Abstract
In mice, the major urinary proteins (MUP) play a key role in pheromonal communication by binding and transporting semiochemicals. MUP-IV is the only isoform known to be expressed in the vomeronasal mucosa. In comparison with the MUP isoforms that are abundantly excreted in the urine, MUP-IV is highly specific for the male mouse pheromone 2-sec-butyl-4,5-dihydrothiazole (SBT). To examine the structural basis of this ligand preference, we determined the X-ray crystal structure of MUP-IV bound to three mouse pheromones: SBT, 2,5-dimethylpyrazine, and 2-heptanone. We also obtained the structure of MUP-IV with 2-ethylhexanol bound in the cavity. These four structures show that relative to the major excreted MUP isoforms, three amino acid substitutions within the binding calyx impact ligand coordination. The F103 for A along with F54 for L result in a smaller cavity, potentially creating a more closely packed environment for the ligand. The E118 for G substitution introduces a charged group into a hydrophobic environment. The sidechain of E118 is observed to hydrogen bond to polar groups on all four ligands with nearly the same geometry as seen for the water-mediated hydrogen bond network in the MUP-I and MUP-II crystal structures. These differences in cavity size and interactions between the protein and ligand are likely to contribute to the observed specificity of MUP-IV.
Keywords: pheromone, X-ray crystallography, major urinary protein, lipocalin, structure
Introduction
The major urinary proteins of the mouse (MUP) sequester, transport, and slowly release pheromones into the environment, playing a key role in social signaling.1–4 The mouse genome encodes for ∼ 38 MUPs5,6 which have been classified into four groups based on nucleotide sequence similarity and tissue-specific expression patterns. Group 1 MUPs are expressed mainly in the liver and kidney and excreted into the urine at high levels.7,8 Group 2 comprise pseudogenes and group 3 isoforms are glycosylated.5,8 The group 4 MUPs are expressed in the lachrymal and salivary glands.5,9 Historically, focus has been on naturally abundant, small, volatile ligands of MUPs, such as 2-sec-butyl-4,5-dihydrothiazole (SBT), 2,3-dehydro-exo-brevicomin (DHB), 6-hydroxy-6-methylheptan-3-one (HMH), and 2,5-dimethylpyrazine (DMP), which have been shown to influence aggression and puberty in mice.10–13 It has also been shown that expression patterns of the 8–14 excreted group 1 isoforms are similar among related mice,5,8,14 which suggests that the MUP protein profile might also play a role in social communication.15–18 The precise roles of the group 4 isoforms are currently unclear. In rodents, pheromones are detected both by the main olfactory bulb and the vomeronasal organ (VNO).1,19 It has been proposed that the nasal MUP isoforms may function in facilitated transport to olfactory and vomeronasal receptors.20 MUP isoform IV (MUP-IV), belongs to group 49 and is the only MUP known to be expressed in the VNO.20 Interestingly, SBT binds to MUP-IV about 20 times more tightly than to any of the five most abundant group 1 isoforms.20
The first X-ray crystal structure of MUP21 confirmed that the mouse MUP belong to the lipocalin superfamily.22 X-ray and NMR structures of MUP-I and MUP-II have been determined in the presence of SBT23,24 as well as other ligands.24–32 However, until now, there have been no published structures of a group 4 isoform. We have determined the X-ray crystal structures of MUP-IV in complex with three mouse pheromones to examine the structural basis of ligand binding specificity.
Results
Overall structural similarity of MUPs
Four MUP-IV crystal structures were solved in the orthorhombic space group using the Protein Data Bank coordinates for MUP-I (PDB code 1i06, Ref. 24) as the search model for molecular replacement. Each of the MUP-IV crystal structures show continuous electron density for residues 10–161; residues 1–9 and 162.† were not observed in the electron density maps. As expected, MUP-IV adopts the canonical lipocalin fold, consisting of an eight-stranded beta barrel enclosing a hydrophobic cavity.22 Overall, the four MUP-IV structures reported here exhibit mainchain root mean square deviations (RMSD) of less than 0.2 Å to each other. MUP-IV is also highly similar to MUP-I24 and MUP-II23 with an overall 1 Å rmsd for all backbone atoms for the pairwise comparisons. The largest differences between these structures occur at the N- and C-termini and at the loops near the single disulfide bond, reaching a maximum of ∼ 5 Å for this region (Fig. 1). This overall similarity in tertiary structure was anticipated, as MUP-IV shares ∼ 75% sequence identity with MUP-I and MUP-II (Fig. 2). However, inside the occluded binding cavity, the sequence identity to MUP-I and MUP-II drops to 65% where the most structurally significant cavity substitutions are an E118 for G, an F54 for L, and an F103 for A.
Figure 1.
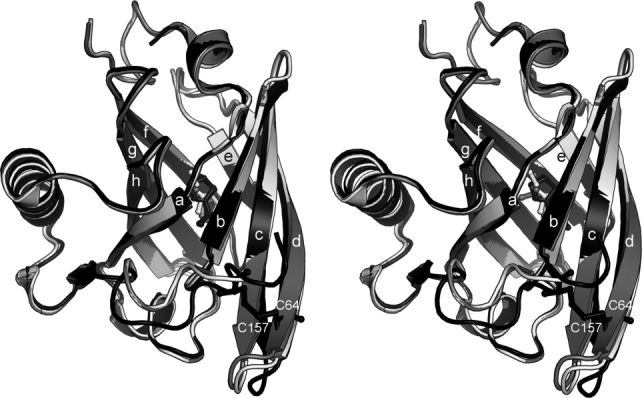
Stereo image showing structure overlay of the SBT complexes of MUP-I (light grey, 1i06, ref. 24), MUP-II (dark grey, 1jv4, ref. 23), and MUP-IV (black). Protein rendered as ribbon diagrams and SBT in ball-and-stick. Strands labeled according to Ref. 24.
Figure 2.
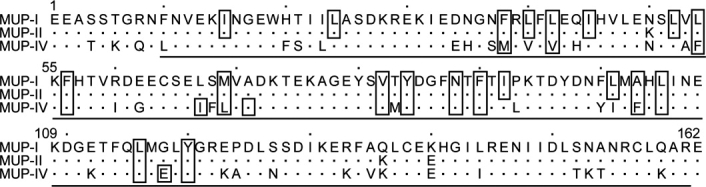
MUP sequence alignment. Dots represent residues that are identical to MUP-I. Boxed residues line the ligand binding pocket, as identified by CASTp.60 Underlined sequence denotes crystallographically observed residues in MUP-IV. Sequence identies: 98% for MUP-II to MUP-I; 74% for MUP-IV to MUP-I; 75% for MUP-IV to MUP-II.
Ligand–protein interactions in MUP-IV
We solved three high-resolution pheromone-bound structures of MUP-IV from crystals soaked with synthetically derived 2-heptanone (HT), DMP, and SBT (Fig. 3). The effects of these pheromones are known; HT extends estrus,33 DMP inhibits puberty,12 and SBT both promotes intermale aggression as well as inducing estrus synchrony.10,11 We also solved the structure for a crystal which was not soaked with pheromone, but which contains a serendipitous ligand. This is consistent with the findings of other groups, where significant difference electron density peaks are observed in the MUP binding pocket even in the absence of exogenously introduced ligand.21,24,26 Data collection and refinement statistics are presented in Table I.
Figure 3.
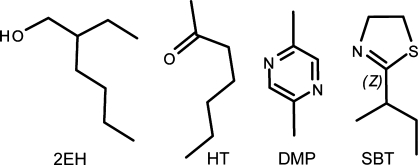
Chemical structures of 2EH (2-ethylhexanol) and the three MUP pheromones soaked into the MUP-IV crystals: DMP (2,5-dimethylpyrazine), HT (2-heptanone) and SBT (2-sec-butyl-4,5-dihydrothiazole).
Table I.
Data Collection and Refinement Statistics
| Dataset | 2EH | HT | DMP | SBT |
|---|---|---|---|---|
| PDB code | 3kfh | 3kfg | 3kfi | 3kff |
| Data collection | ||||
| Wavelength (Å) | 0.90 | 0.90 | 1.54 | 0.90 |
| Resolution (Å) | 25–1.02 | 25–1.43 | 50–1.42 | 25–0.96 |
| Highest Shell (Å) | 1.06–1.02 | 1.48–1.43 | 1.47–1.42 | 0.99–0.96 |
| Cell edges (P212121) (Å) | 47.8, 54.1, 61.4 | 47.7, 53.8, 61.0 | 47.8, 53.4, 61.5 | 47.6, 53.7, 61.2 |
| Mosaicity | 0.49 | 1.20 | 1.20 | 0.56 |
| Total observations | 476,188 | 100,588 | 85,540 | 439,458 |
| Unique observations | 75,259 | 28,552 | 29,102 | 92,249 |
| Redundancy | 6.4 | 3.5 | 2.9 | 4.8 |
| Completeness (%) | 92.0 (47.3) | 96.0 (91.9) | 95.1 (83.2) | 95.7 (73.0) |
| Rmerge | 0.070 (0.303) | 0.099 (0.299) | 0.054 (0.277) | 0.068 (0.302) |
| I/σ | 19.9 (2.2) | 10.4 (2.4) | 16.3 (2.4) | 16.9 (2.4) |
| Wilson B | 8.2 | 16.6 | 20.6 | 5.9 |
| Refinement | ||||
| Program | Shelxl | Refmac5 | Refmac5 | Shelxl |
| Rwork (%) | 13.0 | 16.3 | 14.8 | 13.6 |
| Rfree (%) | 16.2 | 20.3 | 18.1 | 16.4 |
| RMSD bonds (Å) | 0.016 | 0.019 | 0.015 | 0.014 |
| RMSD angles (Å) | 2.30 | 1.80 | 1.51 | 2.10 |
| Ramachandran most favored (%) | 91.5 | 91.5 | 92.3 | 90.8 |
| Allowed (%) | 7.7 | 7.7 | 7.0 | 8.5 |
| No. protein atoms | 1354 | 1312 | 1275 | 1348 |
| No. alt. sidechain atoms | 94 | 58 | 27 | 94 |
| No. alt. mainchain atoms | 12 | 6 | 0 | 6 |
| No. H2O atoms | 270 | 252 | 232 | 344 |
| No. Cl− atoms | 1 | 2 | 2 | 1 |
| No. Ligand atoms | 9 | 17 | 8 | 18 |
| Bav protein (Å2) | 14.7 | 20.1 | 24.8 | 11.3 |
| Bav Solvent (Å2) | 28.2 | 35.6 | 38.2 | 24.8 |
| Bav Ligand (Å2) | 22.0 | 21.0 | 22.8 | 17.6 |
| Modeled ligand | 2EH | HT and HEH | DMP | R-SBT and S-SBT |
2EH, MUP-IV with bound 2-ethylhexanol; HT, MUP-IV soaked with 2-heptanone; DMP, MUP-IV with bound 2,5-dimethylpyrazine; SBT, MUP-IV with bound 2-sec-butyl-4,5-dihydrothiazole. Numbers in parentheses are values for the highest resolution shell.
As previously observed, Y97 occurs near a hairpin turn, falling into a disallowed region of the Ramachandran plot.24
Initial electron density maps for the serendipitous ligand structure are shown in Fig. 4(A). At the 1.0 Å resolution of this data set, the density is well modeled by 2-ethylhexanol (2EH). The origin and binding properties of this endogenous ligand are unknown; however, 2EH is a common flavorant in foodstuffs and is also a byproduct of plastic degradation.34–36 The hydroxyl group of 2EH is located within hydrogen bonding distance to both E118 OE2 and the carbonyl of M38 [Figure 4(E)]. Potential packing contacts for all four ligands are listed in Supporting Information Table I.
Figure 4.
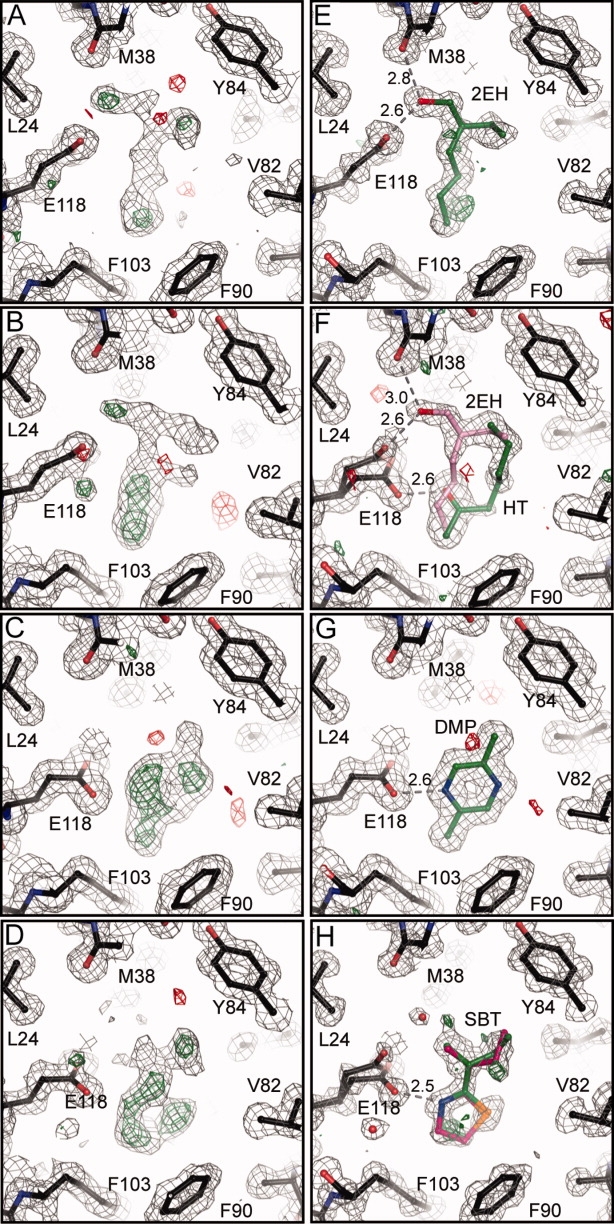
Electron density maps for MUP-IV. A-D: Sigma-A weighted maps after molecular replacement, model rebuilding, and five cycles of restrained positional and isotropic B-factor refinement to 1.5 Å. RW ∼ 28, RF ∼ 30, FOM ∼ 0.77 for all four structures. E-H: Final sigma-A weighted m2Fo-Fc maps. Dotted lines show potential hydrogen bonding contacts. Maps are contoured at 1.0σ and 3.0σ for m2Fo-Fc and mFo-Fc, respectively.
As can be seen in Fig. 4(B), in the 1.4 Å structure solved from the crystal soaked with HT, the electron density for the ligand was somewhat ambiguous. Modeling multiple conformations of HT resulted in difference electron density peaks larger than 3σ around the ligands (Supporting Information Figure 1). Re-examination of the initial electron density maps in comparison with those from the 2EH structure suggested that 2EH might also be present in this structure at partial occupancy. Including both HT and 2EH, each at half occupancy, satisfactorily modeled the observed density [Figure 4(F)]. In this structure, E118 is observed in two rotamer positions, one similar to that observed in the 2EH structure (A), and the second with the plane of the carboxyl rotated about 65° (B). The half-occupancy 2EH is bound in the same orientation and makes the same contacts as in the 2EH structure. The HT carbonyl is positioned at an appropriate distance from the OE2 atom of E118 and the OH atom of Y120 to accepted hydrogen bonds.
In contrast to the HT data, the electron density for the ligand was quite clear in the 1.4 Å DMP-bound structure [Fig. 4(C)]. Here, DMP is well-modeled at full occupancy in a single conformation, with the N4 atom of the pyrazine ring at hydrogen bonding distance to the OE2 atom of E118 [Fig. 4(G)]. In this structure, the sidechain of E118 is in the (B) position. The plane of the DMP ring is perpendicular to and in van der Waals contact with the sidechains of F56 and L105; the methyl groups potentially make packing contact with Y84 and F103 at the top and bottom of the cavity, respectively.
We also solved the structure of MUP-IV complexed with SBT to 1.0 Å resolution. SBT exists as a racemic mixture and it has been shown that each enantiomer binds to MUP-I with equal affinity.37 The electron density for SBT bound to MUP-IV is consistent with at least two positions for the alkyl chain and the peak heights are consistent with the placement of the dihydrothiazole ring nitrogen near E118 and with the sulfur atom oriented toward V82 [Fig. 4(D)]. Based on the electron density peak heights and the local interactions, we modeled both the R and S enantiomers into the density, each at partial occupancy [Fig. 4(H)]. However, a persistent peak at > 5σ of the Fo-Fc electron density maps as well as clear density for an alternative rotamer (A) of E118 led us to model a water molecule at low occupancy near the C9 of SBT. Alternately, we could envision a third orientation of SBT, either flipped 180° about the horizontal or about the vertical axis in Fig. 4(H). However, the resulting maps for each of these models had difference peaks greater than 4σ surrounding the ligand (Supporting Information Figure 1). Ultimately, the observed electron density was sufficiently modeled with two enantiomers of SBT, each at occupancy of 0.4 and a corresponding occupancy of 0.2 for the water near the C9 atom and for the (A) rotamer of E118. In each enantiomer, atom N3 is within hydrogen bonding distance to the OE2 of E118 in the (B) rotamer position. Similar to what we observe for DMP, the dihydrothiazole ring and C6 are perpendicular to and in potential van der Waals contact with the sidechains of F56 and L105. The branched alkyl chain of SBT is in close contact with V40, L116, L69, and Y84.
Discussion
Ligand coordination in MUP-IV
We were somewhat surprised to find that SBT binds to MUP-IV in an inverted orientation relative to MUP-I (Fig. 5). In the first X-ray structure of MUP-I, the endogenous ligand was modeled as SBT in a similar inverted orientation.21 It has since been the consensus, based on higher resolution MUP-I and MUP-II crystal structures23,24 as well as NMR data37 that SBT binds to MUP-I and MUP-II in the orientation shown in Figure 5(C), with the branched sec-butyl group oriented toward the base of the cavity. In contrast, alignment of protein backbone atoms reveals several differences in SBT binding between MUP-IV and MUP-I (or MUP-II). In MUP-IV, the branched sec-butyl group is oriented toward the top of the cavity and the two ordered water molecules that form a hydrogen bond network in MUP-I24 and MUP-II23 are absent. Instead, the carboxyl moiety of E118 (G118 in MUP-I and MUP-II) occupies this space, within hydrogen bonding distance to the N3 atom of SBT [Fig. 5(B)].‡ The sidechain of Y120 is also rotated ∼ 90° relative to its position in MUP-I and MUP-II, such that the OE1 atom of E118 (in both rotamers) is within hydrogen bonding distance of the Y120 hydroxyl.
Figure 5.
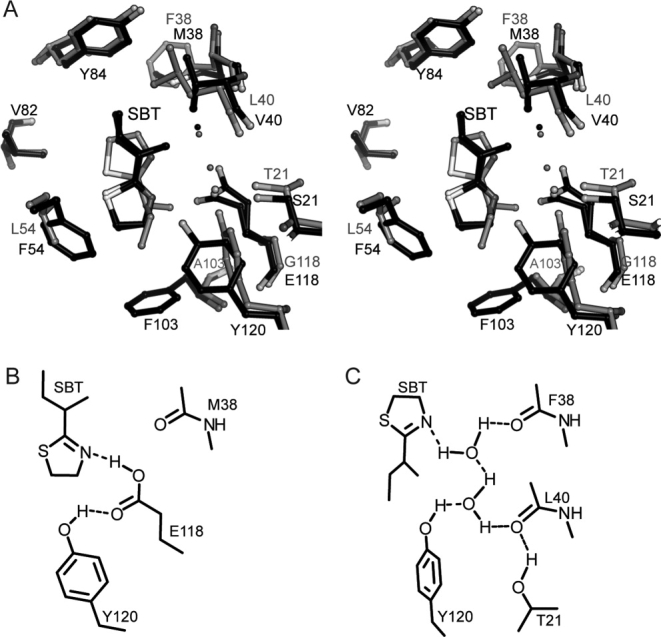
Details of SBT coordination in MUP-IV versus MUP-I. A. Stereoview of structure overlay, protein backbone atoms aligned. The MUP-IV complex is shown in black and the MUP-I complex (1i06, Ref. 24) is shown in gray ball-and-stick representations. B. Hydrogen bonding diagram for the SBT-MUP-IV complex. C. Hydrogen bonding diagram for SBT-MUP-I complex (1i06) adapted with permission from Figure 4, Ref. 24. For clarity, views are rotated 180° about the vertical, relative to Figure 4.
As we observe a low occupancy water molecule near C9 of SBT in almost the same location as one of the ordered cavity waters in MUP-I and MUP-II [Fig. 5(A)], we cannot rule out that a small fraction of SBT may be bound in the upright orientation (see Supporting Information Figure 1H). However, in this position, the distance between the thiazole ring N3 and the E118 carboxyl oxygens is greater than 3.5 Å. Potentially, the low-occupancy water could provide a water-mediated hydrogen bond with E118, likely resulting in weaker binding for this particular orientation. Consequently, the hydrogen bond between the thiazole ring nitrogen and the E118 carboxyl is the most likely explanation for the inverted orientation of SBT observed in our MUP-IV structure.
The water-mediated hydrogen bond network was also observed in the MUP-I structure with HMH (PDB code 1i05).24 Thus, it is of interest to examine the potential differences in the interactions with HMH that can be provided by the two proteins. Although we do not have the structure of MUP-IV with HMH, the interaction of HMH with MUP-I does share features with 2EH and HT binding to MUP-IV (Fig. 6). Manual modeling of HMH onto 2EH/HT in MUP-IV suggests that with a small rotation, the MUP-IV cavity could accommodate the open conformation of HMH with hydrogen bonding interactions between the carbonyl and hydroxyl groups of HMH and the sidechains of E118 and Y120, respectively [Fig. 6(E)]. Inverting the orientation of HMH increases potential steric clashes with F90 and F103 at the base of the MUP-IV cavity (not shown).
Figure 6.
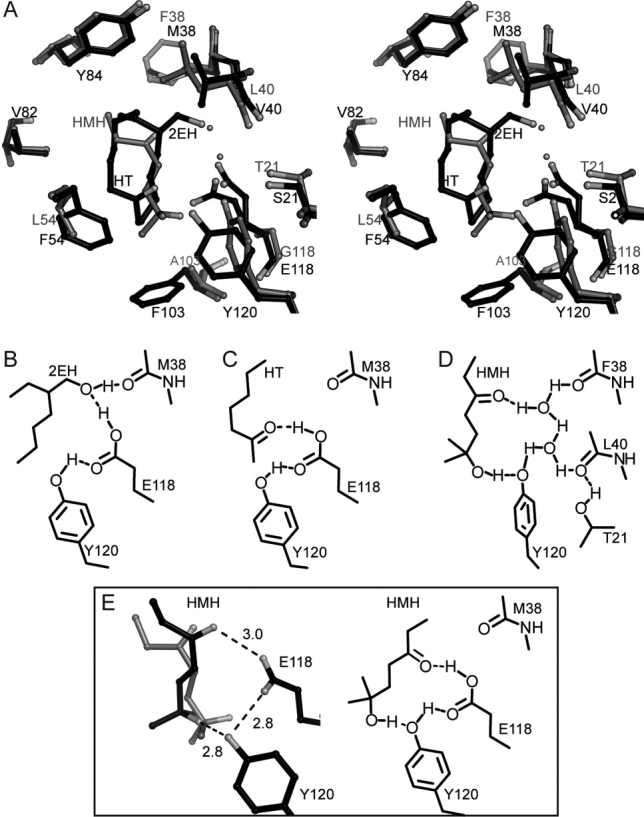
2EH and HT coordination in MUP-IV versus HMH coordination in MUP-I. A. Stereoview of structure overlay, protein backbone atoms aligned. The MUP-IV-HT/HEH complex is shown in black and the MUP-I-HMH complex (1i05, Ref. 24) is shown in gray ball-and-stick representations. B. Hydrogen bonding diagram for 2EH-bound MUP-IV. C. Hydrogen bonding diagram for HT-bound MUP-IV. D. Hydrogen bonding pattern in the the HMH-MUP-I complex (1i05) adapted with permission from Figure 4, Ref. 24. E. Potential fit of HMH in the MUP-IV cavity. Left: HMH was manually rotated from its initial position in MUP-I (gray) to a position which minimizes van der Waals contacts in MUP-IV (black). Right: Potential hydrogen bonding interactions for HMH modeled into the MUP-IV cavity.
Interestingly, in the structures of two pyrazine derivatives bound to MUP-I determined by Bingham et al., the water mediated hydrogen bonding network was not observed.26 An alignment of these MUP-I complexes with our DMP-bound MUP-IV structure shows that in MUP-I, the pyrazine ring is shifted into the space occupied by E118 in MUP-IV [Fig. 7(A)]. This is likely because of the observed hydrogen bonding interaction between the ligand and Y120 [Fig. 7(C)].26 Hence, it appears that the binding mode of pyrazine derivatives also differs between MUP-IV and MUP-I. However, Y120 does play a role in ligand coordination in each of these structures: participating in either a direct or a water-mediated hydrogen bond with the ligand in MUP-I or stabilizing the orientation of E118 in MUP-IV. A general role for Y120 in MUP binding to a variety of ligands has also been noted recently by Pertinhez, et al.32
Figure 7.
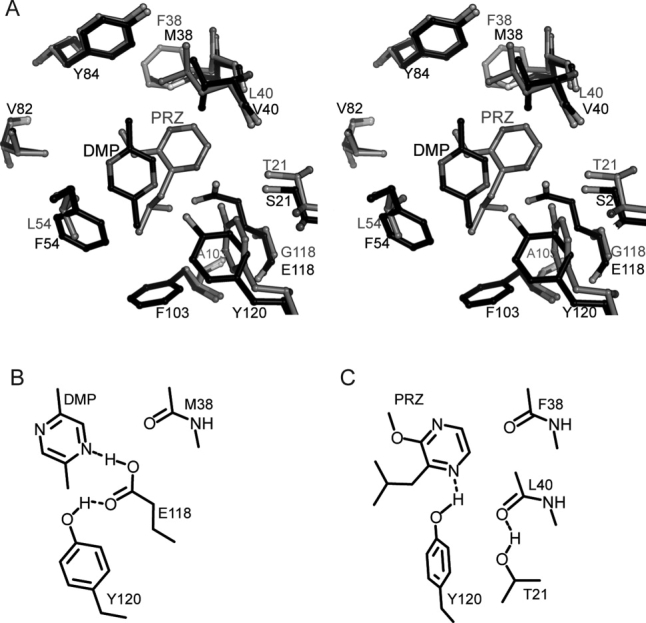
Pyrazine coordination in MUP-IV versus MUP-I. A. Stereoview of structure overlay, protein backbone atoms aligned. MUP-IV complex shown in black and MUP-I complex (1qy1, ref 26) shown in gray ball-and-stick representations. B. Hydrogen bonding diagram for DMP-MUP-IV complex. C. Hydrogen bonding diagram for PRZ-MUP-I complex (1qy1).
The MUP-IV cavity
As observed for other MUP structures,21,23–32,37 each of our MUP-IV ligands reside closer to one side of the cavity, making packing contacts with 9–12 of the 16 residues that line the MUP-IV pocket (Fig. 2, Supporting Information Table I). However, the cavity volume in MUP-IV is ∼ 25% smaller than that of MUP-I or MUP-II (Supporting Information Table II). This is primarily because of three interior substitutions: E118 for G, F54 for L, and F103 for A (Fig. 8). In MUP-I and MUP-II, the base of the cavity is formed by I15, in packing contact with L42, I92, and L101. In MUP-IV, the base of the cavity is formed by F103, F54, and F90, shortening the longest dimension by about 4 Å. Although differently shaped in the regions furthest from the ligand (Fig. 8), all three MUP cavities are roughly the same dimension in the plane perpendicular to the longest dimension, through the center of the ligand (Supporting Information Table II). Increased contact area with ligand in the smaller MUP-IV cavity is likely to contribute to the increased affinity of MUP-IV for SBT when compared with MUP-I and MUP-II.20 On the other hand, the smaller cavity may be less able to accommodate larger ligands such as HMH, possibly contributing to the decreased relative affinity for this ligand.20
Figure 8.
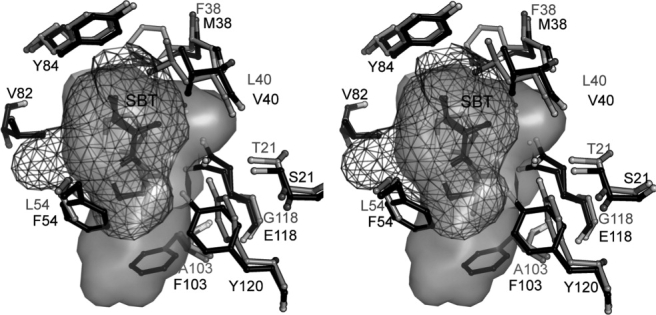
Stereoview of the molecular surface inside the MUP cavity. MUP-IV shown in black ball-and stick representation with a mesh rendering of the cavity surface. MUP-I shown in grey ball-and-stick with the cavity displayed as a solid surface (1i06, Ref. 24).
Implications for thermodynamics of binding
Numerous studies have shown a large favorable enthalpic and small unfavorable entropic contribution to MUP-ligand binding even though the MUP cavity is largely hydrophobic.20,26,27,29,31,40,41 When compared with MUP-I, SBT binding to MUP-IV is characterized by a more favorable enthalpy (by 5 kcal/mol) and less favorable entropy (by 21 cal/mol/K).20 From the known difference in affinities of MUP-I and MUP-IV for SBT,20 we can calculate a ΔΔG of −1.9 kcal/mol in favor of binding to MUP-IV, which is within the range expected for hydrogen bonding interactions. However, there are other factors that are likely to impact ligand binding to MUP-IV when compared with MUP-I. For example, ligand stabilization by hydrogen bonding occurs in both cavities, though based on the polarizability of carboxylate versus water, we would expect the substitution of a direct hydrogen bond with the E118 sidechain in MUP-IV to be more favorable. The smaller cavity is likely to provide a more closely packed environment for ligands, which might contribute favorably to binding energy. Indeed, SBT is less solvent accessible in MUP-IV versus MUP-I or MUP-II (Supporting Information Table II). However, a more tightly packed environment could also result in decreased conformational mobility of ligand and protein, contributing unfavorably to the entropy term. Simultaneously, based on the cavity volume difference of about 100 Å3, it is also reasonable to think that less water may be displaced upon ligand binding to MUP-IV, further contributing to the larger entropy penalty. One caveat here is that the differences in cavity shape and the inverted orientation of SBT make it difficult to draw direct conclusions from comparison of MUP-IV and MUP-I, as even the conserved residues contact different ligand atoms. In addition, if a more closely packed environment underlies the greater entropic cost of binding, we would expect to see a greater entropic penalty for other ligands bound to MUP-IV, not just SBT. However, this is not the case. In fact, the available thermodynamic data show that relative to MUP-I, there is a smaller entropic penalty for binding of HMH and DHB.20 Thus, elucidation of the contributions of cavity substitutions to the thermodynamics of ligand binding to MUP-IV will require additional studies.
Summary
MUPs are members of the lipocalin superfamily, a diverse group of proteins with a common fold and low-sequence identity.42,43 Lipocalins are currently of medical interest because of the allergic nature of excreted lipocalins from some species, including mouse.44,45 Lipocalins are also being engineered to target specific molecules for both medical and biotech applications.46,47 Sequence identity amongst lipocalins is very low, approaching 20%, making it difficult to identify lipocalins based on sequence alone.42,48,49 With respect to the key protein–ligand interactions discussed in this article, inspection of lipocalin sequences in the PFAM database (PF00061)50 shows that at least 16 amino acids occur at the position equivalent to 118 (in MUP-IV), with glutamate showing up most frequently (Supporting Information Figure 2). Conservation is also low at position 120 (Figure S2). Low-sequence conservation of cavity residues simply reflects the diverse nature of ligands accomodated by a common architecture.22,32,42,43,48
In summary, our X-ray crystal structures of MUP-IV elucidate the role of E118 in MUP-IV and reinforce the role of Y120 in ligand binding within the MUP family. In each structure, we observe the carboxyl of E118 (G118 in MUP-I and MUP-II) to form a bifurcated hydrogen bond with the hydroxyl of Y120 and each of the four ligands presented here, replacing the ordered water network observed in MUP-I and MUP-II. Conversely, the hydrogen bond between the E118 and Y120 sidechains likely stabilizes the position of E118 while still allowing the flexibility to interact with the polar groups on a wide variety of ligands. In conjunction with the decreased cavity size because of the F103 for A and F54 for L substitutions, E118 potentially contributes to the increased binding affinity of MUP-IV for SBT in comparison to the group 1 isoforms.
Materials and Methods
Materials
MUP-IV was expressed in E. coli, purified using nickel-affinity chromatography, followed by proteolytic removal of the 6-HIS tag, as previously described.20,37 HT was purchased from Sigma (Sigma-Aldrich, St. Louis, MO) and DMP was purchased from Fluka (Sigma-Aldrich, St. Louis, MO). SBT was synthesized as described previously.37,51 Calcium chloride and ethylene glycol were purchased from Sigma (Sigma-Aldrich, St. Louis, MO), and polyethylene glycol 3350 was purchased from Hampton Research (Aliso Viejo, CA).
Crystallization, data collection, and refinement
MUP-IV crystals were grown by sitting drop vapor diffusion with 0.2–0.5M calcium chloride dihydrate and 20% (w/v) polyethylene glycol 3350. Crystals were soaked in mother liquor with pheromone at 5–10 mM (∼ 30:1 molar ratio) for ∼ 5 min and then cryoprotected with 25% ethylene glycol in the soak solution before flash cooling in an N2 gas stream at –180°C. Data for the structure of MUP-IV with DMP was collected on a home source RU-H2R generator with Raxis IV++ image plate detector. Data for the remaining structures were collected at the advanced photon source at Argonne National Laboratory (Argonne, IL) on the BioCARS beamline 14BMC. To limit overloads in the low-resolution shell for the SBT and 2EH structures, data from two different exposure times were merged. Data were indexed, integrated, and scaled using the HKL2000 program suite52 and cross-validated data sets were created using the CCP4 suite.53 The structure of MUP-IV was solved by molecular replacement in AMoRe54 using the protein coordinates of MUP-I, PDB code 1i06.24 The resulting model was rigid-body refined and subjected to one round of simulated annealing to reduce model bias using the Crystallography and NMR System.55 All models were then refined to ∼ 1.4 Å using Refmac5 (v5.2.0019)56 from within the CCP4i suite. The structures containing SBT and 2EH were then refined to the full resolution (∼ 1 Å) in Shelxl.57 All structures were refined with riding hydrogens and anisotropic individual B-factors. Model building was performed using Coot.58 Validation and structure comparisons were accomplished using the CCP4 suite programs Procheck, LSQKAB, Areaimol, and Baverage.53 Cavity surfaces and volumes were calculated with SwissPDB Viewer (v4.0)59 and CASTp.60 Figures were generated using PyMol (v0.99rc6 for windows).61
Sequence analysis
Lipocalin sequence alignments were obtained from the PFAM database (PF00061).50 The smaller set of 156 representative sequences (April 30, 2010) was used to generate the frequency plot via the WebLogo62 server.
Acknowledgments
We thank Spencer Anderson at BioCARS for helpful suggestions with our data collection strategy. We also thank D. Wiesler for SBT synthesis and S.D. Sharrow for providing purified MUP-IV protein.
Glossary
Abbreviations:
- DMP
2,5-dimethylpyrazine
- 2EH
2-ethylhexanol
- HT
2-heptanone
- MUP
Major urinary protein
- SBT
2-sec-butyl-4,5-dihydrothiazole
Footnotes
Numbering is from the first residue of the mature sequence, thus offset by 16 residues from the gene ORF.
We note that all MUP structures, including those presented here, have been determined at pH 4.8-7.2.21,23,24,26-32 The pKa of the conjugate acid of thiazole and pyrazine derivatives is around pH 2,38,39 thus, the sp2 hybridized nitrogen is less likely to be protonated at the pH of the experiments than is a Glu residue. To satisfy hydrogen bonding potential, we show E118 as protonated at OE2 in the figures. This is a reasonable assumption, given the hydrophobic nature of the MUP cavity and the pH of the crystals.
References
- 1.Brennan PA, Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- 2.Zufall F, Leinders-Zufall T. Mammalian pheromone sensing. Curr Opin Neurobiol. 2007;17:483–489. doi: 10.1016/j.conb.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann KH, Dettner K, Tomaschko KH. Chemical signals in insects and other arthropods: from molecular structure to physiological functions. Physiol Biochem Zool. 2006;79:344–356. doi: 10.1086/499991. [DOI] [PubMed] [Google Scholar]
- 4.Hurst JL, Robertson DHL, Tolladay U, Beynon RJ. Proteins in urine scent marks of male house mice extend the longevity of olfactory signals. Animal Behav. 1998;55:1289–1297. doi: 10.1006/anbe.1997.0650. [DOI] [PubMed] [Google Scholar]
- 5.al-Shawi R, Ghazal P, Clark AJ, Bishop JO. Intraspecific evolution of a gene family coding for urinary proteins. J Mol Evol. 1989;29:302–313. doi: 10.1007/BF02103618. [DOI] [PubMed] [Google Scholar]
- 6.Chamero P, Marton TF, Logan DW, Flanagan K, Cruz JR, Saghatelian A, Cravatt BF, Stowers L. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 7.Clark AJ, Clissold PM, Al Shawi R, Beattie P, Bishop J. Structure of mouse major urinary protein genes: different splicing configurations in the 3'-non-coding region. Embo J. 1984;3:1045–1052. doi: 10.1002/j.1460-2075.1984.tb01925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop JO, Clark AJ, Clissold PM, Hainey S, Francke U. Two main groups of mouse major urinary protein genes, both largely located on chromosome 4. EMBO J. 1982;1:615–620. doi: 10.1002/j.1460-2075.1982.tb01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahan K, Denaro M, Gilmartin M, Shi Y, Derman E. Expression of six mouse major urinary protein genes in the mammary, parotid, sublingual, submaxillary, and lachrymal glands and in the liver. Mol Cell Biol. 1987;7:1947–1954. doi: 10.1128/mcb.7.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novotny M, Harvey S, Jemiolo B, Alberts J. Synthetic pheromones that promote inter-male aggression in mice. Proc Natl Acad Sci USA. 1985;82:2059–2061. doi: 10.1073/pnas.82.7.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jemiolo B, Harvey S, Novotny M. Promotion of the whitten effect in female mice by synthetic analogs of male urinary constituents. Proc Natl Acad Sci USA. 1986;83:4576–4579. doi: 10.1073/pnas.83.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novotny M, Jemiolo B, Harvey S, Wiesler D, Marchlewska-Koj A. Adrenal-mediated endogenous metabolites inhibit puberty in female mice. Science. 1986;231:722–725. doi: 10.1126/science.3945805. [DOI] [PubMed] [Google Scholar]
- 13.Novotny MV, Ma W, Wiesler D, Zidek L. Positive identification of the puberty-accelerating pheromone of the house mouse: the volatile ligands associating with the major urinary protein. Proc Royal Soc, London. 1999;266:2017–2022. doi: 10.1098/rspb.1999.0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mudge JM, Armstrong SD, McLaren K, Beynon RJ, Hurst JL, Nicholson C, Robertson DH, Wilming LG, Harrow JL. Dynamic instability of the major urinary protein gene family revealed by genomic and phenotypic comparisons between c57 and 129 strain mice. Genome Biol. 2008;9:R91.1–R91.16. doi: 10.1186/gb-2008-9-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst JL, Payne CE, Nevison CM, Marie AD, Humphries RE, Robertson DH, Cavaggioni A, Beynon RJ. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414:631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 16.Cheetham SA, Thom MD, Jury F, Ollier WE, Beynon RJ, Hurst JL. The genetic basis of individual-recognition signals in the mouse. Curr Biol. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Sherborne AL, Thom MD, Paterson S, Jury F, Ollier WE, Stockley P, Beynon RJ, Hurst JL. The genetic basis of inbreeding avoidance in house mice. Curr Biol. 2007;17:2061–2066. doi: 10.1016/j.cub.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thom MD, Stockley P, Jury F, Ollier WE, Beynon RJ, Hurst JL. The direct assessment of genetic heterozygosity through scent in the mouse. Curr Biol. 2008;18:619–623. doi: 10.1016/j.cub.2008.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Leinders-Zufall T, Lane AP, Puche AC, Ma W, Novotny MV, Shipley MT, Zufall F. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature. 2000;405:792–796. doi: 10.1038/35015572. [DOI] [PubMed] [Google Scholar]
- 20.Sharrow SD, Vaughn JL, Zidek L, Novotny MV, Stone MJ. Pheromone binding by polymorphic mouse major urinary proteins. Protein Sci. 2002;11:2247–2256. doi: 10.1110/ps.0204202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bocskei Z, Groom CR, Flower DR, Wright CE, Phillips SE, Cavaggioni A, Findlay JB, North AC. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature. 1992;360:186–188. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- 22.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318(Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuser PR, Franzoni L, Ferrari E, Spisni A, Polikarpov I. The X-ray structure of a recombinant major urinary protein at 1.75 a resolution. A comparative study of X-ray and NMR-derived structures. Acta Cryst D. 2001;57:1863–1869. doi: 10.1107/s090744490101825x. [DOI] [PubMed] [Google Scholar]
- 24.Timm DE, Baker LJ, Mueller H, Zidek L, Novotny MV. Structural basis of pheromone binding to mouse major urinary protein (MUP-I) Protein Sci. 2001;10:997–1004. doi: 10.1110/ps.52201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucke C, Franzoni L, Abbate F, Lohr F, Ferrari E, Sorbi RT, Ruterjans H, Spisni A. Solution structure of a recombinant mouse major urinary protein. Eur J Biochem. 1999;266:1210–1218. doi: 10.1046/j.1432-1327.1999.00984.x. [DOI] [PubMed] [Google Scholar]
- 26.Bingham RJ, Findlay JB, Hsieh SY, Kalverda AP, Kjellberg A, Perazzolo C, Phillips SE, Seshadri K, Trinh CH, Turnbull WB, Bodenhausen G, Homans SW. Thermodynamics of binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the major urinary protein. J Am Chem Soc. 2004;126:1675–1681. doi: 10.1021/ja038461i. [DOI] [PubMed] [Google Scholar]
- 27.Barratt E, Bingham RJ, Warner DJ, Laughton CA, Phillips SE, Homans SW. Van der waals interactions dominate ligand-protein association in a protein binding site occluded from solvent water. J Am Chem Soc. 2005;127:11827–11834. doi: 10.1021/ja0527525. [DOI] [PubMed] [Google Scholar]
- 28.Malham R, Johnstone S, Bingham RJ, Barratt E, Phillips SE, Laughton CA, Homans SW. Strong solute-solute dispersive interactions in a protein-ligand complex. J Am Chem Soc. 2005;127:17061–17067. doi: 10.1021/ja055454g. [DOI] [PubMed] [Google Scholar]
- 29.Barratt E, Bronowska A, Vondrasek J, Cerny J, Bingham R, Phillips S, Homans SW. Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor. J Mol Biol. 2006;362:994–1003. doi: 10.1016/j.jmb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 30.Redondo C, Vouropoulou M, Evans J, Findlay JB. Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor. Faseb J. 2007;22:1043–1054. doi: 10.1096/fj.07-8939com. [DOI] [PubMed] [Google Scholar]
- 31.Syme NR, Dennis C, Phillips SE, Homans SW. Origin of heat capacity changes in a “nonclassical” hydrophobic interaction. Chembiochem. 2007;8:1509–1511. doi: 10.1002/cbic.200700281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertinhez TA, Ferrari E, Casali E, Patel JA, Spisni A, Smith LJ. The binding cavity of mouse major urinary protein is optimised for a variety of ligand binding modes. Biochem Biophys Res Commun. 2009;390:1266–1271. doi: 10.1016/j.bbrc.2009.10.133. [DOI] [PubMed] [Google Scholar]
- 33.Jemiolo B, Andreolini F, Xie TM, Wiesler D, Novotny M. Puberty-affecting synthetic analogs of urinary chemosignals in the house mouse, mus domesticus. Physiol Behav. 1989;46:293–298. doi: 10.1016/0031-9384(89)90270-9. [DOI] [PubMed] [Google Scholar]
- 34.Vitali M, Leoni V, Chiavarini S, Cremisini C. Determination of 2-ethyl-1-hexanol as contaminant in drinking water. J AOAC Int. 1993;76:1133–1137. [PubMed] [Google Scholar]
- 35.Nalli S, Horn OJ, Grochowalski AR, Cooper DG, Nicell JA. Origin of 2-ethylhexanol as a voc. Environ Pollut. 2006;140:181–185. doi: 10.1016/j.envpol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Varlet V, Knockaert C, Prost C, Serot T. Comparison of odor-active volatile compounds of fresh and smoked salmon. J Agric Food Chem. 2006;54:3391–3401. doi: 10.1021/jf053001p. [DOI] [PubMed] [Google Scholar]
- 37.Zidek L, Stone MJ, Lato SM, Pagel MD, Miao Z, Ellington AD, Novotny MV. NMR mapping of the recombinant mouse major urinary protein I binding site occupied by the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry. 1999;38:9850–9861. doi: 10.1021/bi990497t. [DOI] [PubMed] [Google Scholar]
- 38.Albert A, Goldacre R, Phillips J. The strength of heterocyclic bases. J Chem Soc. 1948:2240–2249. (Resumed) [Google Scholar]
- 39.Keyworth D. Notes: basicity and ionization constants of some pyrazine derivatives. J Org Chem. 1959;24:1355–1355. [Google Scholar]
- 40.Sharrow SD, Novotny MV, Stone MJ. Thermodynamic analysis of binding between mouse major urinary protein-I and the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry. 2003;42:6302–6309. doi: 10.1021/bi026423q. [DOI] [PubMed] [Google Scholar]
- 41.Sharrow SD, Edmonds KA, Goodman MA, Novotny MV, Stone MJ. Thermodynamic consequences of disrupting a water-mediated hydrogen bond network in a protepheromone complex. Protein Sci. 2005;14:249–256. doi: 10.1110/ps.04912605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flower DR, North CT, Sansom EC. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 43.Grzyb J, Latowski D, Strzalka K. Lipocalins—a family portrait. J Plant Pysiol. 2006;163:895–915. doi: 10.1016/j.jplph.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Virtanen T. Lipocalin allergens. Allergy. 2001;56:48–51. doi: 10.1034/j.1398-9995.2001.00915.x. [DOI] [PubMed] [Google Scholar]
- 45.Salo PM, Jaramillo R, Cohn RD, London SJ, Zeldin DC. Exposure to mouse allergen in U.S. homes associated with asthma symptoms. Environ Health Perspect. 2009;117:387–391. doi: 10.1289/ehp.11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skerra A. Alternative binding proteins: anticalins—harnessing the structural plasticity of the lipocalin ligand pocket to engineer novel binding activities. FEBS J. 2008;275:2677–2683. doi: 10.1111/j.1742-4658.2008.06439.x. [DOI] [PubMed] [Google Scholar]
- 47.Ramoni R, Bellucci S, Grycznyski I, Grycznyski Z, Grolli S, Staiano M, De Bellis G, Micciulla F, Pastore R, Tiberia A, Conti V, Merli E, Varriale A, Rossi M, D'Auria S. The protein scaffold of the lipocalin odorant-binding protein is suitable for the design of new biosensors for the detection of explosive components. J Phys: Condens Matter. 2007;19:395012–395018. [Google Scholar]
- 48.Flower D. Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 1993;2:753–761. doi: 10.1002/pro.5560020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramana J, Gupta D. LipocalinPred: a SVM-based method for prediction of lipocalins. BMC Bioinformatics. 2009;10:445–454. doi: 10.1186/1471-2105-10-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfam version 24.0, Wellcome Trust Sanger Institute in Cambridge, UK http://pfam.sanger.ac.uk/ Pfam sequence database based on UniProt release 15.6 (Swiss-Prot release 57.6 and SP-TrEMBL release 40.6)
- 51.Novotny MV, Xie TM, Harvey S, Wiesler D, Jemiolo B, Carmack M. Stereoselectivity in mammalian chemical communication: male mouse pheromones. Experientia. 1995;51:738–743. doi: 10.1007/BF01941272. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Cryst D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 54.Navaza J. Amore: An automated package for molecular replacement. Acta Cryst A. 1994;50:157–163. [Google Scholar]
- 55.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 57.Sheldrick G. A short history of shelx. Acta Cryst A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Guex N, Peitsch M. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 60.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pymol(tm) Windows version 99. Copyright 2006 DeLano Scientific lnc. Program and documentation can be obtained from http://www.pymol.org/
- 62.Crooks G, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]


