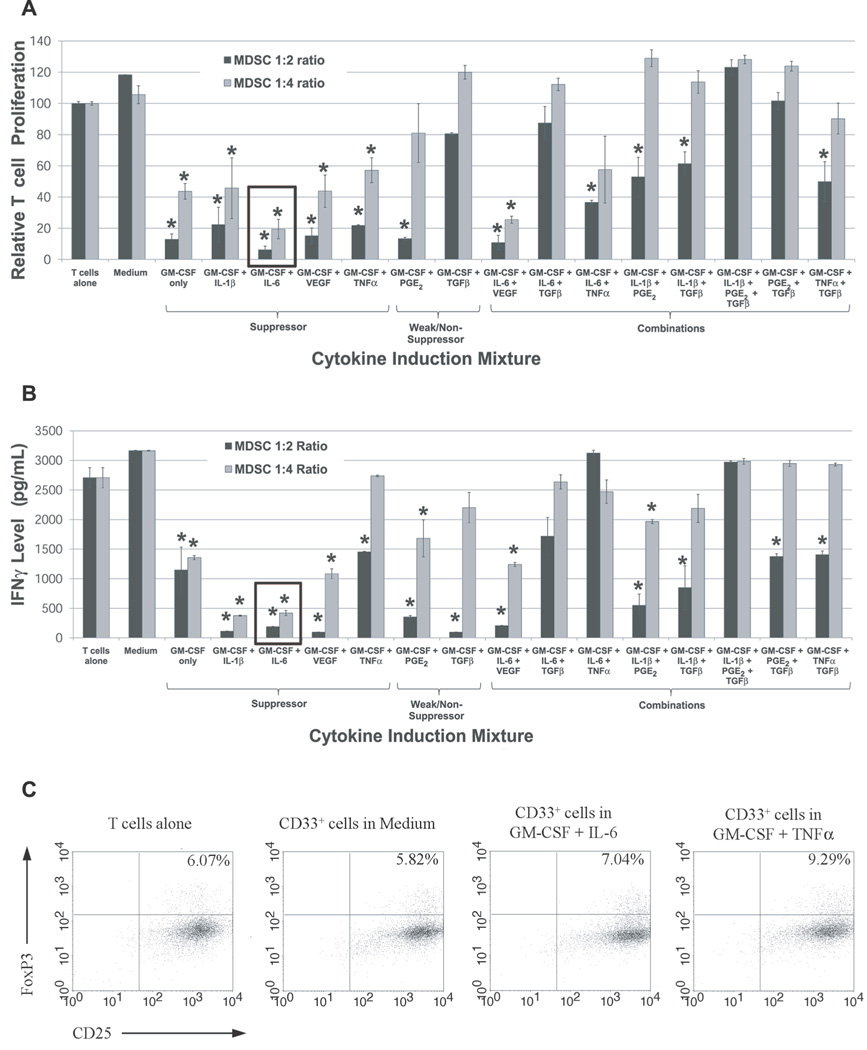
Figure 2. Cytokine-induced CD33+ MDSC demonstrate potent suppressive function.
PBMC from normal donors were cultured for one week in the presence of different cytokine mixtures. CD33+ cells were then isolated and tested for their ability to suppress the (A) proliferation and (B) INFγ production by autologous T cells at ratios of 1:2 and 1:4. CD33+ cells from cultures treated with GM-CSF and IL-6, Il-1β, VEGF, PGE2, or TNFα demonstrated suppressive function. For both graphs, mean is shown with SEM. Conditions with statistically significant decreases in mean T cell proliferation compared to stimulated T cells alone are indicated by an asterisk. (C) Treg expansion by cytokine-induced MDSC in Suppression Assays: the fraction of CD25+FoxP3+ T cells (CFSE-labeled) at the conclusion of a three day Suppression Assay with cytokine-induced CD33+ cells and fresh autologous T cells was analyzed by flow cytometry. Co-cultures with CD33+ cells induced by GM-CSF + IL-6 or GM-CSF + TNFα showed increases in CD25+FoxP3+ T cells relative to stimulated T cells cultured alone or with CD33+ cells from medium-only cultures (n=1).
Results from these studies show the generation of potent CD33+ MDSC after incubation of non-suppressive PBMC with select cytokine combinations. Notably, GM-CSF + IL-6 and GM-CSF + IL-6 + VEGF combinations generated MDSC with the ability to suppress autologous T cell proliferation by 80.6 and 74.5 percent, respectively (mean, n=4), at a 1:4 ratio (Figure 2A). Furthermore, GM-CSF alone, GM-CSF + IL-1β, GM-CSF + TNFα, and GM-CSF + VEGF conditions yielded MDSC with significant suppressive function (range 43–57% suppression at 1:4 ratio). CD33+ cells treated with GM-CSF + PGE2 exhibited weak suppressive ability. While these data suggest a positive role for IL-6, VEGF, IL-1β, TNFα, and GM-CSF in MDSC generation, TGFβ does not appear to have a major role in the promotion of suppressive function in CD33+ cells since TGFβ treatment consistently decreased the potency of other cytokines in the induction of suppressive function (Figure 2A). GM-CSF was found to be critical to maintain myeloid cell viability, as reported previously (24), and was included in all cytokine mixtures, though at lower concentrations than used by Ko et al (10ng/mL versus 50ng/mL) (24). However, even this low level of GM-CSF alone appeared to induce some suppressive function in CD33+ cells, as shown in Figure 2A, and warrants further study as an inducing cytokine for human MDSC.
IFNγ production was evaluated in addition to T cell proliferation to provide a second measure of MDSC suppression (Figure 2B). Consistent with strong inhibition of autologous T cell proliferation, CD33+ cells induced by GM-CSF + IL-6 significantly inhibited IFNγ production in autologous T cells at 1:2 and 1:4 ratios (Figure 2B). CD33+ suppressor cells induced by other cytokine mixtures also inhibited IFNγ expression by autologous T cells, though none to the same extent as GM-CSF + IL-6-generated CD33+ cells. Mean IFNγ levels in responder T cells were statistically lower, though to a lesser extent, in co-cultures with CD33+ cells induced by cytokine mixtures GM-CSF + IL-1β, GM-CSF + PGE2, GM-CSF + VEGF, GM-CSF + IL-6 + VEGF, GM-CSF + IL-1β + PGE2, and GM-CSF alone at ratios of 1:2 and 1:4 (Figure 2B). Although, TGFβ-cultured CD33+ cells did not inhibit T cell proliferation, these co-cultures did demonstrate decreased IFNγ production. This finding suggested that expansion of Treg cells may be occurring from fresh CD3+ cells in the Suppression Assay co-cultures (11,20). To evaluate this possibility, responder T cells from Suppression Assay co-cultures were stained for Treg markers after three days, and the fraction of CD25+FoxP3+ T cells (CFSE-labeled) was measured by flow cytometry (Figure 2C). The fraction of CD25+FoxP3+ from Suppression Assays of CD33+ treated with GM-CSF + IL-6 or GM-CSF + TNFα was slightly increased (7.04% and 9.29%, respectively) compared to stimulated T cells alone (6.07%) and T cells co-cultured with CD33+ cells from medium-only culture (5.82%) (Figure 2C). Suppression Assays with CD33+ cells induced by some other cytokine mixtures showed similar increases in the fraction of CD25+FoxP3+ T cells: GM-CSF + VEGF (8.49%), GM-CSF + IL-1β (7.55%), GM-CSF + TGFβ (6.93%), GM-CSF (9.20%) (data not shown). These results suggest that Treg expansion may contribute to MDSC-mediated T cell suppression in some settings, as reported previously (11,20), though conclusions are limited by the low sample number for these studies.
Morphology of cytokine induced CD33+ MDSC
The morphology of cytokine-induced CD33+ cell populations was examined by Wright-Giemsa staining of cytospin cell preparations (Figure 3A). The CD33+ cells isolated from cytokine-treated PBMC cultures showed remarkably homogeneous morphology and were distinctly different from starting PBMC and freshly isolated CD33+ cells. The morphology of starting PBMC following differential density gradient separation was typical of healthy human donors, showing numerous lymphocytes and mixed monocyte populations with rare granulocytes and eosinophils. Non-induced CD33+ cells isolated from PBMC demonstrated a lymphocyte-depleted monocyte population of small cells, as expected. In contrast, cytokine-induced CD33+ cells were consistently large mononuclear cells with basophilic, granular-appearing cytoplasm (Figure 3A). As reported previously, bi-nucleate cells were commonly observed in CD33+ cells cultured under numerous conditions (27). With cytokine exposure, cells appeared to change from blast-like cells with scant cytoplasm (Figure 3A, panels 1–2) to cytoplasm predominant cells (Figure 3A, panels 3–8). However, no discernable difference was observed in morphology between suppressive and non-suppressive cytokine-treated CD33+ cells by Wright-Giemsa staining.
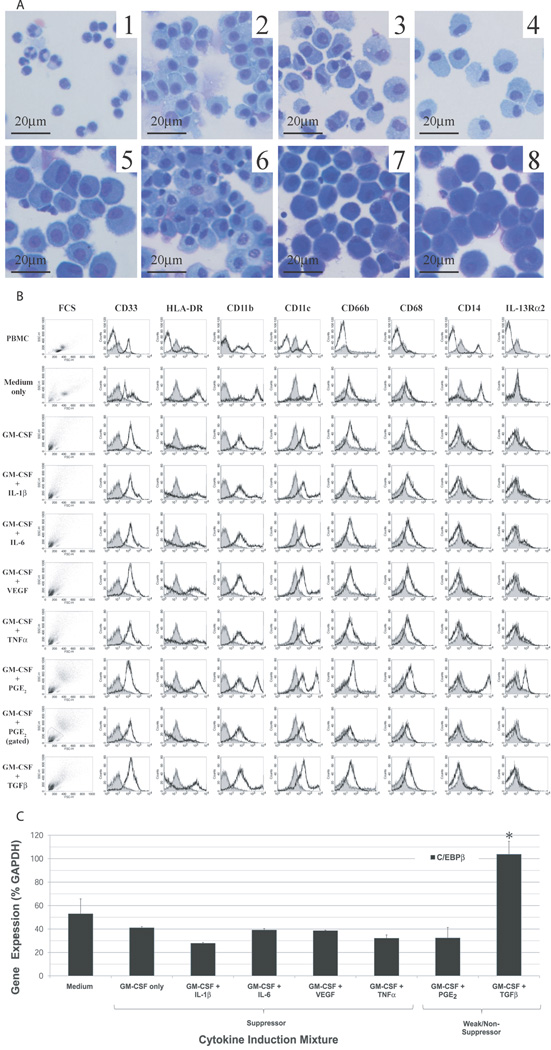
Figure 3. The morphology and phenotype of cytokine-induced CD33+ MDSC resemble tumor-induced MDSC.
(A) Morphology of cytokine-induced CD33+ populations compared to PBMC by Wright-Giemsa staining (x400, original magnification). Starting PBMC population (1) shows small granulocytic and monocytic cells scattered amongst lymphocytes. CD33+ cells isolated after one week culture with complete medium alone (2) or following cytokine induction (3–8) appear as large mononuclear cells with abundant cytoplasm. Non-suppressive cytokine-induced CD33+ cells (3,4) are similar in morphology to suppressive cytokine-induced CD33+ cells, though the cytoplasm of the latter frequently appear more basophilic. (B) Phenotypes of cytokine-induced CD33+ cells compared to whole PBMC and CD33+ cultured in medium alone were determined by flow cytometry. The expression of putative MDSC markers (CD33, CD11b, IL-13Rα2, CD66b) and markers of mature antigen presenting cells (HLA-DR, CD11c, CD14, CD68) was evaluated for each sample (black line) relative to isotype controls (gray). In addition, forward and side scatter analyses of cells were performed to compare size and granularity of cytokine-induced CD33+ cells to controls. For GM-CSF + PGE2 induced CD33+ cells, two discrete populations of cells were noted. For this cytokine mixture, only the granulocytic population (shown gated) expressed a phenotype consistent with human MDSC. (C) Expression of transcription factor C/EBPβ by CD33+ cells induced under different cytokine milieu was evaluated by quantitative RT-PCR techniques. There was a statistically significant difference in mean C/EBPβ gene expression only between GM-CSF + TGFβ and Medium treated CD33+ cells (p <0.05), with no difference amongst any other groups.
Phenotype of cytokine induced CD33+ MDSC resembles tumor-induced MDSC
The surface phenotype of cytokine-treated CD33+ cells was analyzed by flow cytometry and compared to the phenotypes of whole PBMC and CD33+ cells cultured in medium alone (Figure 3B). Flow cytometry analyses of cell forward and side scatter demonstrated a primarily granulocytic CD33+ population induced by cytokines, as compared to the largely monocytic morphology seen in PBMC and medium-only CD33+ cell populations. Staining for CD33 confirmed a high purity of target cells following anti-CD33 magnetic bead labeling and column separation. The reported phenotype of human MDSC is CD33+HLA-DRlowCD11b+CD66b+, with low expression of differentiated macrophage and dendritic cell markers (4,5). CD33+ cells from all cytokine-treated cultures showed similar increases in CD11b and CD66b expression relative to isotype controls, with low to intermediate expression of monocyte/macrophage-associated markers CD68 and CD14. Low expression of antigen presentation protein HLA-DR appeared to distinguish suppressive from non-suppressive cytokine-treated CD33+ cells. A small increase in IL-13Rα2 expression was also observed in suppressive, but not non-suppressive or control, CD33+ cells. For GM-CSF + PGE2 induced CD33+ cells, two discrete populations of cells were noted. For this cytokine mixture, only the granulocytic population (shown gated separately in Figure 3B) expressed a phenotype consistent with human MDSC. Overall, suppressive cytokine-induced CD33+ cells generated by in vitro culture demonstrated a phenotype consistent with that previously reported for human MDSC (4).
Expression of C/EBPβ by cytokine-induced CD33+ cells does not correlate with suppressive function
Expression of transcription factor C/EBPβ by CD33+ cells induced under different cytokine milieu was evaluated by quantitative RT-PCR techniques (Figure 3C). Although C/EBPβ has been proposed as a transcriptional marker up-regulated in activated murine MDSC (15), there was not a statistically significant difference in the expression of this transcription factor between suppressive and non-suppressive human cytokine-induced CD33+ cells (Figure 3C).
Cytokine-induced CD33+ MDSC have up-regulated iNOS, TGFβ, VEGF, and NOX2
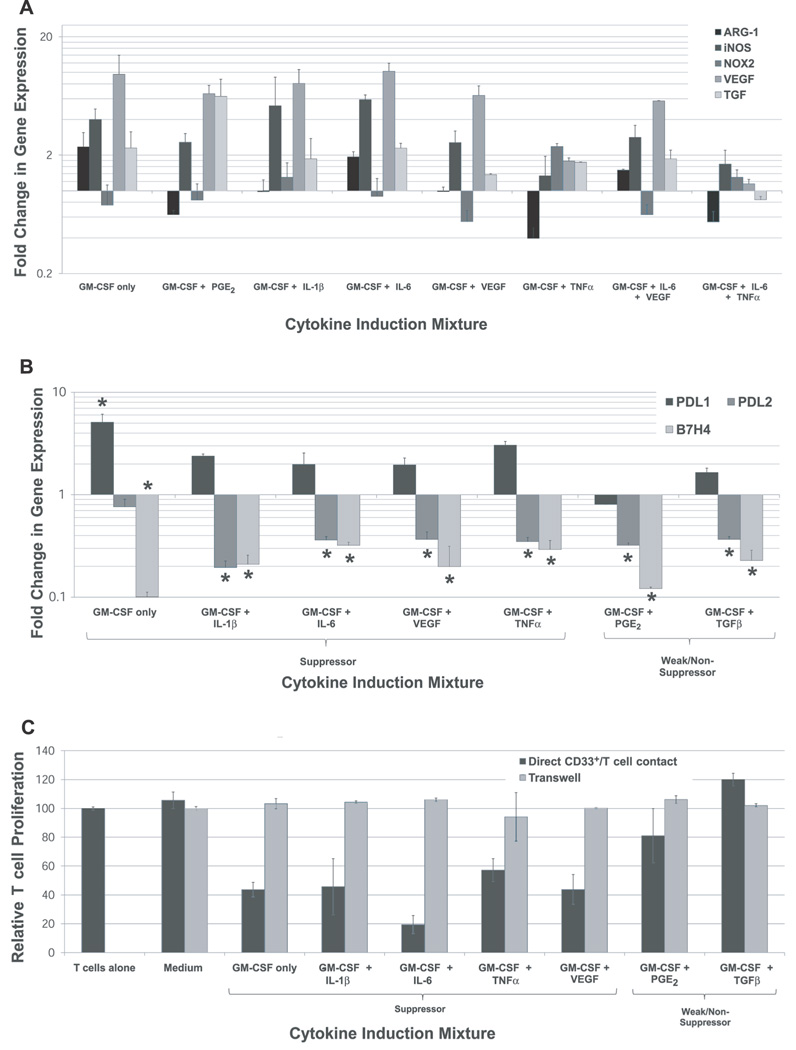
MDSC-mediated suppression of effector T cell responses has been shown to correlate with increased expression of ARG-1, iNOS, NOX2, VEGF, and TGFβ by suppressor cells (16–20). To better characterize the nature of suppressive cytokine-induced CD33+ cells, gene expression of these putative mechanisms of MDSC suppression were evaluated by quantitative RT-PCR techniques (Figure 4A). Cytokine-induced CD33+ suppressor cells demonstrate significant up-regulation of iNOS, NOX2, VEGF, and/or TGFβ compared to freshly isolated, non-induced CD33+ cells. iNOS, VEGF, and TGFβ are consistently up-regulated by various inducing cytokine mixtures, while up-regulation of NOX2 relates most closely with TNFα induction. GM-CSF + IL-6-induced CD33+ cells appeared to mediate their potent suppressive function through ARG-1, iNOS, VEGF, and TGFβ, with minimal contribution from NOX2 (Figure 4A). Also, different cytokine-induced myeloid suppressor cell groups demonstrated various patterns of gene expression (Figure 4A). Previous reports have shown increased expression of inhibitory B7-homologues on murine MDSC derived from some experimental tumor models, though the functional significance of these markers is debated (21,22). Gene expression levels of PDL1 (B7H1), PDL2 (B7H2), and B7H4 were compared between suppressive and non-suppressive cytokine-induced CD33+ cells by quantitative RT-PCR studies (Figure 4B). PDL1 expression was generally observed to be up-regulated in all groups of cytokine-induced CD33+ cells, though the increase was statistically significant only for GM-CSF-induced cells (p<0.05). Cytokine-induced CD33+ cells showed consistent down-regulation of PDL2 and B7H4 genes relative CD33+ from medium-only cultures, with no differences between suppressive and non-suppressive cells (Figure 4B). These results are consistent with reports from experimental tumor models in mice (21,22), and do not support a role for PDL1, PDL2, or B7H4 in suppression mediated by these human cytokine-induced CD33+ cells.
Figure 4. The expression of suppressive genes by cytokine- and tumor-induced CD33+ MDSC varies with the inducing cytokine milieu and the suppression of autologous T cells is contact dependent.
(A) Gene expression of reported suppressive mechanisms in MDSC (ARG-1, iNOS, NOX2, VEGF, TGFβ) in different subsets of cytokine-induced CD33+ MDSC as determined by quantitative RT-PCR techniques. Mean fold change relative to CD33+ cells cultured in medium alone shown, +SEM. (B) Gene expression levels of PDL1 (B7H1), PDL2 (B7H2), and B7H4 were compared between suppressive and non-suppressive cytokine-induced CD33+ cells by quantitative RT-PCR studies. Mean fold change in expression relative to CD33+ from medium-only cultures shown, with SEM. Statistically significant values are indicated by an asterisk. (C) CD33+ MDSC-mediated suppression of autologous T cells is contact dependent. Suppressive cytokine-induced CD33+ cells were co-cultured with fresh T cells isolated from the same donor in a single well or separated by a transwell insert at a 1:4 ratio in a modified MDSC Suppression Assay. Mean T cell proliferation is shown, with SEM. For all transwell samples, mean T cell proliferation in the presence of cytokine-induced CD33+ cells was not statistically significantly different from stimulated T cells alone.
Suppression by human CD33+ MDSC is contact dependent
A variety of suppressive mechanisms have been reported for human and murine MDSC, though it is unclear whether or not they require cell-to-cell contact with target T cells. In our assays, separation of suppressive cytokine-induced CD33+ cells from responder T cells by a transwell inserts (0.4µm pores) was found to abrogate the suppressive effects of all cytokine-induced MDSC (Figure 4C). These data suggest that many, if not all, suppressive mechanisms employed by these cells are contact dependent.
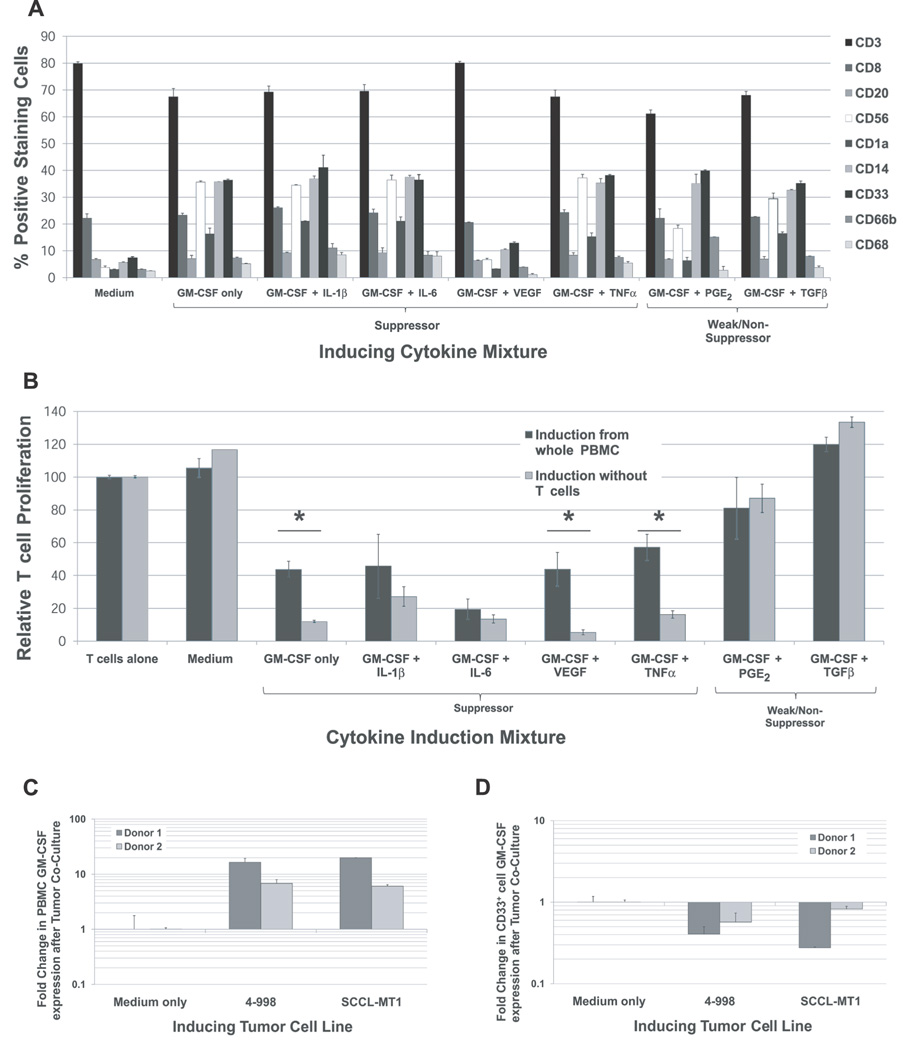
Cytokine induction expands CD56+, CD33+, and CD14+ cell populations
Suppressive CD33+ cells with MDSC-like morphology, phenotype, and gene expression were successfully generated from whole PBMC derived from healthy volunteer donors. To shed light on the cellular context of CD33+ suppressor cell induction, changes in cell types and numbers during cytokine induction were measured by flow cytometry analyses (Figure 5A). All cytokine mixtures, with the exception of GM-CSF + VEGF, showed expansion of CD56+ (natural killer cells (NK)), CD33+ (myeloid), CD14+ (monocytes), and to a lesser extent CD1a+ (DC) cell populations relative to PBMC cultured in medium alone. This effect occurs with GM-CSF alone, suggesting that this cytokine promotes proliferation of cells in the myeloid and granulocyte compartment, consistent with its known function in normal hematopoeisis (Ref 28). Expansion of CD66b+ cells above the levels observed with GM-CSF alone occurred with the addition of IL-1β or PGE2 to cytokine mixtures. T cell numbers and CD4/CD8 ratio appeared to be largely unaffected by cytokine induction. Cytokine mixtures also did not appear to affect the frequency of B cells (CD20+) or macrophages (CD68+). Cytokine mixtures inducing CD33+ suppressor cells did not produce changes in cell types and numbers distinct from those observed for cytokine mixtures that did not induce MDSC.
Figure 5. Cellular context for cytokine induction of CD33+ MDSC from normal donor PBMC.
(A) Changes in cell types and frequencies during cytokine induction were measured by flow cytometry. Significant expansion of CD56+ (NK), CD33+ (myeloid), and CD14+ (monocyte) cell populations was observed in PBMC treated with all cytokine mixtures, with the exception of GM-CSF + VEGF. The addition of IL-1β or PGE2 appeared to increase the frequency of CD66b+ cells beyond that observed with GM-CSF treatment alone. The number and CD4/CD8 ratio of T cells did not appear to be affected by the cytokine combinations examined here, nor did the frequency of B cells (CD20+) or macrophages (CD68+). Cytokine mixtures inducing CD33+ suppressor cells did not produce changes in cell types and numbers distinct from those observed for cytokine mixtures that did not induce MDSC. (B) The suppressive function of CD33+ cells from whole PBMC or from T-cell depleted PBMC cultures treated with cytokine mixtures was compared to determine the requirement of T cells in the induction of MDSC. CD33+ MDSC generated in the absence of T cells demonstrated a comparable suppressive capacity to those generated from whole PBMC for most cytokine mixtures examined. However, for cytokine mixtures GM-CSF alone, GM-CSF + TNFα, and GM-CSF + VEGF, CD33+ generated in the absence of T cells were more suppressive than those generated from whole PBMC (p<0.05). (C and D) To shed light on possible sources of GM-CSF for the induction of suppressor cells in tumor-PBMC co-cultures, gene expression of GM-CSF in PBMC following direct co-culture with inducing tumor cell lines was measured by qRT-PCR techniques and compared to cells cultured in medium alone (mean shown, + SD). GM-CSF expression by PBMC was strongly up-regulated following tumor cell line co-culture. Analysis of GM-CSF expression in the CD33+ cell fraction suggests that the increase in GM-CSF observed is not localized to this subpopulation.
T cells are not needed for cytokine induction of CD33+ MDSC from normal donor cells
T cells comprise a large fraction of PBMC and depletion of this population may increase the yields of CD33+ cells from cytokine-induced cultures. To determine whether T cells were necessary for the induction of suppressive CD33+ cells by the cytokine mixtures used above, cytokine induction experiments were repeated using T cell-depleted PBMC. The suppressive function of CD33+ cells from whole PBMC or from T-cell depleted PBMC cultures treated with cytokine mixes was then compared, as shown in Figure 5B. CD33+ MDSC generated in the absence of T cells demonstrated a comparable suppressive capacity to those generated from whole PBMC for most cytokine mixtures examined. Interestingly, induction studies using GM-CSF alone, GM-CSF + TNFα, and GM-CSF + VEGF showed that CD33+ generated in the absence of T cells were more suppressive than those generated from whole PBMC (p<0.05).
GM-CSF expression by PBMC is up-regulated by co-culture with tumor cell lines
Cytokine induction studies showed that GM-CSF alone or in combination with IL-6, IL-1β, and VEGF generates potent CD33+ suppressor cells. While this cytokine appears to be critical in human myeloid suppressor cell induction, MDSC-inducing tumor cell lines were found to have very low GM-CSF expression (Figure 1 and ELISA data not shown). PBMC, however, do have strong expression of GM-CSF (Figure 1) and thus may provide a source of GM-CSF for MDSC induction in the tumor micro-environment. GM-CSF expression by PBMC in the presence or absence of inducing tumor cell line was measured by quantitative RT-PCR (Figure 5C). These data showed strong up-regulation of GM-CSF production by normal donor PBMC following direct co-culture with MDSC-inducing cell lines 4–998 (osteogenic sarcoma) or SCCL-MT1 (head and neck squamous cell carcinoma), but not culture in medium alone. Analysis of the CD33+ cell fraction showed that the increase in GM-CSF production was not localized to this population (Figure 5D).
DISCUSSION
Immune suppressor cells, including MDSC, contribute to tumor immune tolerance and promote tumor progression. In this study, a group of MDSC-inducing human solid tumor cell lines were evaluated for their expression of factors previously implicated in the generation and activation of MDSC. These data were then used to identify specific cytokine combinations sufficient to induce functionally suppressive myeloid (CD33+) cells from healthy donor PBMC as an in vitro model of tumor-MDSC induction. Characterization of the morphology, phenotype, suppressive capacity, and gene expression of the cytokine-induced suppressive cells showed a population consistent with human MDSC. This study also demonstrated that the induction of CD33+ MDSC from immune competent PBMC by select cytokine combinations occurs in a context of CD33+/CD14+/CD56+ cell expansion and does not require T cells.
A key first step to the present study was the demonstration that human cancer cell lines produced several factors required for the induction of MDSC after co-culture with normal volunteer PBMC. The production of 15 putative MDSC-inducing factors by cancer cell lines was evaluated using previously identified MDSC-inducing and non-inducing human solid tumor cell lines (Figure 1, 13, and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). From these studies it was clear that multiple immune modulatory factors are responsible for MDSC generation, notably IL-6, IL-1β, IDO, COX2, and M-CSF (Figure 1). The notion is consistent with the plethora of reported tumor-derived factors involved in MDSC accumulation and activation in cancer patients and murine tumor models (16–20). This is also supported by the successful generation of suppressive CD33+ cells using six different cytokine mixtures (Figure 2A and B). Immune signature analyses of human cancer biopsies and murine tumor models have shown that the dominant mechanisms of immune evasion and tolerance employed by tumors can vary significantly by tumor histologic type and amongst individual tumors (29). Also, given the significant role of immune suppressor cells in preventing autoimmunity and physiologic tempering of immune responses in trauma and sepsis, it is reasonable that redundant pathways have evolved for the generation of these cells. Thus the identification of several cytokines that were over-expressed by tumor cell lines in association with MDSC induction is not surprising. Several cytokine genes were seen to be consistently down-regulated by MDSC-inducing cancer cell lines, notably GM-CSF and TGFβ (Figure 1). The absence of GM-CSF expression by inducing tumor cell lines is puzzling initially in light of the demonstrated ability of GM-CSF alone or in combination with other cytokines to induce CD33+ suppressor cells (Figures 1 and 2). GM-CSF, in conjunction with IL-4, has long been recognized to promote DC development in vitro (28). As reported previously by others, we noted low levels of GM-CSF greatly enhanced myeloid cell viability in culture and expanded CD33+ cells (24 and Figures 5A and 5B). GM-CSF may derive from stromal cells or PBMC that traffic to the tumor site in response to tumor cell contact. This hypothesis is supported by the strong upregulation of GM-CSF expression by PBMC observed following direct co-culture with tumor cell lines (Figure 5C). Further studies on the source of this cytokine within the tumor microenvironment are warranted by its important role in human MDSC induction demonstrated here. TGFβ was also consistently down-regulated in MDSC-inducing cancer cell lines, consistent with the findings in this study that TGFβ does not promote, and may even antagonize, the generation of suppressive CD33+ cells (Figure 2). The authors acknowledge that gene expression studies may not directly correlate to the protein levels of some tumor derived factors, and thus, there may be additional immune modulatory factors produced by tumors not identified here. In particular, TGFβ detection is challenged by its ubiquitous expression by many cell types, the prevalence of inactive TGFβ protein, and the rapid uptake of active TGFβ by cells (30). Nonetheless, this study of human cancer cell lines (Figure 1) provided the basis for the development of successful in vitro cytokine-based methods for MDSC induction described in this report (Figure 2). Additionally, this study identified IL-6 and GM-CSF as potential therapeutic targets to abrogate tumor-driven accumulation of MDSC.
Using cancer cell line MDSC-induction as a guide, six cytokine combinations were identified for the in vitro induction of potent CD33+ MDSC-like suppressor cells from normal donor PBMC: GM-CSF, GM-CSF + IL-1β, GM-CSF + IL-6, GM-CSF + PGE2, GM-CSF + TNFα, and GM-CSF + VEGF (Figure 2). Of these cytokine mixtures, IL-6 and GM-CSF consistently generated the most suppressive CD33+ cells, as measured by the ability to suppress proliferation of, and IFNγ production by, autologous T cells at different ratios (Figure 2). While hypoxia was not directly examined in this study, IL-6 and TNFα have been shown to increase hypoxia inducible factor-1α (HIF-1α) expression even in the absence of hypoxia (31). Thus, this study is in agreement with previous reports suggesting a role for hypoxia and HIF-1α in IL-6 induced MDSC (32). These results confirm the importance of IL-6 as a therapeutic target for cancer immunotherapy and MDSC-inhibition and provide a means to study specific tumor-generated MDSC subtypes.
Wright-Giemsa staining and flow cytometry analyses of cytokine-induced CD33+ cells and freshly isolated CD33+ cells demonstrate a shift from small, monocytic cells to a homogeneous population of large mononuclear cells with abundant basophilic, granular cytoplasm (Figure 3A). As reported previously for tolerogenic DC (27), bi-nucleated cells were commonly observed in CD33+ cells cultured under different conditions. Importantly, these studies demonstrate that cell morphology alone does not distinguish between CD33+ MDSC and non-suppressive CD33+ cells. This study concurs with previous reports from cancer patients and murine tumor models in reporting a granulocytic MDSC population (by flow cytometry analyses, Figure 3B) which may derive from the expansion and conversion of monocytic precursor cells (33). In characterizing the surface phenotype of cytokine-induced MDSC, this study reports on the expression of several MDSC-associated surface markers not previously examined together (Figure 3B). These results corroborate CD66b and CD11b as markers of human MDSC in conjunction with CD33 positivity (4,5). Importantly, a comparison of cytokine-induced CD33+ cells with and without suppressive function highlights differential expression of HLA-DR and IL-13Rα2 (Figure 3B). These two phenotypic features, HLA-DRlow and IL-13Rα2int, may provide ways to distinguish between suppressive and non-suppressive myeloid cells, in much the same way that FoxP3+ and CD39+ may be used to distinguish CD4+CD25+ Treg from activated T effector cells (34). Transcription factor C/EBPβ has also been suggested as a correlate to suppressive ability in murine MDSC (15), but this study did not find a statistically significant difference in mean C/EBPβ expression between functionally suppressive and non-suppressive cytokine-induced human CD33+ cells (Figure 3C).
CD33+ MDSC induced by GM-CSF and IL-6 mediated potent suppression of autologous T cell proliferation and IFNγ production. Suppressive myeloid cells were also generated by cytokine mixtures of IL-1β, VEGF, TNFα, and PGE2, with GM-CSF added to support cell viability, but the resulting suppressor cells were noticeably less potent (Figure 2A and B). In characterizing the suppressive function of cytokine-induced MDSC, this study evaluated the expression of putative suppression genes and the requirement for cell contact with target cells. As reported previously for murine MDSC, suppression of non-antigen specific autologous T cell responses by human CD33+ MDSC was contact dependent (35). This study did not evaluate antigen-specific suppression and future studies in this area are warranted. In vitro-generated suppressive MDSC exhibited increased expression of iNOS, NOX2, TGFβ, and VEGF (Figure 4A). Interestingly, slight variations in the gene expression profile of, or Treg expansion by, suppressive CD33+ cells occurred with different inducing cytokines (Figure 4A and Figure 2C). IL-6 and GM-CSF induced MDSC appeared to exert strong suppression through up-regulation of ARG-1, iNOS, VEGF, and TGFβ, with minimal contribution from NOX2 expression (Figure 4A). This diversity reflects the variation in suppressive mechanisms reported for MDSC from cancer patients and murine tumor models (16–20) and further suggests the existence of MDSC subpopulations. As noted above, MDSC-inducing tumor cell lines were observed to up-regulate expression of different factors that may be responsible for MDSC generation (Figure 1), which may explain the presence of different MDSC populations amongst tumors. In this study, the generation of suppressive CD33+ MDSC using multiple distinct cytokine mixtures also supports the existence of diverse MDSC subpopulations (Figure 2).
The MDSC Suppression Assay described in this study tested the ability of CD33+ cells to inhibit T cell proliferation and IFNγ production in the presence of IL-2 (100IU/mL) and CD3/CD28 stimulation. This method is similar to suppression assays frequently used to evaluate the function of Treg cells (34). It is important to note that many investigators use antigen presenting cell populations as feeder cells in Treg suppression assays. Just as Treg may be expanded from responder CD3+ T cell populations and contribute to suppression in MDSC Suppression Assays (Figure 2C), Treg induction of MDSC from antigen presenting cells present in Treg Suppression Assays may contribute to inhibition of T cells responses. As demonstrated in this study, CD33+ cells may be induced from normal donor cells by select cytokines and mediate potent suppression of autologous T cells (Figure 2A and B).
Previously, we reported the in vitro generation of human CD33+ MDSC from PBMC by direct co-culture with select human solid tumor cell lines (13 and manuscript submitted for publication: Lechner et al. Induction and functional characterization of human myeloid suppressor cells by PBMC and tumor cell line co-culture). In this study, cytokines IL-6 and GM-CSF, and secondarily GM-CSF in combination with IL-1β, PGE2, TNFα, or VEGF, are shown to be sufficient for the induction of functionally suppressive CD33+ cells from healthy donor PBMC. MDSC are not typically present in healthy individuals (2), and the ability to generate these suppressor cells in vitro from non-suppressive populations represents an important discovery. Suppressor cells induced by IL-6 and GM-CSF should provide an important model for studying tumor-associated human MDSC and may enable the generation of tolerogenic myeloid cells for the adoptive immunotherapy of autoimmune disease. The cytokine-induction combinations identified here allow derivation of human MDSC from a starting population of common, healthy PBMC. This is in contrast to reports of in vitro generation of murine MDSC populations from embryonic, splenic, or bone marrow derived cells (36) – cell fractions that are less readily available from human donors. While many similarities exist between murine and human immune systems, studies in Treg highlight the need to be cognizant of interspecies differences in surface markers and biology of immune suppressor cells (34,37). Thus, the discovery of human-specific MDSC induction methods is a valuable tool for the further study of these suppressor cells. Based upon these results, future investigations are warranted to study the potential of IL-6 and GM-CSF as therapeutic targets for the inhibition of MDSC induction in cancer patients.
ACKNOWLEDGEMENTS
The authors thank Dixon Gray for flow cytometry support and Clive Taylor for microscopy support. The authors also acknowledge Carolina Megiel, Brigid Bingham, and Nathan Feng for assistance with screening of tumor cell lines by RT-PCR.
Financial Support: National Institutes of Health Training Grant award 3T32GM067587-07S1 and Cancer Therapeutics Laboratories, Inc. (Los Angeles, CA).
Abbreviations used in this paper
- ARG-1
arginase-1
- C/EBP
CCAAT/enhancer-binding proteins
- CFSE
5- (and 6-) carboxyfluorescein diacetate succinimidyl ester
- c-kit L
c-kit ligand or stem cell factor
- COX2
cyclo-oxygenase 2
- FLT3L
fms-related tyrosine kinase 3 ligand
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GM-CSF
granulocyte-macrophage colony stimulating factor
- HIF-1α
hypoxia inducible factor-1 alpha
- iDC
immature dendritic cells
- iNOS
indoleamine 2,3-dioxygenase; inducible nitric oxide synthase
- IFNγ
interferon gamma
- IL
interleukin
- M-CSF
macrophage colony stimulating factor
- MDSC
myeloid-derived suppressor cells
- NOX2
NADPH oxidase
- PBMC
peripheral blood mononuclear cells
- PGE
prostaglandin E2
- Treg
regulatory T cells
- TGFβ
transforming growth factor beta
- TNFα
tumor necrosis factor alpha
- VEGF
vascular endothelial growth factor-a
Footnotes
Authorship: M.G.L. developed the methods, screened the tumor cell lines, performed morphology, phenotype, suppression, and gene expression studies, and wrote the paper. D.J.L. assisted with qRT-PCR, flow cytometry, and morphology studies, edited the manuscript, and designed the figures for the paper. A.L.E. supervised the studies, provided morphologic and phenotypic analyses, and assisted with data interpretation. All authors reviewed the paper.
REFERENCES
- 1.Stewart TJ, Abrams SI. How tumours escape mass destruction. Oncogene. 2008;27:5894–5903. doi: 10.1038/onc.2008.268. [DOI] [PubMed] [Google Scholar]
- 2.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Ugel S, Delpozzo F, Desantis G, Papalini F, Simonato F, Sonda N, Zilio S, Bronte V. Therapeutic targeting of myeloid-derived suppressor cells. Curr Opin Pharmacol. 2009;9:470–481. doi: 10.1016/j.coph.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munera V, Popovic PJ, Bryk J, Pribis J, Caba D, Matta BM, Zenati M, Ochoa JB. Stat 6-dependent induction of myeloid derived suppressor cells after physical injury regulates nitric oxide response to endotoxin. Ann Surg. 2010;251:120–126. doi: 10.1097/SLA.0b013e3181bfda1c. [DOI] [PubMed] [Google Scholar]
- 7.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilkovitch D, Lopez DM. The liver is a site for tumor-induced myeloid-derived suppressor cell accumulation and immunosuppression. Cancer Res. 2009;69:5514–5521. doi: 10.1158/0008-5472.CAN-08-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusmartsev S, Gabrilovich DI. Effect of tumor-derived cytokines and growth factors on differentiation and immune suppressive features of myeloid cells in cancer. Cancer Metastasis Rev. 2006;25:323–331. doi: 10.1007/s10555-006-9002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, Cheng Z, Shah SV, Wang GJ, Zhang L, Grizzle WE, Mobley J, Zhang HG. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumitriu IE, Dunbar DR, Howie SE, Sethi T, Gregory CD. Human dendritic cells produce TGF-beta 1 under the influence of lung carcinoma cells and prime the differentiation of CD4+CD25+Foxp3+ regulatory T cells. J Immunol. 2009;182:2795–2807. doi: 10.4049/jimmunol.0712671. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues JC, Gonzalez GC, Zhang L, Ibrahim G, Kelly JJ, Gustafson MP, Lin Y, Dietz AB, Forsyth PA, Yong VW, Parney IF. Normal human monocytes exposed to glioma cells acquire myeloid-derived suppressor cell-like properties. Neuro Oncol. 2010;12:351–365. doi: 10.1093/neuonc/nop023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lechner MG, Megiel C, Liebertz DJ, Epstein AL. Myeloid-derived suppressor cell induction and mechanisms of suppression in 100 human solid tumor cell lines. Keystone Symposium Molecular and Cellular Biology and Immune Escape in Cancer. 2010 abstract: 182. [Google Scholar]
- 14.Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/EBP beta regulatory domains in specifying MCP-1 and IL-6 transcription. Mol Immunol. 2007;44:1384–1392. doi: 10.1016/j.molimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Bronte V. The transcription factor C/EBPβ is a master regulator of tumor-induced, myeloid-dependent immune suppression. Keystone Symposium Molecular and Cellular Biology and Immune Escape in Cancer. 2010 abstract: 144. [Google Scholar]
- 16.Bak SP, Alonso A, Turk MJ, Berwin B. Murine ovarian cancer vascular leukocytes require arginase-1 activity for T cell suppression. Mol Immunol. 2008;46:258–268. doi: 10.1016/j.molimm.2008.08.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, Padhya T, McCaffrey TV, McCaffrey JC, Gabrilovich DI. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donkor MK, Lahue E, Hoke TA, Shafer LR, Coskun U, Solheim JC, Gulen D, Bishay J, Talmadge JE. Mammary tumor heterogeneity in the expansion of myeloid-derived suppressor cells. Int Immunopharmacol. 2009;9:937–948. doi: 10.1016/j.intimp.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010;70:68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serafini P, Mgebroff S, Noonan K, Borrello I. Myeloid-derived suppressor cells promote cross-tolerance in B cell lymphoma by expanding regulatory T cells. Cancer Res. 2008;68:5439–5449. doi: 10.1158/0008-5472.CAN-07-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Zeng B, Zhang Z, Zhang Y, Yang R. B7-H1 on myeloid-derived suppressor cells in immune suppression by a mouse model of ovarian cancer. Clin Immunol. 2008;129:471–481. doi: 10.1016/j.clim.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 22.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaraj S, Gabrilovich DI. Tumor escape mechanism governed by myeloid-derived suppressor cells. Cancer Res. 2008;68:2561–2563. doi: 10.1158/0008-5472.CAN-07-6229. [DOI] [PubMed] [Google Scholar]
- 24.Ko JS, Zea AH, Rini BI, Ireland JL, Elson P, Cohen P, Golshayan A, Rayman PA, Wood L, Garcia J, Dreicer R, Bukowski R, Finke JH. Sunitinib mediates reversal of myeloid-derived suppressor cell accumulation in renal cell carcinoma patients. Clin Cancer Res. 2009;15:2148–2157. doi: 10.1158/1078-0432.CCR-08-1332. [DOI] [PubMed] [Google Scholar]
- 25.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35(Database issue):D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha P, Clements VK, Fulton AM, Ostrand-Rosenberg S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007;67:4507–4513. doi: 10.1158/0008-5472.CAN-06-4174. [DOI] [PubMed] [Google Scholar]
- 27.Dong R, Adams S, Bouma G, Eddouda A, Chana P, Moulding D, Duncan A, Anderson J. Newly identified multinuclear cells in cultured human dendritic cells possess the phenotype of regulatory DC. Keystone Symposium Molecular and Cellular Biology and Immune Escape in Cancer. 2010 abstract: 171. [Google Scholar]
- 28.Morse MA, Zhou LJ, Tedder TF, Lyerly HK, Smith C. Generation of dendritic cells in vitro from peripheral blood mononuclear cells with granulocyte-macrophage-colony-stimulating factor, interleukin-4, and tumor necrosis factor-alpha for use in cancer immunotherapy. Ann Surg. 1997;226:6–16. doi: 10.1097/00000658-199707000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadun RE, Sachsman SM, Chen X, Christenson KW, Morris WZ, Hu P, Epstein AL. Immune signatures of murine and human cancers reveal unique mechanisms of tumor escape and new targets for cancer immunotherapy. Clin Cancer Res. 2007;13:4016–4025. doi: 10.1158/1078-0432.CCR-07-0016. [DOI] [PubMed] [Google Scholar]
- 30.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 31.Garimella PM, Wellman TL, Wong C, Lounsbury KM. Cytokine regulation of HIF-1 in ovarian cancer. Proc Amer Assoc Cancer Res. 2006;2026:1859. [Google Scholar]
- 32.Gabrilovich DI. Regulation of myeloid-derived suppressor cells in cancer. Keystone Symposium Molecular and Cellular Biology and Immune Escape in Cancer. 2010 abstract: 142. [Google Scholar]
- 33.Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, De Baetselier P, Van Ginderachter JA. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 34.Colombo MP, Piconese S. Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nature Reviews Cancer. 2007;7 doi: 10.1038/nrc2250. 880-877. [DOI] [PubMed] [Google Scholar]
- 35.Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z, French DL, Ma G, Eisenstein S, Chen Y, Divino CM, Keller G, Chen SH, Pan PY. Development and Function of Myeloid-Derived Suppressor Cells Generated from Mouse Embryonic and Hematopoietic Stem Cells. Stem Cells. 2010;28:620–632. doi: 10.1002/stem.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie M. Immunology uncaged. Science. 2010;327:1573. doi: 10.1126/science.327.5973.1573. [DOI] [PubMed] [Google Scholar]