Abstract
Inteins are the protein equivalent of introns. They are remarkable and robust single turnover enzymes that splice out of precursor proteins during post-translational maturation of the host protein (extein). The Deinococcus radiodurans Snf2 intein is the second member of the recently discovered Class 3 subfamily of inteins to be characterized. Class 3 inteins have a unique sequence signature: (a) they start with residues other than the standard Class 1 Cys, Ser or Thr, (b) have a noncontiguous, centrally located Trp/Cys/Thr triplet, and (c) all but one have Ser or Thr at the start of the C-extein instead of the more common Cys. We previously proposed that Class 3 inteins splice by a variation in the standard intein-mediated protein splicing mechanism that includes a novel initiating step leading to the formation of a previously unrecognized branched intermediate. In this mechanism defined with the Class 3 prototypic Mycobacteriophage Bethlehem DnaB intein, the triplet Cys attacks the peptide bond at the N-terminal splice junction to form the class specific branched intermediate after which the N-extein is transferred to the side chain of the Ser, Thr, or Cys at the C-terminal splice junction to form the standard intein branched intermediate. Analysis of the Deinococcus radiodurans Snf2 intein confirms this splicing mechanism. Moreover, the Class 3 specific Block F branched intermediate was isolated, providing the first direct proof of its existence.
Keywords: intein, protein splicing, branched intermediate, mechanism, protein processing
Introduction
Intein-mediated protein splicing is a post-translational process where the intervening intein sequence is removed from a protein precursor and the flanking protein fragments (termed exteins) are ligated to form the mature extein protein. The mechanism is self-catalyzed by the combined action of the intein and the first C-extein residue, which act as a single turnover enzyme. The process results in cleavage of the peptide bonds at the intein N- and C-termini and formation of a new peptide bond between the exteins. Understanding the mechanism of protein splicing has led to the development of numerous methods of protein semisynthesis, control of protein expression, and other biotechnology applications. To date, over 500 inteins have been identified in archaea, eubacteria, and eukaryotes (see the InBase online intein database at http://www.neb.com/neb/inteins.html).1 There are four conserved motifs present in all intein splicing domains (Fig. 1).2–5 Intein residues are referred to using the Block letter and the Block position separated by a colon.
Figure 1.
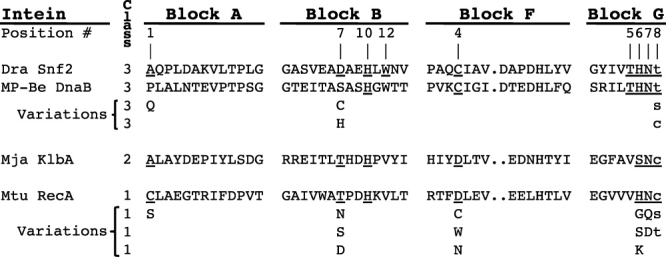
The four protein splicing domain motifs are depicted for the three classes of inteins. The position number in each Block for residues known to be important for catalysis is indicated. Conserved positions are populated by groups of residues that have similar properties except for the Block B His (B:10), which is almost invariant.1–5 An exemplary sequence is shown for each intein class. The most common residue found at selected positions is underlined and the common variations for these positions are shown. Note that no covariation amongst positions is implied in the rows listing the variations. To date, only 1 putative Class 3 intein (Bvi IcmO) has a Cys in the +1 position. Intein residues are in upper case using the single letter amino acid code and the +1 C-extein residue is in lower case. The periods represent a gap in some inteins due to a variable sized loop in Block F. Numbering of intein residues starts with 1 at the intein N-terminus and +1 at the C-extein N-terminus. Residues are referred to in the text using the Block letter and the Block position separated by a colon. An alternate Block nomenclature exists: Block A = N1, Block B = N3, Block F = C2, and Block G = C1.3 See InBase for a complete list of intein sequences and block sequences.1
Inteins were recently divided into three classes based on sequence signatures and splicing mechanisms.5 Class 1 inteins splice by the “standard” reaction scheme (Fig. 2).6,7 Splicing is initiated when the intein N-terminal Ser1 or Cys1 undergoes an N-[S/O] acyl migration to form the linear (thio)ester intermediate (II). Although Thr1 mutants were shown to splice, Thr1 has not been observed in nature.1,8 In step 2, the first residue of the C-extein (Cys+1, Ser+1, or Thr+1) cleaves the N-terminal splice junction (thio)ester bond, transferring the N-extein to its side chain while forming the Block G branched intermediate (BI) (III), so called because the branch point is in intein Block G (Fig. 1). The Block G BI is then resolved by Asn cyclization, which results in C-terminal splice junction cleavage releasing the free intein with a C-terminal succinimide ring (IV) and the ligated exteins bound by a (thio)ester bond (V). Asp and Gln can undergo similar cyclization reactions.6,7,9–11 The intein penultimate His assists Asn cyclization.6,7 A native peptide bond is formed between the exteins (VI) after a spontaneous [S/O]-N acyl shift. Slow hydrolysis of the succinimide ring yields Asn (VII) or isoasparagine. Mutations and foreign extein residues often reduce splicing and result in off-pathway single or double splice junction cleavage in the absence of splicing (Fig. 3). Theoretically, the observed amount of on-pathway or off-pathway cleavage at a specific splice junction can change due to either a change in the reaction rate at that splice junction or a change in the reaction rate at an earlier or later step in the splicing pathway. For example, the observation of increased N-terminal cleavage products could be the result of a direct increase in the rate of off-pathway N-terminal cleavage. Alternatively, this could be indirectly caused by a decrease in the rate of C-terminal cleavage that results in increased susceptibility of stalled (thio)ester intermediates II or III to off-pathway N-terminal cleavage.
Figure 2.
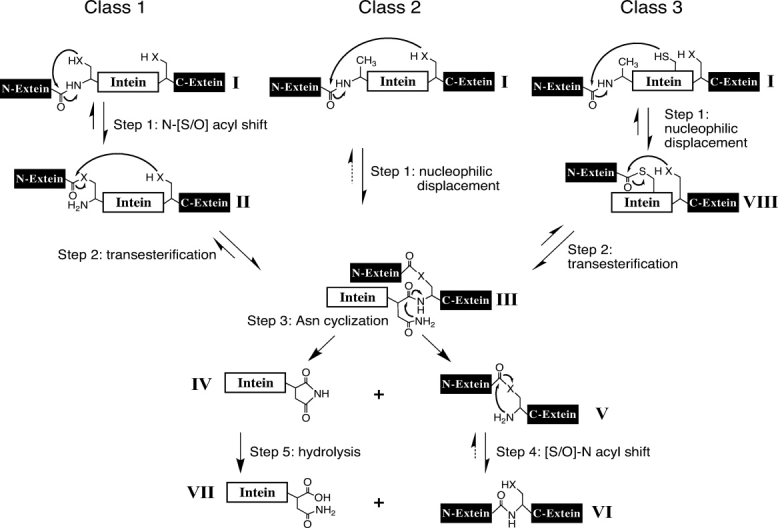
The splicing mechanisms for the three classes of inteins. The majority of inteins follow the Class 1 intein-mediated protein splicing mechanism, which consists of four coordinated nucleophilic displacement reactions. Class 2 and 3 inteins lack an N-terminal Cys, Ser, or Thr and, therefore, cannot perform the initial acyl shift required by Class 1 inteins. In Class 2 inteins, Cys+1 directly attacks an amide bond at the N-terminal splice junction to form the standard Block G branched intermediate. Known examples of Class 2 inteins are the KlbA inteins.12 The Class 3 intein reaction scheme also consists of four coordinated nucleophilic displacements but includes two branched intermediates: (Step 1) the Cys in Block F (Cys323 in the Dra Snf2 intein) attacks the peptide bond at the N-terminal splice junction, forming the Block F branched intermediate; (Step 2) the N-extein is transferred to the side chain of the +1 residue (Thr+1 in the Dra Snf2 intein) by a transesterification reaction resulting in the formation of the same Block G branched intermediate as in Class 1 or 2. Once the Block G branched intermediate is formed, the remainder of the splicing reaction is the same in all inteins. Residues within the intein assist these enzymatic reactions. However, in all classes, there is variability in the residues and positions within the intein that facilitate each reaction. Tetrahedral intermediates are not shown. “X” represents the sulfur or oxygen atoms in the side chains of Ser, Thr, or Cys and dashed arrows represent chemically feasible reverse reactions that have not been experimentally verified.
Figure 3.
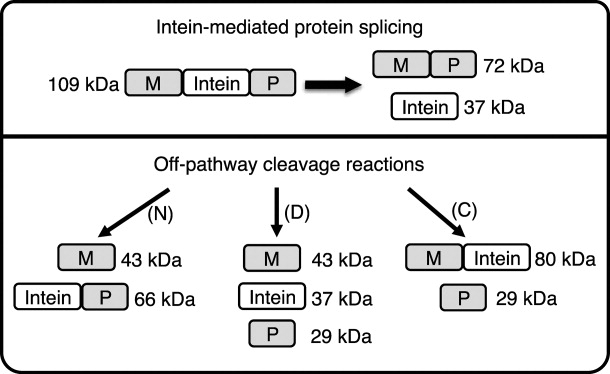
A schematic representation of splicing and off-pathway cleavage reactions for the Dra Snf2 intein (I) in a model precursor consisting of MBP as the N-extein (M) and a fragment of paramyosin (P) as the C-extein. Protein splicing yields ligated MP and free intein, while off-pathway N-terminal splice junction cleavage (N) yields M + IP and off-pathway C-terminal splice junction cleavage (C) yields MI + P. Cleavage of the (thio)ester linkage in branched intermediates also yields M + IP. Some mutations result in double cleavage (D) at both splice junctions to yield M + I + P. Molecular weights of the Dra Snf2 intein MIP precursor and potential products are listed.
A growing number of atypical inteins are being identified that are unable to generate the Class 1 linear thioester intermediate (II) because they start with residues other than Cys, Ser, or Thr (Fig. 1).1–5 Two variations in the intein-mediated protein splicing mechanism were previously described, which involve direct attack on an amide bond at the N-terminal splice junction to generate a BI.5–7,12,13 In Class 2 inteins, exemplified by the Methanococcus jannaschii (Mja) KlbA intein,12,13 the Block G BI (III) is formed by nucleophilic attack of Cys+1 on the peptide bond at the N-terminal splice junction (Fig. 2). In Class 3 inteins, exemplified by the Mycobacteriophage Bethlehem (MP-Be) DnaB intein,5 a Block F BI (VIII) is initially formed by nucleophilic attack of the Block F position 4 (F:4) Cys on the peptide bond at the N-terminal splice junction (Fig. 2). However, the proposed Block F BI was never observed in the MP-Be DnaB intein.5 A reversible transesterification reaction moves the N-extein to the side chain of the +1 residue to generate the standard Block G BI (III). The remainder of the splicing mechanism is the same for all three intein classes once the Block G BI (III) is formed. Class 2 and 3 inteins have overcome the barrier to direct attack of an amide bond at the N-terminal splice junction that is present in Class 1 inteins.
Differences amongst the three classes of inteins are also evident in the specific residues that populate certain positions in conserved motifs (Fig. 1) and the mechanistic roles that they play.1–7 Putative Class 3 inteins can be identified by class-specific sequence features: (a) the absence of a Cys, Ser, or Thr N-terminal nucleophile at A:1, (b) a noncontiguous WCT triplet at B:12, F:4, and G:5, and (c) Thr+1 or Ser+1 as the first C-extein residue at G:8 instead of the more common Cys+1 (Fig. 1).1,5 Only 1 putative Class 3 intein, the Burkholderia vietnamiensis G4 (Bvi) IcmO intein, has Cys+1. The Cys in the WCT triplet is the branch point for the Block F BI (VIII).5 This position is most often Asp in Class 1 and 2 inteins.1 Block B residues activate the N-terminal splice junction for on-pathway cleavage during BI formation and off-pathway N-terminal cleavage. The B:10 His is essential for splicing and N-terminal cleavage, while mutation of the more variable B:7 residue in Class 1 and 2 inteins usually reduces splicing and N-terminal cleavage, but does not block it.5–7,12–14
The essential characteristics of Class 3 inteins are the mechanistic properties specific to this subfamily of inteins as determined in the prototypic MP-Be DnaB intein: (a) cleavage at the N-terminal splice junction in the absence of all standard Class 1 and Class 2 N- and C-terminal splice junction nucleophiles at A:1, G:7, and G:8, (b) activation of the N-terminal splice junction by a Block B motif that includes the conserved His (B:10) and the WCT triplet Trp (B:12), (c) breakdown of native but not denatured Block G BI (III) in response to thiols or Cys despite an ester linkage at the Block G branch point, (d) an absolute requirement of the WCT triplet Cys at F:4 for splicing, (e) both a Block F BI (VIII) and a Block G BI (III), and (f) nonconservative mutation of the F:4 Cys prevents formation of both the Block F BI and the Block G BI, as well as off-pathway N-terminal cleavage.
It is important to determine whether these properties of the MP-Be DnaB intein are specific to this intein or if they can be generalized to other members of Class 3. The Ala1 Deinococcus radiodurans (Dra) Snf2 intein has the Class 3 intein signature sequences. Although splicing of the Dra Snf2 intein was previously described, the mechanism was not characterized.15 This study examines the reactivity of the Dra Snf2 intein and confirms that the properties listed above are common to the class. Furthermore, the Class 3 Block F BI (VIII) was detected for the first time.
Results and Discussion
Mutagenesis of conserved residues in the Dra Snf2 intein
The 343 amino acid Dra Snf2 intein splices in vivo in a model precursor consisting of the Escherichia coli maltose binding protein (MBP or M) as the N-extein and a fragment of Dirofilaria immitis paramyosin (P) as the C-extein.15 In this study, expression of this MIP precursor was induced with 1 mM IPTG (isopropyl β-d-thiogalactoside) for 2 h at 37°C (37°C sample) or for 2 h at 37°C followed by overnight incubation at 15°C (15°C sample). Such temperature shifts allow mutated inteins that are marginally misfolded or aggregated at 37°C to refold at 15°C with the help of the cell's refolding machinery. However, grossly misfolded proteins are not rescued by temperature shifts nor are enzymes with mutations in essential catalytic residues. The wild type Dra Snf2 intein spliced to completion in vivo to yield MP and I under both conditions (Fig. 4 and Table I).
Figure 4.
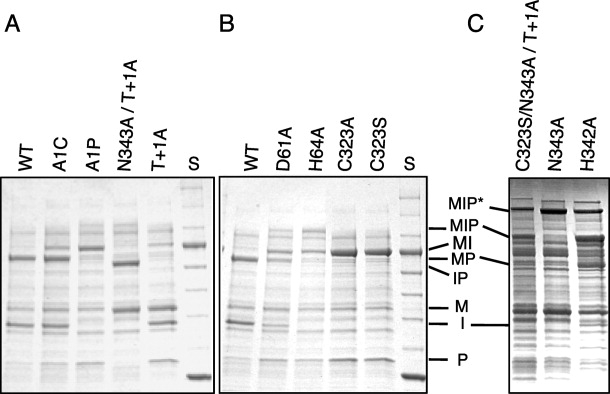
The effects of mutating conserved Dra Snf2 intein residues were examined. Mutated MIP precursors were expressed at 37°C for 2 h followed by 15°C overnight. Soluble proteins were electrophoresed in SDS-PAGE followed by staining with Coomassie Blue. Mutations are listed above each lane. Part A: splice junction residues, Part B: mutations in Blocks B and F, and Part C: mutations yielding branched intermediates. Multiple mutations in the same protein are separated by a slash. Abbreviations as in Figure 3 plus WT, wild type Dra Snf2 intein and “S,” 10–250 kDa Protein Ladder (250, 150, 100, 80, 60, 50, 40, 30, 25 kDa, where 80 and 25 kDa are darker bands).
Table I.
The Effects of Mutating Splice Junction Proximal Dra Snf2 Intein Residues
| Residue | Mutation | Temp (°C) | Splice (MP+I) | Precursor (MIP) | N-cleavea (M+IP) | C-cleavea (MI+P) | DC b (M+I+P) | Branch c (MIP*) |
|---|---|---|---|---|---|---|---|---|
| Block A | ||||||||
| WT | 15/37 | ++++ d | ||||||
| Ala1 | Pro | 37 | ++++ | |||||
| Pro | 15 | + | ++++ | |||||
| Ser | 15/37 | ++++ | + | |||||
| Cys | 37 | +++ | ++ | + | ||||
| Cys | 15 | ++++ | + | + | ||||
| Block G | ||||||||
| His342 | Ala | 15 | + | +++ | ++ (G) | |||
| Asn343 | Ala | 15/37 | ++++ (G) | |||||
| Thr+1 | Ala | 37 | + | + | +++ | |||
| Ala | 15 | + | ++++ | |||||
| Cys | 37 | ++++ | + | |||||
| Cys | 15 | ++++ | ||||||
| Ser | 15/37 | ++++ | + | |||||
| eMultiple mutations | ||||||||
| N343/ T+1 | A/A | 15 | + | ++++ | ||||
| C323/ N343 / T+1 | S/A/A | 15 | ++ | ++ (F) | ||||
N-cleave and C-cleave = off-pathway single splice junction cleavage reactions. Abbreviations as in Figure 3.
Double cleavage at both splice junctions.
F = Block F BI (VIII) and G = Block G BI (III).
Key to the relative amount of MIP substrate and products: blank, none detected; +, <10%; ++, 10–50%; +++, 51–90%; ++++, >90%.
Multiple mutations in the same precursor are separated by a slash.
Substitutions of the Dra Snf2 N-terminal Ala1 reduced splicing and off-pathway N-terminal cleavage to varying degrees, which resulted in increased amounts of off-pathway C-terminal cleavage [Fig. 4(A) and Table I]. Replacement of Ala1 by Pro yielded precursor at 37°C and C-terminal cleavage products at 15°C. Ala1Ser spliced efficiently at both temperatures, while Ala1Cys yielded both spliced and C-terminal cleavage products. These results were similar to those observed after mutation of the Mja KlbA (Ala1) and MP-Be DnaB (Pro1) intein N-termini in that some mutations reduced the reactivity of the N-terminal splice junction, while others did not.5,12,13 However, the relative amounts of products observed with each amino acid were usually specific to the intein, which is also true for all positions tested in this study.
The following Dra Snf2 intein C-terminal splice junction proximal residues were mutated (Fig. 4 and Table I): the intein penultimate His342 (G:6), the intein C-terminal Asn343 (G:7), and the C-extein Thr+1 (G:8). Substitution of Asn343 by Ala yielded mainly BI, confirming that Asn343 is required for resolution of the Block G BI and off-pathway C-terminal cleavage. Mutation of His342 to Ala yielded precursor and BI, indicating that this penultimate His facilitates Asn cyclization. Mutation of Thr+1 to Ala yielded double cleavage products (M + I + P) while conservative substitution by Cys or Ser permitted splicing. N-terminal cleavage still occurred when Asn343Ala was combined with Thr+1Ala because the Block F Cys mediates N-terminal cleavage in Class 3 inteins. Thus, the Dra Snf2 intein is neither Class 1 as it begins with Ala1 nor Class 2 as Thr+1 is not required for N-terminal cleavage.
All inteins use Block B residues to activate the N-terminal splice junction. The residues populating the conserved Block B positions in the Dra Snf2 intein are Asp61 (B:7), His64 (B:10), and Trp66 (B:12). His64 is only required for N-terminal reactions as C-terminal cleavage was efficient at 15°C when this residue was mutated [Fig. 4(B), Supporting Information Figure 1 and Table II]. In the MP-Be DnaB intein, the B:10 His65 was required for both N- and C-terminal reactions.5 Asp is not commonly found at B:7 although it is present in a few other inteins.1–5 Mutation of Asp61 to Ala inhibited reactions at the N-terminal splice junction reducing the amount of spliced products and increasing the amount of C-terminal cleavage products [Fig. 4(B) and Table II]. Asp61 could be replaced by Ser or Thr (residues normally present at this position in Class 1 and 2 inteins) with minimal effects on splicing (Supporting Information Fig. 1 and Table II). These results are markedly different than those observed with the MP-Be DnaB intein, where substitution of Ser62 (B:7) by a residue unlikely to assist catalysis (Ala) had no detectable effect.5 The modeled structure of the MP-Be DnaB intein indicates that the Ser62 hydroxyl forms a hydrogen bond with the Pro1 amide nitrogen.5 This interaction likely helps to stabilize the position of Pro1 for catalysis. However, Pro has a much lower degree of flexibility than other amino acids and, therefore, might not need the assistance of Ser62 to maintain proper alignment at the intein active site, thus explaining why loss of the B:7 residue did not significantly decrease splicing of the MP-Be DnaB intein. Crystal and NMR structures reveal that the B:7 residue also plays a key role in positioning the intein N-terminal splice junction residues for catalysis in Class 1 and Class 2 inteins.13,16,17 These same structures suggest a catalytic role for the B:7 residue in some inteins.13,17 The diversity of effects caused by mutation of the B:7 residue thus reflects different functional roles for this residue in individual inteins. The Dra Snf2 intein uses positions B:7, B:10, and B:12 to conformationally minimize and chemically activate the N-terminal splice junction, expanding the repertoire of catalytically important residues in Class 3.
Table II.
The Effects of Mutating Conserved Dra Snf2 Intein Block B and Block F Residues
| Residue | Mutation | Temp (°C) | Splice (MP+I) | Precursor (MIP) | N-cleavea (M+IP) | C-cleavea (MI+P) |
|---|---|---|---|---|---|---|
| Block B | ||||||
| Asp61 | Ala | 37 | ++++ b | |||
| Ala | 15 | ++ | ++ | |||
| Ser | 37 | +++ | ++ | |||
| Ser | 15 | ++++ | ||||
| Thr | 15/37 | ++++ | ||||
| His64 | Ala | 37 | ++++ | |||
| Ala | 15 | ++ | +++ | |||
| Asn | 37 | ++++ | ||||
| Asn | 15 | ++ | +++ | |||
| Trp66 | Ala | 15/37 | ++++ | |||
| Tyr | 37 | ++++ | ||||
| Tyr | 15 | ++++ | ||||
| Block F | ||||||
| Cys323 | Ala | 15/37 | + | ++++ | ||
| Asp | 37 | ++++ | ||||
| Asp | 15 | ++++ | ||||
| Thr | 37 | ++++ | ||||
| Thr | 15 | ++++ | ||||
| Ser | 37 | +++ | ++ | |||
| Ser | 15 | ++++ | ||||
| Gly | 15/37 | + | ++++ | |||
| Glu | 37 | ++++ | ||||
| Glu | 15 | ++++ | ||||
The Class 3 WCT triplet Trp66 residue was also crucial for splicing in the Dra Snf2 intein, as substitution by Ala yielded MIP precursor at both temperatures (Table II). However, the more conservative substitution of Trp66 to Tyr spliced to completion at 15°C but yielded precursor at 37°C (Supporting Information Fig. 1 and Table II). Such temperature dependent enzyme activity generally suggests that the mutation caused conformational instability or effected protein folding. Lower temperatures are thought to improve reactivity by stabilizing the protein structure. The B:12 Trp participated in hydrophobic packing with surrounding residues in the modeled MP-Be DnaB intein structure, which improves conformational stability.5 The B:12 Trp might also activate N-terminal cleavage by participating in a cation-π interaction with the Block B His that could stabilize the positive charge generated when this His is protonated during the splicing reaction.5 The Dra Snf2 intein data support these hypotheses.
According to the splicing mechanism proposed for the prototypic MP-Be DnaB intein, the F:4 Cys should be essential in all Class 3 inteins. The following Cys323 (F:4) substitutions were made in the Dra Snf2 intein: Ala, Asp, Thr, Ser, Gly, and Gln [Fig. 4(B), Supporting Information Fig. 2 and Table II]. These substitutions completely block splicing and N-terminal cleavage. However, C-terminal cleavage was observed at 15°C for all mutants, whereas only Ser, Ala, and Gly mutants resulted in C-terminal cleavage at 37°C. Similar results were obtained with the MP-Be DnaB intein,5 although the degree of C-terminal cleavage was greater in the Dra Snf2 intein F:4 mutants.
Characterization of the branched intermediate that accumulates in the Asn343Ala and His342Ala mutants
Mutation of Asn343 to Ala blocks branch resolution by C-terminal cleavage and results in BI accumulation [Fig. 4(C)]. The slowly migrating BI band yielded two residues per cycle of Edman degradation that correspond to the N-terminus of the N-extein (M) and the intein (I). It also reacted with antisera directed against the MBP and paramyosin exteins (data not shown). After affinity purification of the Asn343Ala BI sample over Amylose resin, mass spectroscopy indicated that the slowly migrating band had the same molecular mass as the linear precursor. These results confirm that the slowly migrating band is a BI.
When an intein BI accumulates, the (thio)ester linkage is usually alkaline labile at elevated temperatures.5,18–20 The affinity purified Asn343Ala BI was incubated overnight in vitro at 4°C, room temperature, 30°C or 37°C, pH 6 or 9, and in the presence or absence of 50 mM DTT [Fig. 5(A) and data not shown]. The Asn343Ala BI was stable under all conditions at pH 6 and stable at 4°C or room temperature at pH 9. The Asn343Ala BI was labile at 30 and 37°C at pH 9 independent of DTT, as demonstrated by its decay to M + IP. The Dra Snf2 intein Block G BI has an ester linkage at its Thr+1 branch point, but decay of this BI was stimulated by DTT, as was the case with the MP-Be DnaB intein. Under these mild conditions, DTT should not enhance cleavage of an ester bond. However, DTT dependent decay of the ester-linked Block G BI (III) would occur if it were in equilibrium with the DTT sensitive thioester-linked Block F BI (VIII) (Fig. 2). Increased breakdown of Class 3 Block G BIs in response to thiols and interconversion between the two types of BI is thus a common feature of Class 3 inteins.
Figure 5.
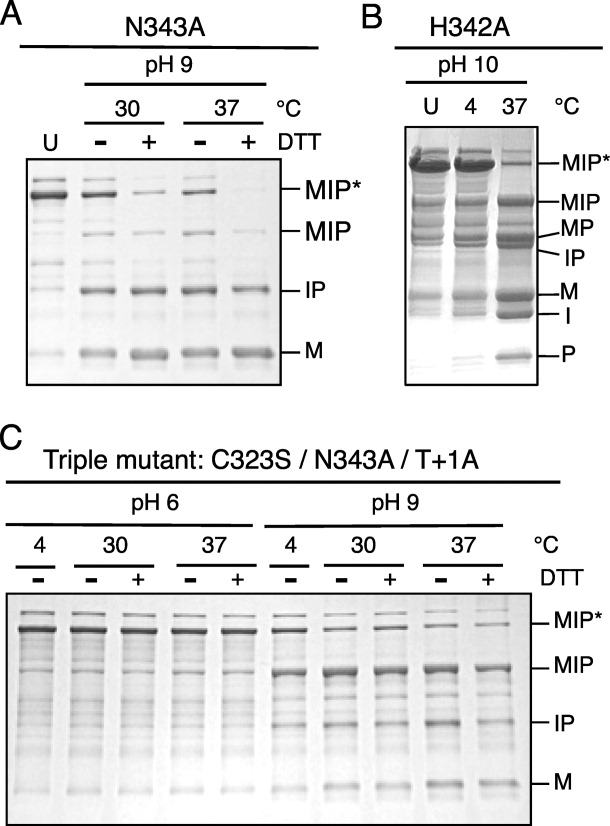
Analysis of Dra Snf2 intein branched intermediates in vitro. Branched intermediates (MIP*) accumulated in three different Dra Snf2 intein mutants: Part A, the C-terminal Asn343 was mutated to Ala, Part B, the penultimate His342 was mutated to Ala, and Part C, a triple mutant (Cys323Ser/ Asn343Ala/ Thr+1Ala). Samples were purified by affinity chromatography on Amylose resin and adjusted to pH 6, 9, or 10. Samples were then incubated overnight at the indicated temperatures to examine the breakdown of the BI (thio)ester linkage to yield M + IP in response to pH, temperature or DTT. The time zero sample has a “U” above the lane. Samples were electrophoresed in SDS-PAGE and stained with Coomassie blue. The Asn343Ala and triple mutants were incubated in presence (+) or absence (−) of 50 mM DTT; the His342Ala mutant was incubated in presence of 50 mM DTT. Abbreviations as in Figure 4.
A BI also accumulated when the intein penultimate His342 was mutated to Ala [Fig. 4(B)]. The His342Ala BI was stable at pH 5 when incubated overnight at 4 and 37°C (data not shown). Unlike the BIs from the Asn343Ala and the triple mutant (see below), the His342Ala Block G BI can proceed forward to form spliced products. The ester linkage can also be cleaved yielding M + IP or the Block G BI can revert back to the Block F BI, which can revert back further to the linear precursor. All of these reactions occurred when purified His342Ala BI was incubated overnight at 37°C, pH 10 yielding MIP, MP + I, M + IP, and M + I + P [Fig. 5(B)]. These off-pathway and reverse reactions are consistent with a reduced rate of Asn cyclization in the absence of His342. The presence of double cleavage products (M + I + P) indicates that either Cys323 can cleave the N-terminal splice junction in MI after C-terminal cleavage has occurred or Asn cyclization occurred after the ester bond in the BI was cleaved, since MP forms if Asn cyclization occurs before cleavage of this ester bond. It is unlikely that M and P formed from the ester linked MP (V) as the spontaneous [O/S]-N acyl rearrangement that forms the peptide bond between the exteins is very rapid.21 Reversion of the Block G BI to MIP at pH 10 was also observed in the Pyrococcus GB-D Pol intein and is, therefore, a common feature of all classes of inteins.20
Another difference between the MP-Be DnaB intein and the Dra Snf2 intein was seen in the role of their penultimate His residues. Mutation of the MP-Be DnaB intein penultimate His340 had only a minimal effect on splicing or C-terminal cleavage.5 This difference may relate to the observation that the Block B His assists C-terminal cleavage in the MP-Be DnaB intein but not in the Dra Snf2 intein. In the MP-Be DnaB intein, the Block B His may replace or complement the facilitating interaction normally provided by the penultimate His.
Trapping the Block F branched intermediate
Historically, intein BIs accumulate when the branch point is an ester rather than a thioester,18,19 so a triple mutant consisting of Cys323Ser, Asn343Ala, and Thr+1Ala was constructed. A slowly migrating band accumulated in this triple mutant [Fig. 4(C)]. This band yielded two residues per cycle of Edman degradation, which corresponded to the N-terminus of the N-extein (M) and the intein (I). It also reacted with antisera directed against the extein domains (data not shown). Molecular weight determination by mass spectroscopy revealed that it had the same mass as the linear precursor. As Thr+1 was mutated to Ala, this band could not be the Block G BI (III). Instead, the triple mutation captured the elusive Block F BI (VIII). The strategy of mutating Cys323 to Ser probably trapped the Block F BI in the Dra Snf2 intein triple mutant because the ester, once formed, is chemically stable under mild conditions while a thioester is not. Similar results were observed in a Class 1 intein, where Cys+1 did not permit BI accumulation, but Cys+1Ser did permit accumulation.19
The triple mutant BI was purified over Amylose resin and then incubated in vitro in the presence or absence of 50 mM DTT at pH 6 or 9 and at 4, 30 or 37°C [Fig. 5(C) and data not shown]. The Block F BI from the triple mutant was stable at pH 6. However at pH 9, reversion of the triple mutant Block F BI back to MIP occurred at all temperatures tested, including 4°C. DTT treatment of the triple mutant Block F BI had no detectable effect on breakdown of its ester linkage, further indicating that no other thioester intermediate exists when the F:4 Cys323 is mutated.
An essential characteristic of Class 3 inteins is the absolute requirement for splicing of a Cys at position 4 in Block F because this Cys serves as the nucleophile that initiates the splicing reaction. Although Ser could substitute for this Cys to form the Block F BI in the triple mutant, the relative rate of BI formation to Asn cyclization was too slow to allow splicing in the Cys323Ser single mutant where C-terminal cleavage dominated.
Materials and Methods
Cloning, mutagenesis, and protein expression
The MIP precursor consists of the 343 amino acid Dra Snf2 intein surrounded by the E. coli maltose-binding protein (MBP or M) as the N-extein and the ΔSal fragment of D. immitis paramyosin (P) as the C-extein.15 In addition, the intein was flanked by three native Dra Snf2 residues (Leu-Gly-Lys) at the N-terminal splice junction and Thr-Val-Gln-Thr instead of the native Dra Snf2 C-extein residues Thr-Leu-Gln-Thr at the C-terminal splice junction. Amino acid substitutions were introduced by site-directed mutagenesis using the Phusion Site-Directed Mutagenesis Kit (New England BioLabs, Ipswich, MA) or the Quikchange Kit (Stratagene, La Jolla, CA) as described by the manufacturers. Each clone was sequenced by the New England BioLabs DNA sequencing core facility.
Protein expression was induced with 1 mM IPTG in T7 Express cells (New England BioLabs, Ipswich, MA) at an OD600 of 0.6–0.7 for 2 h at 37°C and then harvested (37°C sample) or further induced for 20 h at 15°C (15°C sample). Cell pellets were lysed by sonication in Buffer A (20 mM Na2HPO4, pH 7, with 0.5 M NaCl). The protein splicing activities of mutant constructs were assessed by SDS-PAGE of soluble cell lysates and by Western blot analysis with anti-MBP or anti-Paramyosin sera as described previously.5 The 10–250 kDa Protein Ladder (250, 150, 100, 80, 60, 50, 40, 30, 25, 20, 15, 10 kDa, where 80 and 25 kDa are darker bands) was used as the size standard (New England BioLabs, Ipswich, MA). Selected samples were purified by affinity chromatography over Amylose resin in Buffer A at pH 7 or pH 6 and eluted with 10 mM Maltose in Buffer A at the same pH (New England BioLabs, Ipswich, MA). Soluble lysates or purified protein were boiled for 5 min in SDS-PAGE sample buffer with 5 mM DTT (New England BioLabs, Ipswich, MA), loaded onto 10% or 10%–20% Tris-Glycine SDS-PAGE gels (Invitrogen, Carlsbad, CA) and either stained with Coomassie Blue or transferred to nitrocellulose for Western blot analysis. The Western blots helped to distinguish between MI, MP, and IP, which have a similar relative mobility.
Investigation of branched intermediates
Amylose purified samples of Block F and Block G BIs were incubated overnight in vitro at various temperatures, pH values, and plus or minus 50 mM DTT. The pH of BI samples was changed by addition of pH 5 or pH 10 sodium phosphate buffer. Asn343Ala samples were incubated at 4°C, room temperature or 37°C at pH 6 or 9. Triple mutant samples were incubated at 4, 30, and 37°C at pH 6 or 9. His342Ala samples were incubated at 4°C, room temperature or 37°C at pH 6 or 10. Time zero samples before incubations were directly boiled in SDS-PAGE sample. To prevent decay of the BI in pH 9 samples during preparation for electrophoresis, the pH of these samples was adjusted to pH 6.5–7 immediately before heating by adding an equal volume of 0.5 M sodium phosphate buffer (pH 4.3). Samples including BIs from the Asn343Ala and the triple mutant had their mass determined by electrospray ionization time-of-flight mass spectroscopy (ESI-TOF MS) and were subjected to N-terminal sequencing as described.5
Conclusions
This study generalizes the properties of Class 3 inteins beyond the MP-Be DnaB intein. The Dra Snf2 intein uses positions B:7, B:10, and B:12 to conformationally minimize and chemically activate the N-terminal splice junction and the penultimate His to activate C-terminal cleavage. Compared to the Class 3 prototype, the MP-Be DnaB intein, the Dra Snf2 intein expands the repertoire of catalytically important Class 3 intein residues to include B:7 and the penultimate His. More importantly, this study proves the existence of the Block F BI (VIII), thus confirming the dual BI mechanism proposed for Class 3 inteins. Our understanding of protein splicing mechanisms has led to a multitude of intein-based applications. Understanding this new intein-mediated protein splicing mechanism can further these pursuits. Variations in protein splicing mechanisms are more easily tolerated because the enzyme and substrate are linked in one precursor molecule. Therefore, inteins may provide a window into the process of converting one enzyme active site to another, which is more difficult in standard enzyme systems. Inteins continue to fascinate us and provide mechanistic surprises.
Acknowledgments
The authors thank Manoj Cheriyan (NEB), Jack Benner (NEB), and Maggie Johnson (University of Alberta) for helpful discussions, Casey Madinger (NEB) and Jack Benner for mass spectroscopy determinations, and Don Comb (NEB) for support and encouragement.
Statement for Broader Audience: Inteins mediate self-excision from precursors using the same tricks that all enzymes use. This article analyzes the second member of intein Class 3, generalizing the properties of the prototype. It further proves the proposed Class 3 splicing mechanism that includes two branch intermediates by capturing the class specific branch intermediate for the first time. Novel inteins provide tools for understanding enzyme function and for biotechnology applications such as protein semisynthesis.
References
- 1.Perler FB. InBase: the intein database. Nucl Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pietrokovski S. Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 1994;3:2340–2350. doi: 10.1002/pro.5560031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietrokovski S. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 1998;7:64–71. doi: 10.1002/pro.5560070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perler FB, Olsen GJ, Adam E. Compilation and analysis of intein sequences. Nucl Acids Res. 1997;25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tori K, Dassa B, Johnson MA, Southworth MW, Brace LE, Ishino Y, Pietrokovski S, Perler FB. Splicing of the Mycobacteriophage Bethlehem DnaB inteidentification of a new mechanistic class of inteins that contain an obligate block F nucleophile. J Biol Chem. 2010;285:2515–2526. doi: 10.1074/jbc.M109.069567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulus H. Inteins as enzymes. Bioorg Chem. 2001;29:119–129. doi: 10.1006/bioo.2001.1203. [DOI] [PubMed] [Google Scholar]
- 7.Mills KV, Perler FB. The mechanism of intein-mediated protein splicing: variations on a theme. Protein Pept Lett. 2005B;12:751–755. doi: 10.2174/0929866054864337. [DOI] [PubMed] [Google Scholar]
- 8.Hodges RA, Perler FB, Noren CJ, Jack WE. Protein splicing removes intervening sequences in an archaea DNA polymerase. Nucl Acids Res. 1992;20:6153–6157. doi: 10.1093/nar/20.23.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amitai G, Dassa B, Pietrokovski S. Protein splicing of inteins with atypical glutamine and aspartate C-terminal residues. J Biol Chem. 2004;279:3121–3131. doi: 10.1074/jbc.M311343200. [DOI] [PubMed] [Google Scholar]
- 10.Mills KV, Manning JS, Garcia AM, Wuerdeman LA. Protein splicing of a Pyrococcus abyssi intein with a C-terminal glutamine. J Biol Chem. 2004;279:20685–20691. doi: 10.1074/jbc.M400887200. [DOI] [PubMed] [Google Scholar]
- 11.Mills KV, Dorval DM, Lewandowski KT. Kinetic analysis of the individual steps of protein splicing for the Pyrococcus abyssi PolII intein. J Biol Chem. 2005;280:2714–2720. doi: 10.1074/jbc.M412313200. [DOI] [PubMed] [Google Scholar]
- 12.Southworth MW, Benner J, Perler FB. An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. EMBO J. 2000;19:5019–5026. doi: 10.1093/emboj/19.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson MA, Southworth MW, Herrmann T, Brace L, Perler FB, Wuthrich K. NMR structure of a KlbA intein precursor from Methanococcus jannaschii. Protein Sci. 2007;16:1316–1328. doi: 10.1110/ps.072816707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki M, Nogami S, Satow Y, Ohya Y, Anraku Y. Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. A random mutagenesis study of the entire Vma1-derived endonuclease sequence. J Biol Chem. 1997;272:15668–15674. doi: 10.1074/jbc.272.25.15668. [DOI] [PubMed] [Google Scholar]
- 15.Southworth MW, Perler FB. Protein splicing of the Deinococcus radiodurans strain R1 Snf2 intein. J Bacteriol. 2002;184:6387–6388. doi: 10.1128/JB.184.22.6387-6388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizutani R, Nogami S, Kawasaki M, Ohya Y, Anraku Y, Satow Y. Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA1-derived endonuclease bearing the N and C-terminal propeptides. J Mol Biol. 2002;316:919–929. doi: 10.1006/jmbi.2001.5357. [DOI] [PubMed] [Google Scholar]
- 17.Klabunde T, Sharma S, Telenti A, Jacobs WR, Jr, Sacchettini JC. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat Struct Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 18.Xu MQ, Comb DG, Paulus H, Noren CJ, Shao Y, Perler FB. Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong S, Williams KS, Wotkowicz C, Xu MQ. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J Biol Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- 20.Xu MQ, Southworth MW, Mersha FB, Hornstra LJ, Perler FB. In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 21.Shao Y, Paulus H. Protein splicing: estimation of the rate of O-N and S-N acyl rearrangements, the last step of the splicing process. J Pept Res. 1997B;50:193–198. doi: 10.1111/j.1399-3011.1997.tb01185.x. [DOI] [PubMed] [Google Scholar]


