Abstract
Fatty acid binding proteins (FABP) have been characterized as facilitating the intracellular solubilization and transport of long-chain fatty acyl carboxylates via noncovalent interactions. More recent work has shown that the adipocyte FABP is also covalently modified in vivo on Cys117 with 4-hydroxy-2-nonenal (4-HNE), a bioactive aldehyde linked to oxidative stress and inflammation. To evaluate 4-HNE binding and modification, the crystal structures of adipocyte FABP covalently and noncovalently bound to 4-HNE have been solved to 1.9 Å and 2.3 Å resolution, respectively. While the 4-HNE in the noncovalently modified protein is coordinated similarly to a carboxylate of a fatty acid, the covalent form show a novel coordination through a water molecule at the polar end of the lipid. Other defining features between the two structures with 4-HNE and previously solved structures of the protein include a peptide flip between residues Ala36 and Lys37 and the rotation of the side chain of Phe57 into its closed conformation. Representing the first structure of an endogenous target protein covalently modified by 4-HNE, these results define a new class of in vivo ligands for FABPs and extend their physiological substrates to include bioactive aldehydes.
Keywords: oxidative stress, crystal structure, 4-HNE, fatty acid binding protein
Introduction
Obesity-linked insulin resistance and a variety of metabolic comorbidities are associated with inflammation and have been causally linked to an increase in reactive oxygen species (ROS) such as superoxide anion, hydrogen peroxide, and hydroxyl radicals.1–4 Of the various deleterious effects of ROS, particularly hydroxyl radicals, oxidation of polyunsaturated fatty acids (PUFA) and phospholipids in membranes followed by Hock cleavage generates a family of lipid-derived reactive aldehydes with the most common being malondialdehyde and 4-hydroxy-2-nonenal (also known as (2E,4R)-4-hydroxynon-2-enal or 4-HNE).5–7 4-HNE is an α,β-unsaturated 9 carbon aldehyde that undergoes Michael addition reactions with the side chains of cysteine, histidine and lysine residues. As such residues are frequently used in catalysis, alkylation reactions typically result in enzyme inactivation followed by targeted degradation leading to overall net loss of function.8
Grimsrud et al. recently discovered in murine adipose tissue that the adipocyte fatty acid binding protein (AFABP also known as aP2) is the soluble protein most highly modified by 4-HNE.9 Approximately 6–8% of the AFABP in adipose tissue of obese mice is covalently modified on Cys117 by 4-HNE and results in decreased affinity of the protein for fatty acids.9 AFABP facilitates the diffusion of fatty acids in the adipocyte and mediates transport of fatty acids derived from triacylglycerol breakdown.10–12 Molecular disruption of AFABP in mice leads to reduced lipolysis and mild obesity as well as the surprising finding of an anti-diabetic and anti-atherogenic phenotype.12–17 The latter result has spurred on the development of a variety of AFABP targeted small molecule drugs that may be efficacious in unlinking obesity from diabetes and atherogenesis.18,19
Like other intracellular lipid-binding proteins, the structures of AFABP in both its apo and several different holo forms consist of a β-barrel formed by 10 antiparallel β-strands (referred to as A-J) defining an interior water filled binding cavity that serves as the ligand-binding site.20–23 One end of the β-barrel is closed by the packing of protein side chains while the other end is partially restricted by a helix-turn-helix motif that serves as the portal for ligand entry/exit. The ligand-binding cavity contains several structurally conserved water molecules that are present in both apo- and holo-protein. The helix-turn-helix motif of AFABP and other FABP isoforms are important for fatty acid transfer from FABPs to membranes24 and four surface-exposed charged residues in this motif represent a protein-protein interaction domain.25,26
Physiological ligands for AFABP are long-chain fatty acids and carboxylate derivatives that bind to the protein noncovalently through acyl chain hydrophobic and entropic interactions and salt bridges between the carboxylate and Arg126 and Tyr128. AFABP binds avidly to a variety of long-chain fatty acids (Kd ≈ 5-50 nM) with the affinity declining substantially when the fatty acid is less than 12 carbons.27,28 The discovery that AFABP is modified by 4-HNE in vivo represents a major shift in our understanding of AFABP biology. Firstly, 4-HNE has only 9 carbons and is shorter than any ligands previously believed to interact with the adipocyte fatty acid binding protein in vivo. Secondly, 4-HNE does not possess a carboxyl group but is an aldehyde. Thirdly, AFABP and 4-HNE interact through a covalent linkage. Taken together, the above observations suggest that the cellular functions and specificity of AFABP is much broader than previously anticipated and provides new interpretation of the anti-diabetes function of AFABP inhibitors. Herein we report the crystal structure and analysis of AFABP with noncovalently bound 4-HNE and covalently modified with 4-HNE. To the best of our knowledge this is the first conformation of an endogenously modified AFABP as well as the first structure of 4-HNE covalently modifying an endogenous target protein.
Results
Enrichment of 4-HNE modified AFABP
Bacterially derived apo mouse AFABP was modified with 4-HNE for 30-70 minutes and the reactions quenched by addition of β-mercaptoethanol. The modified protein was applied to a long-chain acyl carboxylate affinity column and eluted with 1M NaCl.29 Protein in the bound and unbound fractions was analyzed by mass spectrometry (Fig. 1) where covalent 4-HNE modification was accompanied by a ∼156 Da increase in the detected molecular mass of the protein. The bound fraction contained approximately 90% 4-HNE modified protein while a sample not subject to affinity chromatography represented a 70:30 mixture of modified and unmodified AFABP, respectively. Crystals were obtained from protein prepared without or with affinity enrichment and represented two forms; one with partial occupancy, noncovalently bound 4-HNE and one with covalently bound 4-HNE, respectively.
Figure 1.
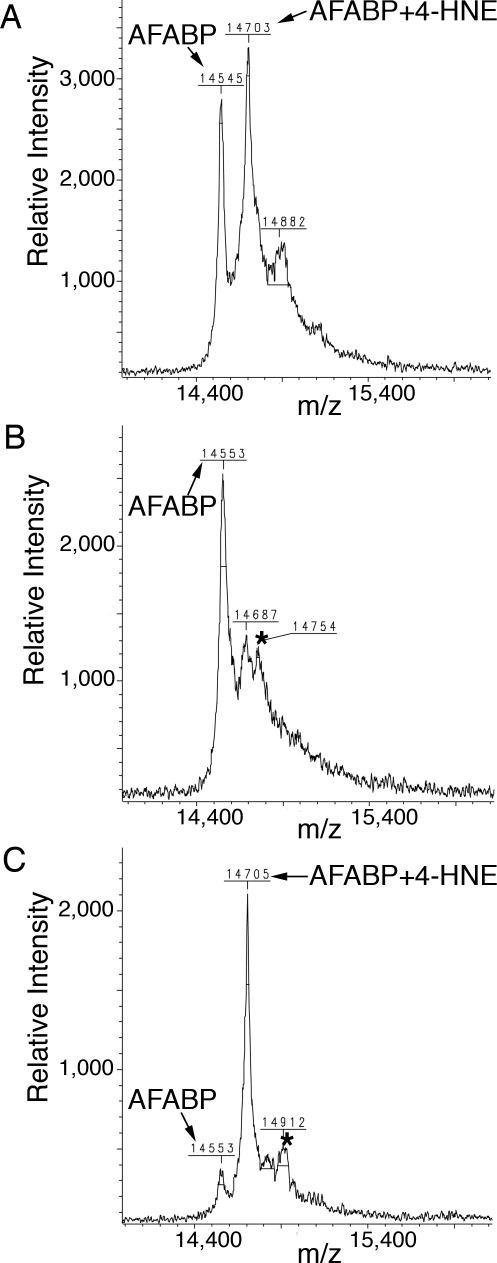
Enrichment of covalently 4-HNE modified AFABP. Shown are mass spectrometric analysis spectra of (A) sample after modification of AFABP with 4-HNE, (B) flow through fraction from the long-chain fatty acid affinity column, (C) eluted fraction from the long-chain fatty acid affinity column. Unmodified AFABP as well as AFABP modified with 4-HNE is indicated with arrows. * indicates a matrix adduct. Errors in the m/z values are within the mass accuracy of the instrument used.
Overall structure and 4-HNE binding
Crystal structures were analyzed for AFABP containing noncovalently and covalently bound 4-HNE. Statistics for the diffraction data and subsequent model refinement are shown in Table I. The overall folds for both forms are very similar to previously solved structures of AFABP with fatty acid ligands, but present unique conformational changes in a number of regions (Fig. 2). The overall fold consists of 10 anti-parallel β-strands arranged into a β-barrel and two α−helices forming a lid to the ligand-binding cavity where 4-HNE is located. The ligand-binding cavity also contains ordered water molecules, some of which are conserved with respect to the family of known structures.
Table I.
X-ray Data Collection and Refinement Statistics
| Covalent 4-HNEa | Noncovalent 4-HNEa | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 50.3, 81.3, 92.7 | 77.6, 93.2, 49.1 |
| Resolution (Å) | 20.0–1.9 (1.94–1.90)* | 50.0–2.3 (2.34–2.30)* |
| Rsym (%) | 6.8 (45.6)* | 6.5 (33.2)* |
| I/σI | 20.3 (3.2)* | 16.6 (2.9)* |
| Completeness (%) | 97.4 (100.0)* | 93.7 (73.5)* |
| Redundancy | 4.4 (4.4)* | 3.6 (3.2)* |
| Refinement | ||
| Resolution (Å) | 19.86–1.9 | 19.97–2.30 |
| No. reflections | 33276 | 7381 |
| Rwork / Rfree (%) | 19.7/22.8 | 21.4/26.3 |
| No. atoms | ||
| Protein | 2039 | 1022 |
| Ligand/ion | 22/5 | 11/21 |
| Water | 365 | 91 |
| B-factors (Å2) | ||
| Protein | 23.8 | 32.7 |
| Ligand/ion | 31.0/16.3 | 64.7/68.3 |
| Water | 37.1 | 45.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.016 |
| Bond angles (°) | 1.323 | 1.160 |
Data were collected from one crystal.
Values in parentheses are at resolution limit.
Figure 2.
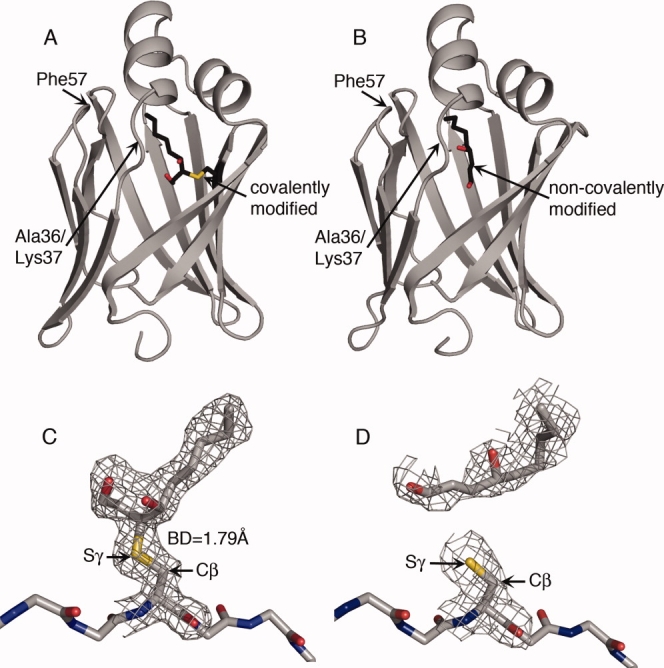
The structure of AFABP with covalently and noncovalently bound 4-HNE. The cartoons illustrate the overall structure of AFABP (A) covalently or (B) noncovalently modified with 4-HNE. AFABP is displayed in cartoon representation with the 4-HNE molecule located in the ligand-binding cavity represented in sticks. The position of the Ala36/Lys37 region and Phe57 is indicated. Electron density surrounding the 4-HNE molecule and Cys117 of AFABP in the (C) covalent and (D) noncovalent forms are compared. 4-HNE, Cys117, and the AFABP backbone around Cys117 are shown in stick representation. The 2|FO|-|FC| maps are contoured at 1.0 σ. The Cβ and Sγ atoms of Cys117 are indicated in the figure. The bond distance (BD) between C3 of the covalently bound 4-HNE and Sγ of Cys117 is given. Carbons are colored grey or black, oxygens red, nitrogens blue, and sulfurs yellow.
Dimerization
For the covalent 4-HNE structure there are two molecules in the asymmetric unit cell and they are related by a local two fold symmetry axis. The root mean square deviation (RMSD) between the Cα atoms in the A and B chain is 0.182 Å. The total solvent accessible surface areas for chain A and chain B are 7145 and 7294 Å2, respectively, as calculated using Surface Racer 5.030 with a probe radius of 1.4 Å. The total solvent accessible surface area for the dimer in the asymmetric unit cell is 12905 Å2 resulting in a large contact area between the two monomers of 1535 Å2. This suggests that the AFABP with 4-HNE covalently attached could potentially form a biological dimer since crystallographic contact areas are usually small. Residues 44-73, corresponding to β-strands C, D, and E, are located in the likely dimerization interface. Three holo-AFABP structures have been proposed to lead to dimerization formation previously based on crystal packing.31,32 These ligands are linoleic acid, troglitazone and 1-anilinonaphthalene-8-sulfonate (ANS). The noncovalent form crystallizes with one molecule in the asymmetric unit cell. This monomer has a total accessible surface area of 6587 Å2. However, a plausible dimer can be found in the crystal packing that closely resembles the dimer found in the covalent form of the protein with a RMSD of the Cα atoms in the covalent and noncovalent 4-HNE dimers of 0.507 Å. The monomers in the noncovalent dimer are related by a crystallographic two-fold symmetry axis. The total solvent accessible surface area of this dimer is 11611 Å2 leading to that 1563 Å2 of the surface area is lost in the dimer compared to two individual monomers, a number very similar to the area lost in the covalent form. Despite the prediction that AFABP may form a dimer, size exclusion chromatography revealed that apo-AFABP and HNE-modified AFABP chromatograph as monomeric proteins and not a dimeric assembly (results not shown). This point is further evaluated in the Discussion.
4-HNE modification
Analysis of the covalently modified AFABP revealed that the chemical modification from the α,β-unsaturated aldehyde results in a covalent bond to the sulfur atom of side chain of Cys117. The result is a protein-thioether bond [Fig. 2(C)]. Consistent with the mass spectrometry results (Fig. 1) the electron density for the ligand is as strong as the electron density for most of the residues in the protein suggesting that essentially all of the protein was covalently modified. In the structure, the sulfur atom of Cys117 is covalently attached to 4-HNE in an S-configuration at carbon 3 (C3) leading to an R-configuration at C4. To ensure that the chirality of C3 was evaluated correctly, C3 was modeled in an R-configuration and one additional round of refinement performed. This generated more negative electron density in the Fo-Fc omit map, especially in the A chain, consistent with the conclusion that C3 adopts an S-configuration. In contrast to the covalent 4-HNE structure, 4-HNE in the noncovalent form is too far away from the side chain sulfur of Cys117 to be covalently attached [Fig. 2(D)]. In addition, the electron density of the ligand in this noncovalent form is much weaker compared to the density for side chains in the protein suggesting that substitution was not stoichiometric.
Ligand coordination
Since 4-HNE is attached covalently to Cys117 the ligand does not reach as far into the binding cavity as does a fatty acid. In the covalent 4-HNE structure the ligand aldehyde is positioned to far away (4.69 Å) to allow direct interaction with Tyr128 that has been identified previously to coordinate fatty acid carboxyl groups together with Arg12621 [Fig. 3(A,C)]. The distance between the aldehyde of 4-HNE and Arg126 is 4.47 Å, while the distance to the fatty acid oleic acid (PDB ID 1LID) is 2.77 Å. Instead, there is an additional water molecule in the covalent 4-HNE structure located 2.69 Å from the aldehyde of the ligand that is not present in the structure with oleic acid. This water molecule forms hydrogen bonds with both the free aldehyde and with Arg126 thereby bridging the two [Fig. 3(A)].
Figure 3.
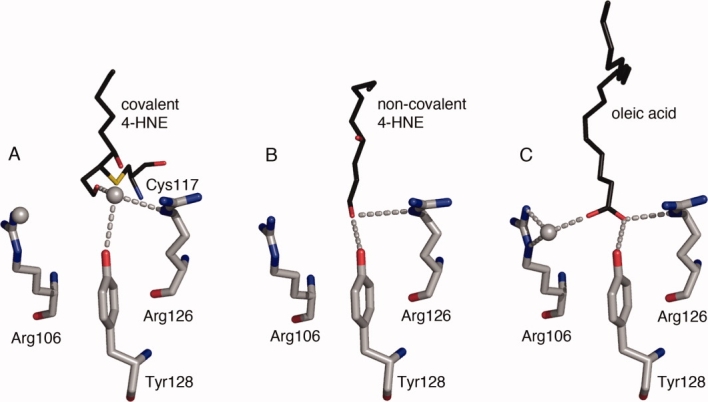
Coordination of the 4-HNE aldehyde function. The free aldehyde of covalently attached 4-HNE is located about 4.5 Å away from Arg126 and Tyr128, and instead of being directly coordinated to these residues a water bridges the aldehyde to Arg126 and Tyr128. Residues Arg106, Arg126 and Tyr128 are represented as sticks in grey and (A) covalent 4-HNE, (B) noncovalent 4-HNE and (C) oleic acid (PDB ID 1LID) are shown in black. Oxygen and nitrogen functional groups are colored in red and blue, respectively. Water molecules are depicted as grey spheres. Hydrogen bonds are indicated by dashed lines. The water molecule on the left in the covalent 4-HNE structure is hydrogen bonding with Arg106.
In contrast to the covalent 4-HNE structure, the noncovalent 4-HNE bound form reveals that the ligand aldehyde extends as far into the ligand-binding cavity as does the carboxyl group of a fatty acid, and the aldehyde aligns well with the carboxyl oxygen of a fatty acid facing Arg126 and Tyr128. This permits the aldehyde to interact directly with both Arg126 and Tyr128 [Fig. 3(B)] without the need for any bridging water molecules. In addition to Arg126 and Tyr128, Arg106 has also been implicated in the coordination of oleic acid by indirectly interacting with the fatty acid carboxylate via a water molecule21 [Fig. 3(C)]. This water molecule is absent in the noncovalent structure. In the covalent structure it is present and forms a hydrogen bond to Arg106 but it is located approximately 5 Å away from the free aldehyde of 4-HNE preventing it from forming an interaction.
The hydrophobic contacts between AFABP and the hydrocarbon chain of 4-HNE are similar in the covalent and noncovalent structure and also exhibit some similarity to those utilized for oleic acid binding (Table II). Since oleic acid is longer than 4-HNE (18 C compared to 9 C), it protrudes farther from the head group coordination site than does 4-HNE. Consistent with this, the acyl chain of 4-HNE is positioned more than 4.5 Å away from Thr29 (helix 2), Val32 (helix 2), M40 (β-strand B), K58 (loop between βC-D) and A75 (loop between βE-F), while the acyl chain oleic acid is within 4.5 Å from these residues. In contrast, the hydrocarbon chain of covalently bound 4-HNE is in close proximity to E116, while the hydrocarbon chains of oleic acid and the noncovalent ligand are not. The covalent 4-HNE bound form is also contacting Ile104 as is oleic acid but the noncovalent 4-HNE is farther away from this residue. In addition to Tyr128, the noncovalent 4-HNE (but not the covalently bound form) is within 4.5 Å distance from Ala33. These results suggest in sum that even though mostly similar contacts are made between AFABP and the three ligands evaluated, ligand specific interactions do take place.
Table II.
Residues Within 4.5 Å From the Different Ligands
| Residue | Covalent 4-HNE | Noncovalent 4-HNE | Oleic acid |
|---|---|---|---|
| F16 (s.c) | X | X | X |
| Y19 (s.c) | X | X | X |
| M20 (s.c) | X | X | X |
| V25 (s.c) | X | X | X |
| T29 (s.c, m.c) | X | ||
| V32 (s.c) | X | ||
| A33 (s.c, m.c) | X | X | |
| M40 (s.c) | X | ||
| F57 (s.c) | X | X | X |
| K58 (s.c) | X | ||
| A75 (s.c, m.c) | X | ||
| D76 (s.c) | X | X | X |
| R78 (s.c) | X | X | X |
| I104 (s.c) | X | X | |
| V115 (s.c) | X | X | X |
| C117 (s.c, m.c) | X | X | X |
| R126 (s.c) | X | X | X |
| Y128 (s.c) | X | X |
Structures were analyzed with PyMol (DeLano Scientific). s.c and m.c refer to side chain and main chain atoms, respectively.
Peptide flip
In some previously solved crystal structures of apo- and holo-AFABP a peptide flip can be observed centering on the amide bond between Ala36 and Lys37.21 This peptide flip is accompanied by a perturbation of the adjacent residues leading to alterations in the orientation of both main chain and side chain atoms. As shown in Figure 4, the covalent 4-HNE and noncovalent 4-HNE structures reveal a similar flip in the peptide bond relative to the apo-AFABP structure and with the ligand linoleic acid. The flip of the amide bond between Ala36 and Lys37 leads to a rotation of approximately 180° in the backbone carbonyl group of Ala36; similar to that found in the AFABP structure with oleic acid.21 In the apo-AFABP structure the Ala36 carbonyl group cannot hydrogen bond to any residue while in the linoleic acid structure a water molecule is located within hydrogen bonding distance. In both the covalent and noncovalent 4-HNE structures, as well as the structure with oleic acid, the carbonyl group of Ala36 forms a hydrogen bond with a water molecule (2.48, 2.81 and 2.61 Å, respectively). Interestingly this water molecule is also within hydrogen bonding distance of the guanidinium group of Arg126, the residue that coordinates the carboxyl group of fatty acids. This water molecule is present in the apo structure and can form a hydrogen bond to the nitrogen backbone atom of Ala36 although the distance is slightly increased (3.15 Å) compared to when Ala36 undergo the peptide flip.
Figure 4.
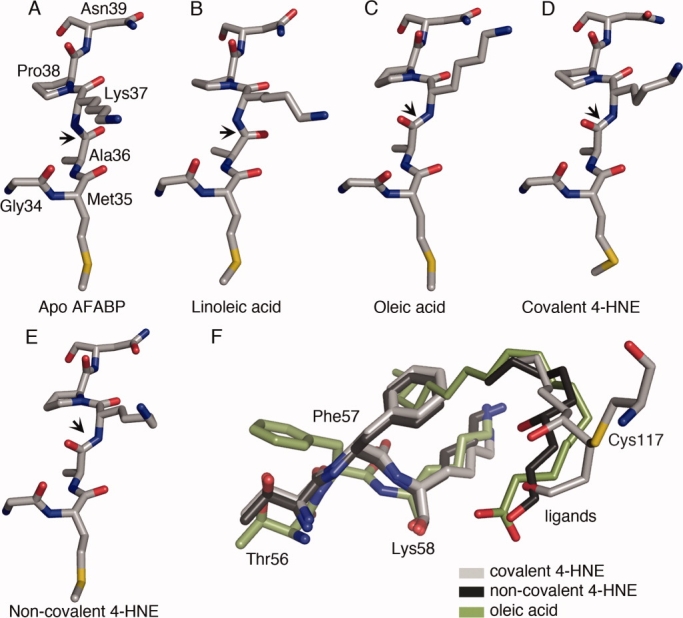
The peptide flip around Ala36 and Lys37. Residue Gly34 to Asn39 are shown in stick representation in (A) apo AFABP (PDB ID 1LIB), (B) AFABP with linoleic acid (PDB ID 2Q9S), (C) AFABP with oleic acid, (D) AFABP with covalently attached 4-HNE, and (E) noncovalent 4-HNE. The Ala36/Lys37 peptide bonds are indicated by arrows. Carbon atoms are colored grey, oxygen atoms red, nitrogen atoms blue and sulfur atoms are colored yellow. (F) The portal region of AFABP and the orientation of Phe57. The structures illustrated are AFABP with oleic acid shown in green, AFABP with covalent 4-HNE in grey and noncovalent 4-HNE in black. The side chains of Thr56, Phe57, and Lys58 are shown in stick representation and ligands in line representation.
The portal amino acid Phe57
An important and potentially regulatory feature in previous crystal structures of AFABP with different ligands concerns Phe57. Phe57 is positioned in a loop region between β-strand C and D, facing the helical domain, and its side chain can adopt two distinct conformations referred to as open or closed.21 This residue has been proposed to act as a gating function affecting access from the external environment to the ligand-binding cavity.33 In the apo structure the side chain of Phe57 is oriented inwardly in what is referred to as the closed position, but when bound with some long-chain fatty acids, the ω-terminus of the bound lipid forces rotation of Phe57 outwardly in what is referred to as the open conformation.21 In both the covalent and noncovalent 4-HNE structures, the side chain of Phe57 adopts the closed form [Fig. 4(F)]. Since 4-HNE is only nine carbons long, it does not protrude as far from the head group coordination towards the portal region as does a long-chain fatty acid allowing the rotation of Phe57 into the closed conformation.
Discussion
Herein we report the structure of AFABP with 4-HNE bound in the ligand-binding cavity. 4-HNE is produced endogenously in the cell as a product of lipid oxidation and this is, to the best of our knowledge, the first structure of AFABP covalently modified with an endogenous ligand and the first structure of 4-HNE covalently modifying an endogenous binding protein. However, this is not the first report of FABPs binding to ligands covalently. Previous studies have suggested that the liver isoform of FABP can bind metabolites of the genotoxic carcinogens 2-acetylaminofluorene and aminoazo dyes in a covalent manner.34,35 Based on MALDI-TOF and LC/MS-MS analysis, it was previously concluded that 4-HNE bound covalently to Cys117 of AFABP9 and our structural results herein confirm that finding. All structures of AFABP, solved to date are more or less superimposable and this was also the case for the AFABP covalently or noncovalently modified with 4-HNE. The overall structures consist of a β-barrel and a helix-turn-helix motif that define an interior cavity that functions as the ligand-binding site. There is an extensive hydrogen-bonding network in the cavity involving both ordered water molecules and amino acid residues and in both structures solved herein, 4-HNE is positioned in the large ligand-binding cavity of AFABP with the free aldehyde group oriented farthest away from the cavity portal and the hydrocarbon tail oriented towards the opening. The modification fills most of the cavity's empty volume and makes further uptake of fatty acids unlikely. This is consistent with the previous observation that the affinity for fatty acids is decreased in the modified protein.9
The attachment of 4-HNE to Cys117 occurs through a Michael addition reaction whereby the sulfur atom of Cys117 acts as a nucleophile and attacks C3 of 4-HNE, thereby leading to a covalent interaction. (R)-4-HNE was used for the binding reactions and as such, the chirality at C4 is not affected by the modification. Upon reduction of the carbon-carbon double bond between C2 and C3 of 4-HNE, a new chiral center is generated in the modification process at C3 that adopts an S configuration. It has been demonstrated previously that 4-HNE attached to an amino acid residue in a protein can be stabilized by undergoing cyclization to form a hemiacetal.7 The electron density of the covalent structure is ambiguous in this matter and does not exclude this possible orientation. Since C3 adopts the S configuration, the distance between the hydroxyl group of 4-HNE and the carbonyl aldehyde is 2.08 Å. This distance would be increased if C3 were to adopt an R configuration thereby eliminating possible hemiacetal formation.
Fatty acid carboxylates in the ligand-binding cavity of AFABP are coordinated through hydrogen bonds with the guanidinium group of Arg126 and the hydroxyl group of Tyr128.21 Structural analysis of AFABP covalently modified with 4-HNE demonstrates that due to the Cys117 4-HNE thioether bond, the aldehyde function is positioned too far away from Arg126 and Tyr128 for direct interaction. To compensate for this distance, an extra water molecule is located at a similar position as the fatty acid carboxylate that bridges the free aldehyde oxygen to the guanidinium group of Arg126 thereby allowing an indirect interaction. In contrast, the noncovalent 4-HNE reaches farther into the ligand-binding cavity and is located at a position equivalent to a carboxylate in a fatty acid. This permits direct interactions between the free aldehyde oxygen and the side chains of both Arg126 and Tyr128 in a similar manner as a fatty acid carboxylate. In both structures, the hydrocarbon chain of 4-HNE adopts an orientation similar to the hydrocarbon chain of oleic acid and the close contacts between the ligands and the protein are highly conserved. The 4-HNE chains are straighter than oleic acid, resulting in C9 of the covalent 4-HNE ending at the same position as C10 of oleic acid and C9 in the noncovalent structure co-localizing with C11 of oleic acid. However, 4-HNE in the covalent form, 4-HNE in the noncovalent form and oleic acid all make a few unique interactions with the protein, as summarized in Table II.
Previously all studies on AFABP have shown it to function as a monomer in solution. However, recently fluorescence anisotropy and small angle x-ray scattering have indicated that the protein in its apo form can exist as a homodimer.31 The crystal lattice packing of the apoprotein suggests that this potential homodimerization occur through interactions involving the helix-turn-helix motif. The same crystal lattice packing was obtained for AFABP complexed with several different ligands such as oleic acid, palmitic acid and hexadecanoic acid. Gillilan et al. observed a shift in crystal lattice packing when certain ligands are used (troglitazone, linoleic acid and 1,8-ANS) to a dimer form characterized by monomers interacting through β-sheets instead of the helix-turn-helix motifs.31 Based on crystal lattice packing and calculation of contact interface AFABP bound either covalently or noncovalently to 4-HNE is predicted to adopt a β-sheet dimer form. However, sedimentation equilibrium analysis indicates that the apoprotein is a monomer and if any dimer exists, the Kd for monomer–monomer interaction is quite high, minimally millimolar (Hellberg, Xu, and Bernlohr, unpublished data). Moreover, size exclusion chromatography supports a monomeric form of the protein in both the apo form and 4-HNE modified form. As stated previously, the affinity of AFABP for fatty acids is decreased upon 4-HNE-modification and it is therefore unlikely that modified AFABP bind additional fatty acids.9 It remains undetermined if 4-HNE-modified AFABP facilitates heterotypic protein-protein association as does AFABP bound with fatty acid, which interacts with the hormone sensitive lipase (HSL)25 and Janus kinase 2 (JAK2)36 and regulates cellular metabolism and signalling.
A major conclusion reached from analysis of a variety of AFABP structures is that the side chain of Phe57 can be rotated inwardly or outwardly to adopt a closed or an open conformation, respectively, in response to different ligands. This residue is located in a loop region adjacent to the entrance of the ligand-binding cavity and has been the subject of considerable debate as to its function as a gatekeeper for ligand entry/exit.23,33,37 The side chain shift from a closed orientation to an open orientation is seen with some ligands and not with others. One possible explanation is that the favored conformation is the closed position and that steric hindrance from a bound lipid forces Phe57 to rotate to the open conformation. Consistent with this, Phe57 is oriented inwardly in the apo structure, with bound linoleic acid and arachidonic acid, as well as in both the noncovalent and the covalent 4-HNE structures. Linoleic acid and arachidonic acid adopt a bent conformation when bound to AFABP and are accommodated in the ligand-binding cavity without nearing Phe57. As 4-HNE is only 9 carbons long it is also deep inside the ligand-binding cavity enabling Phe57 inward rotation. In contrast, bound oleic acid and palmitic acid protrude out of the ligand-binding cavity and consequently Phe57 is rotated into its open form. In this model, Phe57 does not serve as a molecular gatekeeper but the position of its side chain is defined somewhat by the type of ligand bound.
AFABP has been implicated in the development of insulin resistance since mice deficient of this protein exhibit improved insulin sensitivity along with decreased inflammation.13,17,38 The previous work of Grimsrud et al. and Bennaars-Eiden et al. have suggested that a major function for FABPs is as antioxidant proteins functioning to bind to and sequester reactive aldehydes from other proteins.9,39 In this capacity, AFABP fatty acid binding activity would be potentially diminished. Since AFABP is perhaps the most abundant protein in adipocytes and as such, loss of a small fraction due to 4-HNE modification would not likely affect fatty acid trafficking significantly. In contrast, loss of AFABP in the null mice would eliminate a major antioxidant protein and likely lead to increased covalent modification of secondary targets in adipocytes. Similarly, high affinity small molecule pharmacologic agents that target AFABP may have the identical effect, to inhibit the anti-oxidant activity of the FABPs and in turn, lead to covalent modification of secondary targets. If modification of such targets were anti-inflammatory, metabolic improvement may be realized. While the identification of such secondary targets remains speculative, previous studies by Karin and coworkers have reported that IκB kinase is covalently inactivated by Michael addition adducts resulting in diminished inflammatory signaling.40 Future studies will be needed to assess this possibility.
Materials and Methods
Purification, 4-HNE modification and crystallization of AFABP
Mouse AFABP was expressed in E. coli BL21(DE3) pLysS cells and purified as described previously with minor modifications.41 Briefly, native protein was purified using a combination of acid and protamine sulfate fractionation followed by delipidation with hydroxyalkoxypropyl dextran resin and gel filtration chromatography using Superdex G-75. 4-HNE modification of AFABP was conducted as previously described.9 The extent of modification was analyzed using a Bruker Biflex III matrix-assisted laser desorption ionization (MALDI) with time of flight (TOF). AFABP with 4-HNE noncovalently bound was concentrated to 10 mg/mL and dialyzed in 12.5 mM HEPES pH 7.5, 1 mM DTT. The sitting drop vapor diffusion method with 1 μL protein solution and 1 μL well solution was utilized to obtain crystals. The best crystal was found in conditions containing 0.1 M HEPES, 1.6 M NaH2PO4/K2HPO4 pH 7.5. The crystals grew within 2 weeks in 18 °C.
To optimize the purification seme, a long-chain fatty acid affinity column29 was added in the purification procedure followed by 4-HNE modification in room temperature for 70 minutes. The sample was dialyzed and loaded onto the long-chain fatty acid affinity column to enrich for modified protein, which was eluted with 1 M NaCl. Samples from the modification process were analyzed with MALDI-TOF. The eluted fraction, containing covalently modified protein, was concentrated, dialyzed and crystallized as described for the noncovalently bound 4-HNE protein above. The best crystal of AFABP covalently modified with 4-HNE grew in 1.9 M NaH2PO4/K2HPO4 pH 7.5. Crystals were obtained after 4 days in 18 °C.
X-ray data collection and processing
Crystals were frozen in the well solution supplemented with 20% glycerol as a cryoprotectant. Diffraction data were collected at Argonne Advanced Photon Source (APS) at beamline 23-ID-B for the noncovalently modified AFABP and in house using a Rigaku MicroMax 007HF generator with a Cu anode and a Rigaku Raxis IV++ detector for the covalent form of AFABP. Collected data were integrated and scaled using the HKL2000 software.42 Table I shows statistics from the data collection.
Structure determination and refinement
Molecular replacement was used to solve the structures of both noncovalent and covalently modified AFABP. AFABP bound to palmitic acid (PDB ID 1LIE) with the ligand removed was used as a probe for the noncovalent structure while a modified version of the same structure was used for the covalently modified structure. Refinement was performed using Refmac5 in the CCP4 suite and manual model building was done in Coot.43–45 Analysis and figure preparation were performed using PyMol.46
Coordinates
The coordinates and structure factors for the structures have been deposited in the Protein Data Bank. The accession number for AFABP with noncovalently bound 4-HNE is 3JSQ, and for AFABP covalently modified with 4-HNE 3JS1.
Acknowledgments
The authors thank the members of the Bernlohr laboratory for helpful comments and suggestions during the preparation of this manuscript. They thank Todd Markowski at the Center for Mass Spectrometry and Proteomics and the University of Minnesota Supercomputing Institute. They also thank Professor Matthew Picklo of the Department of Pharmacology, Physiology, and Therapeutics at the University of North Dakota School of Medicine and Health Science for supplying purified R-HNE. The diffraction data were collected at Argonne Advanced Photon Source (APS) at beamline 23-ID-B and in Kahlert Structural Biology Laboratory and they thank Ed Hoeffner for technical assistance and Medora Huseby for her help during the data collection process.
References
- 1.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eriksson JW. Metabolic stress in insulin's target cells leads to ROS accumulation—a hypothetical common pathway causing insulin resistance. FEBS Lett. 2007;581:3734–3742. doi: 10.1016/j.febslet.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Grattagliano I, Palmieri VO, Portincasa P, Moschetta A, Palasciano G. Oxidative stress-induced risk factors associated with the metabolic syndrome: a unifying hypothesis. J Nutr Biochem. 2008;19:491–504. doi: 10.1016/j.jnutbio.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Tinahones FJ, Murri-Pierri M, Garrido-Sanchez L, Garcia-Almeida JM, Garcia-Serrano S, Garcia-Arnes J, Garcia-Fuentes E. Oxidative stress in severely obese persons is greater in those with insulin resistance. Obesity (Silver Spring) 2009;17:240–246. doi: 10.1038/oby.2008.536. [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 6.Schneider C, Tallman KA, Porter NA, Brash AR. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J Biol Chem. 2001;276:20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 7.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 8.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Storch J, Thumser AE. The fatty acid transport function of fatty acid-binding proteins. Biochim Biophys Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 11.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 12.Furuhashi M, Fucho R, Gorgun CZ, Tuncman G, Cao H, Hotamisligil GS. Adipocyte/macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J Clin Invest. 2008;118:2640–2650. doi: 10.1172/JCI34750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 14.Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS. Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology. 2000;141:3388–3396. doi: 10.1210/endo.141.9.7637. [DOI] [PubMed] [Google Scholar]
- 15.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med. 2001;7:699–705. doi: 10.1038/89076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor γ and IκB kinase activities. J Biol Chem. 2005;280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertzel AV, Smith LA, Berg AH, Cline GW, Shulman GI, Scherer PE, Bernlohr DA. Lipid metabolism and adipokine levels in fatty acid-binding protein null and transgenic mice. Am J Physiol Endocrinol Metab. 2006;290:E814–823. doi: 10.1152/ajpendo.00465.2005. [DOI] [PubMed] [Google Scholar]
- 18.Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vaillancourt E, Kono K, Babaev VR, Fazio S, Linton MF, Sulsky R, Robl JA, Parker RA, Hotamisligil GS. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature. 2007;447:959–965. doi: 10.1038/nature05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertzel AV, Hellberg K, Reynolds JM, Kruse AC, Juhlmann BE, Smith AJ, Sanders MA, Ohlendorf DH, Suttles J, Bernlohr DA. Identification and characterization of a small molecule inhibitor of fatty acid binding proteins. J Med Chem. 2009;52:6024–6031. doi: 10.1021/jm900720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcelino AM, Smock RG, Gierasch LM. Evolutionary coupling of structural and functional sequence information in the intracellular lipid-binding protein family. Proteins. 2006;63:373–384. doi: 10.1002/prot.20860. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z, Bernlohr DA, Banaszak LJ. The adipocyte lipid-binding protein at 1.6-Å resolution. Crystal structures of the apoprotein and with bound saturated and unsaturated fatty acids. J Biol Chem. 1993;268:7874–7884. [PubMed] [Google Scholar]
- 22.LaLonde JM, Bernlohr DA, Banaszak LJ. X-ray crystallographic structures of adipocyte lipid-binding protein complexed with palmitate and hexadecanesulfonic acid. Properties of cavity binding sites. Biochemistry. 1994;33:4885–4895. doi: 10.1021/bi00182a017. [DOI] [PubMed] [Google Scholar]
- 23.Storch J, McDermott L. Structural and functional analysis of fatty acid-binding proteins. J Lipid Res. 2009;50:S126–S131. doi: 10.1194/jlr.R800084-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corsico B, Cistola DP, Frieden C, Storch J. The helical domain of intestinal fatty acid binding protein is critical for collisional transfer of fatty acids to phospholipid membranes. Proc Natl Acad Sci USA. 1998;95:12174–12178. doi: 10.1073/pnas.95.21.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AJ, Thompson BR, Sanders MA, Bernlohr DA. Interaction of the adipocyte fatty acid-binding protein with the hormone-sensitive lipase: regulation by fatty acids and phosphorylation. J Biol Chem. 2007;282:32424–32432. doi: 10.1074/jbc.M703730200. [DOI] [PubMed] [Google Scholar]
- 26.Smith AJ, Sanders MA, Juhlmann BE, Hertzel AV, Bernlohr DA. Mapping of the hormone-sensitive lipase binding site on the adipocyte fatty acid-binding protein (AFABP). Identification of the charge quartet on the AFABP/aP2 helix-turn-helix domain. J Biol Chem. 2008;283:33536–33543. doi: 10.1074/jbc.M806732200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J Biol Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- 28.Kane CD, Bernlohr DA. A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene 8-sulfonic acid. Anal Biochem. 1996;233:197–204. doi: 10.1006/abio.1996.0028. [DOI] [PubMed] [Google Scholar]
- 29.Buelt MK, Xu Z, Banaszak LJ, Bernlohr DA. Structural and functional characterization of the phosphorylated adipocyte lipid-binding protein (pp15) Biochemistry. 1992;31:3493–3499. doi: 10.1021/bi00128a025. [DOI] [PubMed] [Google Scholar]
- 30.Tsodikov OV, Record MT, Jr., Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 31.Gillilan RE, Ayers SD, Noy N. Structural basis for activation of fatty acid-binding protein 4. J Mol Biol. 2007;372:1246–1260. doi: 10.1016/j.jmb.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ory JJ, Banaszak LJ. Studies of the ligand binding reaction of adipocyte lipid binding protein using the fluorescent probe 1, 8-anilinonaphthalene-8-sulfonate. Biophys J. 1999;77:1107–1116. doi: 10.1016/S0006-3495(99)76961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins AE, Hockenberry JA, Nguyen T, Bernlohr DA. Testing of the portal hypothesis: analysis of a V32G, F57G, K58G mutant of the fatty acid binding protein of the murine adipocyte. Biochemistry. 2002;41:2022–2027. doi: 10.1021/bi015769i. [DOI] [PubMed] [Google Scholar]
- 34.Blackburn GR, Andrews JP, Rao KV, Sorof S. An early event associated with liver carcinogenesis involving loss of a polypeptide that binds carcinogen. Cancer Res. 1980;40:4688–4693. [PubMed] [Google Scholar]
- 35.Blackburn GR, Schnabel SJ, Danley JM, Hogue-Angeletti RA, Sorof S. Principal polypeptide target of carcinogen at the beginning of liver carcinogenesis by three carcinogens. Cancer Res. 1982;42:4664–4672. [PubMed] [Google Scholar]
- 36.Thompson BR, Mazurkiewicz-Munoz AM, Suttles J, Carter-Su C, Bernlohr DA. Interaction of adipocyte fatty acid-binding protein (AFABP) and JAK2: AFABP/aP2 as a regulator of JAK2 signaling. J Biol Chem. 2009;284:13473–13480. doi: 10.1074/jbc.M900075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson MA, Bernlohr DA. Analysis of a series of phenylalanine 57 mutants of the adipocyte lipid-binding protein. Biochemistry. 1998;37:10980–10986. doi: 10.1021/bi980507a. [DOI] [PubMed] [Google Scholar]
- 38.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–50702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- 40.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IκB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 41.Xu ZH, Buelt MK, Banaszak LJ, Bernlohr DA. Expression, purification, and crystallization of the adipocyte lipid binding protein. J Biol Chem. 1991;266:14367–14370. [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. In: Carter C, Sweet R, editors. Methods in enzymology, macromolecular crystallography, part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMol molecular graphics system. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]


