Abstract
The emergence of drug-resistant malaria parasites is the major threat to effective malaria control, prompting a search for novel compounds with mechanisms of action that are different from the traditionally used drugs. The immunosuppressive drug FK506 shows an antimalarial activity. The mechanism of the drug action involves the molecular interaction with the parasite target proteins PfFKBP35 and PvFKBP35, which are novel FK506 binding protein family (FKBP) members from Plasmodium falciparum and Plasmodium vivax, respectively. Currently, molecular mechanisms of the FKBP family proteins in the parasites still remain elusive. To understand their functions, here we have determined the structures of the FK506 binding domain of Plasmodium vivax (PvFKBD) in unliganded form by NMR spectroscopy and in complex with FK506 by X-ray crystallography. We found out that PvFKBP35 exhibits a canonical FKBD fold and shares kinetic profiles similar to those of PfFKBP35, the homologous protein in P. falciparum, indicating that the parasite FKBP family members play similar biological roles in their life cycles. Despite the similarity, differences were observed in the ligand binding modes between PvFKBD and HsFKBP12, a human FKBP homolog, which could provide insightful information into designing selective antimalarial drug against the parasites.
Keywords: FK506 binding proteins, peptidylprolyl cis-trans isomerase, Plasmodium vivax, NMR spectroscopy, X-ray crystallography
Introduction
Malaria is a still threatening disease in many developing countries, responsible for almost 2 million deaths annually.1 Human malaria is caused by infection with one of four related parasitic protozoan species: Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae which are transmitted by the female Anopheles gambiae mosquito vector, where P. falciparum and P. vivax are two major malaria causing species.
P. vivax is the major cause of malaria outside Africa, mainly in Asia and the Americas and is responsible for one third of total malaria cases that occur worldwide each year.2 The parasite has been neglected for a long time due to the lack of a system that would allow the continuous propagation of the parasite in the laboratory except in non-human primates.3 Despite its low fatality rate, P. vivax is associated with severe and chronic clinical symptoms and often causes relapses months after a primary infection has cleared.4 Moreover, the emerging resistance to currently available antimalarial drugs in P. vivax prompt a search for novel therapeutics against the parasite.5
The immunosuppressive drug FK506 is a 23-membered macrolide lactone that binds to the peptidylprolyl cis-trans isomerase (PPIase) domain (or referred to as FK506 binding domain or FKBD) of FK506-binding proteins (FKBPs).6 FKBPs constitute a large family of conserved proteins across many organisms and assist protein folding by catalyzing the conversion of cis- and trans- rotamers of the peptidylprolyl amide bond of protein substrates. They are also involved in other cellular processes such as transcription, translation, cell cycle, and protein assembly.7 Although binding of FK506 to FKBPs results in the inhibition of their PPIase enzymatic activity, its immunosuppressive activity has been demonstrated to occur for the human FKBP12 protein (HsFKBP12) through an indirect mechanism that involves calcineurin, a Ca2+/calmodulin-dependent phosphatase. HsFKBP12/FK506 binary complex binds to calcineurin and inhibits the dephosphorylation of the NF-AT transcription factor, impairing its translocation from the cytoplasm to the nucleus and leading to a temporary suppression of T- and B-cell functions.8,9 Thus, FK506 and a new generation of its synthetic analogues are routinely prescribed to transplant recipients to prevent graft rejection. Apart from their immunosuppressive actions, other clinical effects of these drugs may involve alternative mechanisms, indicating that FK506 has potentially multiple targets in various tissues and organisms. Using P. falciparum parasite culture, FK506 was shown to have anti-malarial activity.10 The search for molecular targets of FK506 in the parasite led to the identification and extensive characterization of PfFKBP35, a member of the FKBP family in P. falciparum,11–15 which is also conserved in P. vivax, named as PvFKBP35.
Like PfFKBP35, PvFKBP35 is a 35 kDa protein with an N-terminal FK506-binding domain, a tripartite tetratricopeptide repeats (TPR) domain, and a Ca2+/calmodulin-binding module (CaM) at its C-terminus. We have already shown that TPR domain of PfFKBP35 binds to C-terminal pentapeptide of PfHsp9016 and found that it shows similar interaction pattern like Hop (Hsp organizing protein) and PP5 (Serine/threonine-protein phosphatase) protein but different from FKBP52. Here, we have studied FK506 binding domain of Plasmodiun vivax with a view to assist the design of new class of FK506 analogues or novel inhibitor molecules that would be both more specific towards the parasite enzyme and also devoid of immunosuppressive activity, in this study we have determined the free FKBD of PvFKBP35 (referred to as PvFKBD) by solution NMR spectroscopy and in complex with FK506 by X-ray crystallography, respectively. We also examined its PPIase activity profile, binding mode with FK506 and the effect of PvFKBD/FK506 complex on calcineurin inhibition. We have compared structural and biochemical properties of PvFKBD with HsFKBP12 which is having only one FK506 binding domain unlike HsFKBP52, which contains two copies of FK506 binding domains one active (FKBD1) and the other inactive (FKBD2), showing unique catalytic and calcineurin inhibitory profiles.17 Therefore, HsFKBP12 provides a good comparison model to compare PvFKBP35 which also contains one FKBD. Our NMR structure of PvFKBD showed that this is a canonical FKBP with similar structural fold like other FKBPs.18 Crystal structure of PvFKBD in complex with FK506 revealed that PvFKBD shows tight binding to FK506 like PfFKBD,12 the homologous FKBD domain from P. falciparum (referred to as PfFKBD) and their modes of interaction with FK506 were similar. However, differences in the ligand binding mode were seen between HsFKBP12 and PvFKBD. Biochemical studies of both parasites FKBDs showed canonical rotamase activity and FKBD/FK506-mediated calcineurin inhibitory profiles. These results indicate that the parasite FKBP family members play a similar biological role in the Plasmodium parasites, and the differences observed between the binding pockets of HsFKBP12 and parasites FKBD might provide insights into designing a novel non-immunosuppressive antimalarial drug against these two parasites.
Results and Discussion
NMR solution structure of PvFKBD
The NMR solution structure was determined based on a total of 2734 nontrivial distance constraints (Table I). The ensemble of 20 low-energy structures is shown in Figure 1(A). The average root-mean-square-deviation (RMSD) values relative to the mean coordinate of 20 representative conformers were 0.58 ± 0.11 Å for the backbone atoms and 0.99 ± 0.09 Å for heavy atoms for residues 5–125. PvFKBD shares the canonical FKBP fold.11,20 There are no distance violations greater than 0.3 Å or dihedral-angle violations greater than 5°. The structure of PvFKBD consists of six antiparallel β-strands and a short central α-helix [Fig. 1(B)]. Overall the active site pocket of PvFKBD resembles those of PfFKBD and HsFKBP12 and contains the conserved aromatic residues (Y43, W77, and F117) that are involved in binding of FK506. Overlay of N, C′ and Cα atoms showed that backbone RMSDs between HsFKBP12 and PvFKBD, HsFKBP12 and PfFKBD, PfFKBD and PvFKBD are 1.3 Å, 1.5 Å, 1.5 Å, respectively [Fig. 1(C)]. Although PfFKBD and PvFKBD show similar topology, differences were observed in the β3/β4 and β5/β6 loop regions.
Table I.
Structural Statistics of PvFKBD
| Crystallographic structural statistics |
| Data collection |
| Space group: P 1 21 1 |
| Unit cell |
| a, b, c (Å): 55.9, 40.9, 56.9 |
| α, β, γ (°): 90.0, 109.5, 90.0 |
| Resolution (Å): 21.22–1.66 (1.75–1.66)a |
| Completeness: 98.4 (90.8)a |
| Redundancy: 4.9 (4.6)a |
| No. of measured reflections: 137829 |
| No. of unique reflections: 28288 |
| I/σ(I): 30.4 (8.2)a |
| Rmerge (%): 2.9 (15.0)a |
| Refinement |
| No. of reflections (working set/test set): 26851/1425 |
| R factor (Rwork/Rfree): 0.188/0.233 |
| No. of atoms |
| Protein/ligand/water: 1910, 57, 261 |
| Average B factors (Å2) |
| Protein/ligand/water: 19.11, 15.67, 34.80 |
| Average R.M.S. deviations |
| Bond lengths (Å): 0.014 |
| Bond angles (°): 1.502 |
| Ramachandran statistics (%)b |
| Most favored: 94.6 |
| Additionally allowed: 5.4 |
| Generously allowed: 0.8 |
| Disallowed: 0.0 |
| NMR structural statistics |
| Experimental restraints |
| Unambiguous NOE restraints |
| Total: 2434 |
| Short range, |i–j|<1: 1304 |
| Medium range, 1<|i–j|<5: 288 |
| Long range, |i–j|>=5: 842 |
| Dihedral angle restrains |
| All: 102 |
| ϕ: 51 |
| Ψ: 51 |
| Hydrogen bond restrains: 72 |
| No. of violations |
| Restraint violations > 0.3 Å: 0 |
| Dihedral angle violations > 5°: 0 |
| Average R.M.S. deviations (Å) |
| Backbone atoms: 0.58 ± 0.11 |
| All heavy atoms: 0.99 ± 0.09 |
| Ramachandran statistics (%)bc |
| Most favored: 82.3 |
| Additionally allowed: 17.3 |
| Generously allowed: 0.4 |
| Disallowed: 0.0 |
Values in parentheses refer to the corresponding values of the highest resolution shell.
The geometry of the final model was checked with PROCHECK.19
Average RMSD to mean for top 20 structures and Ramachandran plot were for residues 5–125.
Figure 1.
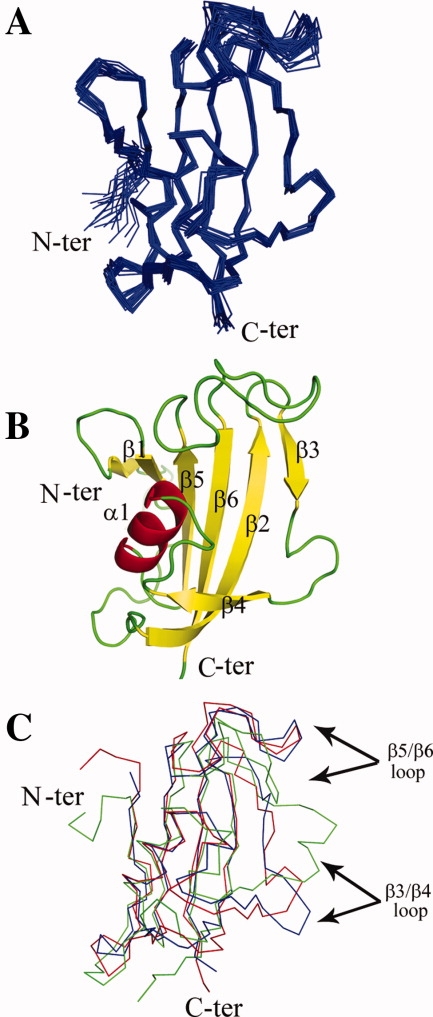
NMR solution structure of PvFKBD. (A) Backbone ensemble of 20 conformers with lowest energy is shown superposed. (B) Ribbon drawing of FKBD of PvFKBP35 (PDB ID: 2KI3). (C) An overlay of the backbone heavy atom trace (N, C′, Cα) of the NMR structures of HsFKBP12 (Blue), PfFKBD (green), PvFKBD (Red) is shown. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Crystal structure of PvFKBD-FK506 complex
The crystal structure of PvFKBD complexed with FK506 afforded a comparison with the NMR solution structure in free form and a closer look at the mode of FK506 binding in the active site. A total of 242 residues, one FK506 molecule and a total of 261 water molecules were included in the final model which was refined to R and Rfree values of 18.8% and 23.3%, respectively, at a resolution of 1.66 Å against data collected from a single native crystal (Table I). Both molecules in the asymmetric unit are nearly identical in structure and related to each other by a non-crystallographic two-fold axis, superimposing with a root mean square deviation of 0.40 Å for 121 α-carbon atoms. Two monomers of PvFKBD assemble to form a covalent dimer by one intermolecular disulfide bond formed between residues C105. Some other hydrophobic and hydrophilic interactions are formed between loops β3-β4 and β5-β6 (Supporting Information Fig. S1A). Hydrogen bonds between main chain amide and carbonyl oxygen atoms involve residues G104 and G106 of one PvFKBD molecule and residues G106 and G104 of the other molecule (Supporting Information Fig. S1B). Side chain of H67 also makes hydrogen bond with main chain of R60 (Supporting Information Fig. S1B). But we noticed that in solution and in dissolved crystals there is no formation of disulfide bond, indicating that the disulfide bond is a crystallization artifact. FK506 binds in a shallow cavity between the α-helix and the β-sheet. Loops comprising residues 56–63, 69–74, and 95–114 flank the binding pocket [Fig. 2(A)]. The binding pocket is lined with conserved hydrophobic residues Y43, F64, V73, I74, W77, Y100, and F117. The indole ring of the strictly conserved W77 in the α-helix serves as the platform for the pipecolinyl ring of FK506 [Fig. 2(B)]. The buried surface of 655 Å2 is comparable to the value found for FK506 in complex with FKBD domain of P. falciparum12 [Fig. 3(A)]. The hydrogen bonds observed in the interaction between PfFKBD and FK506 were also observed in the complex between PvFKBD and FK506 (Table II). Overall PvFKBD reveals a fold similar to that of PfFKBD. Pairwise superimposition of binding pocket of PvFKBD and PfFKBD revealed that both proteins have similar interactions with FK506 [Fig. 3(B)]. Taken together, our data indicate that the Plasmodium parasite FKBP family members share similar molecular mechanism and biological roles in their life cycles.
Figure 2.
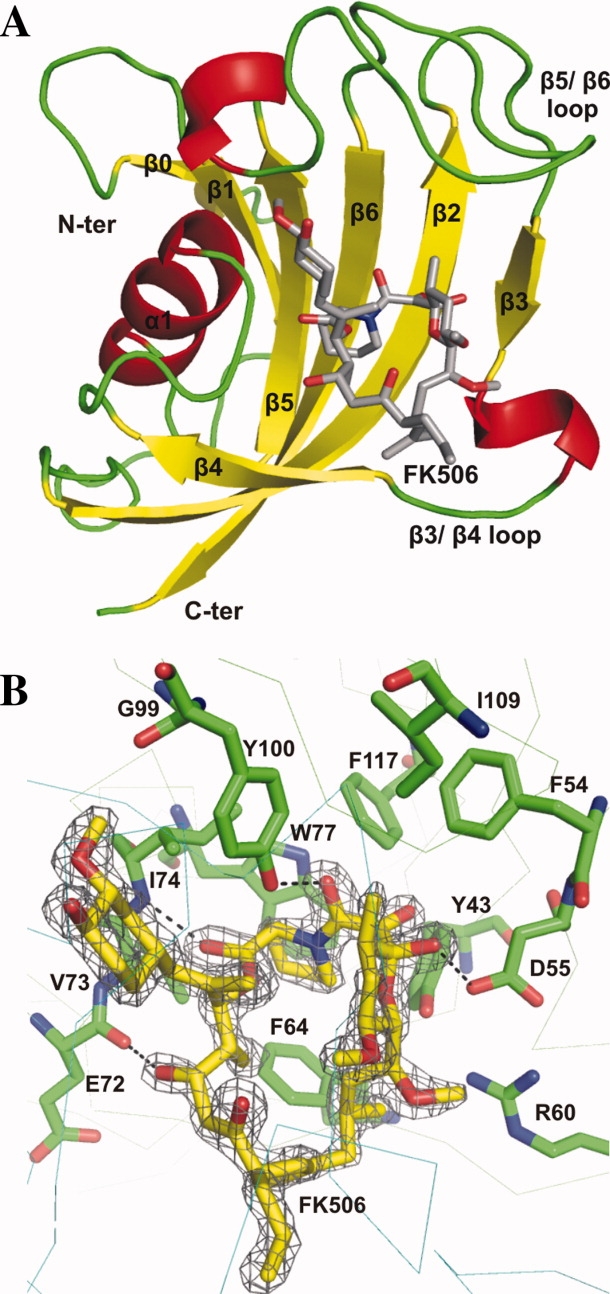
X-ray crystallographic structure of FKBD domain of P. vivax in complex with FK506. (A) The overall structure in complex with FK506 is shown as a ribbon diagram. FK506 ligand is shown with residue side chains depicted in a stick representation (carbon colored grey, nitrogen colored blue, and oxygen colored red). (B) The 2Fo-Fc electron density map encountered at the 1.0 σ level at the region of ligand FK506. The residues near the FK506 are labeled and displayed as stick model in element colors (green, C; blue, N; red, O). Hydrogen bonds are represented by black dashed lines. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 3.
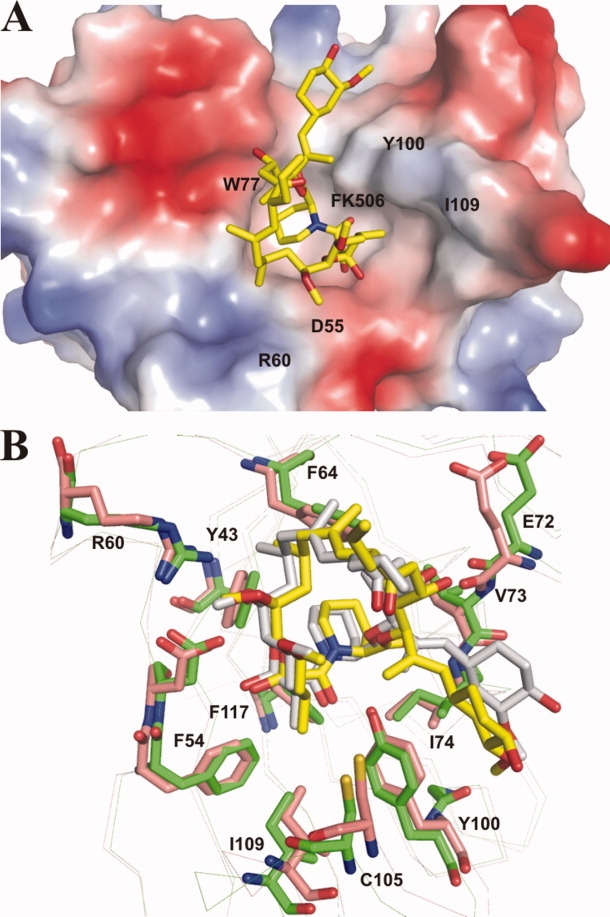
FK506 binding site of PvFKBD. (A) Electrostatic surface representation of the residues near the FK506 binding sites in PvFKBD. The position of bound FK506 is shown as stick model in element colors (yellow, C; blue, N; red, O). Positive and negative charges are in blue and red, respectively. (B) Superimposition of the FK506 binding site of PvFKBD and PfFKBD. Residues of PvFKBD and PfFKBD are labeled and colored in green and red respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table II.
Interactions between FK506 and PvFKBD
| Nonpolar contacts | |||
|---|---|---|---|
| PvFKBD |
|||
| FK506 region | Molecule | Residues | |
| C1 | A | V73, I74, Y100 | |
| C2-N7 | A | Y43, F64, V73, W77, Y100 | |
| C8-C9 | A | Y43, F54, D55, Y100, F117 | |
| C10-C14 | A | Y43, D55, Y100, I109 | |
| C15-C17 | A | Y43, D55, R60, F64 | |
| C18-C23 | B | Y43, D55, F64, W77 | |
| C24-C26 | A | F64, E72 | |
| C27-C34 | A | G49, Y100 | |
| B | C105, G106, E107, S108, I109 | ||
| Hydrogen bonds | |||
| Ligand | PvFKBD | Donor-acceptor distance (Å) | |
| C1 carbonyl | A | I74 N | 2.88 |
| C8 carbonyl | A | Y100 OH | 2.60 |
| C10 hydroxyl | A | D55 Oβ | 2.84 |
| C24 hydroxyl | A | E72 carbonyl | 3.33 |
| C32 hydroxyl | B | C105 carbonyl | 2.70 |
| C32 hydroxyl | B | S108N | 3.13 |
Ligand-induced conformational changes of PvFKBD
The RMSD value calculated by superimposition of the main chains of apoenzyme PvFKBD and FK506-bound PvFKBD is 1.40 Å for 121 α-carbon atoms. Most of the secondary structural elements of apoenzyme were superimposed in FK506-bound PvFKBD without major differences. However, differences were seen in the loops connecting strand β3–β4, and strand β5–β6 (Fig. 4). To examine the ligand-induced conformational changes, 2D 1H-15N HSQC NMR spectroscopy was performed in the absence and presence of FK506. As shown in Figure 5(A), chemical shift perturbations were observed in and around the active site pocket specially in Y43, D55, F64, V73, I74, W77, Y100, and F117 residues [Fig. 5(B)], suggesting that binding pocket is similar in solution and in the crystal [Fig. 5(C)]. Chemical perturbations were also found in the β3/β4 (S57, N61), α1/β5 (C80, V81), and β5/β6 loops (Y100, N112) [Fig. 5(A)], indicating that the conformational changes occurring in these regions are due to the complex formation (Fig. 4). In addition, the heteronuclear 1H-15N NOE measurements for the dynamics of backbone amide of PvFKBD suggest that some of residues are rigidified in the presence of FK506 [Fig. 6(A)], specifically residues present in β3/β4 loop (N61), α1/β5 (V81) loop, and β5/β6 loop (N112) [Fig. 6(B)], giving further evidence to support the conformational changes in PvFKBD upon the complex formation with FK506. But the ligand-induced conformational change of PvFKBD was not as significant as what was observed with PfFKBD/FK506 complex formation.12
Figure 4.
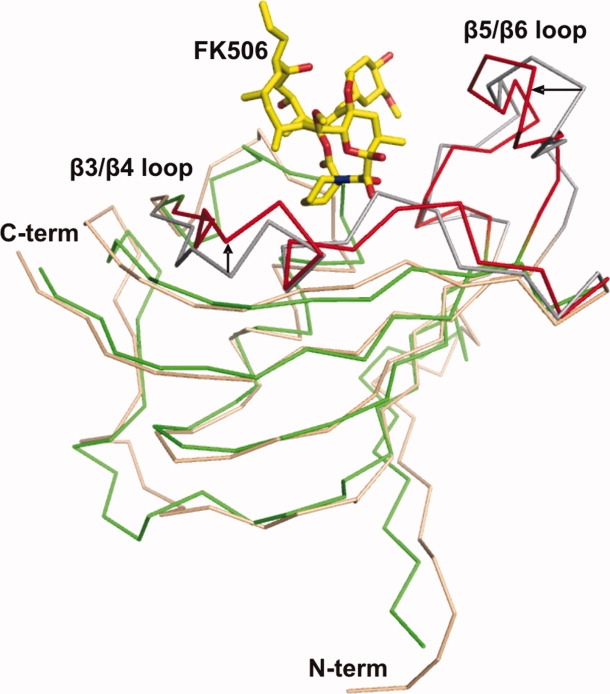
Superimposition of the backbone traces of apoenzyme PvFKBD colored in pink (β3-β4 and β5-β6 loops colored in gray) and FK506-bound PvFKBD colored in green (β3-β4 and β5-β6 loops colored in red). Regions showing differences are highlighted with arrows and labeled. FK506 ligand is shown as yellow sticks. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 5.
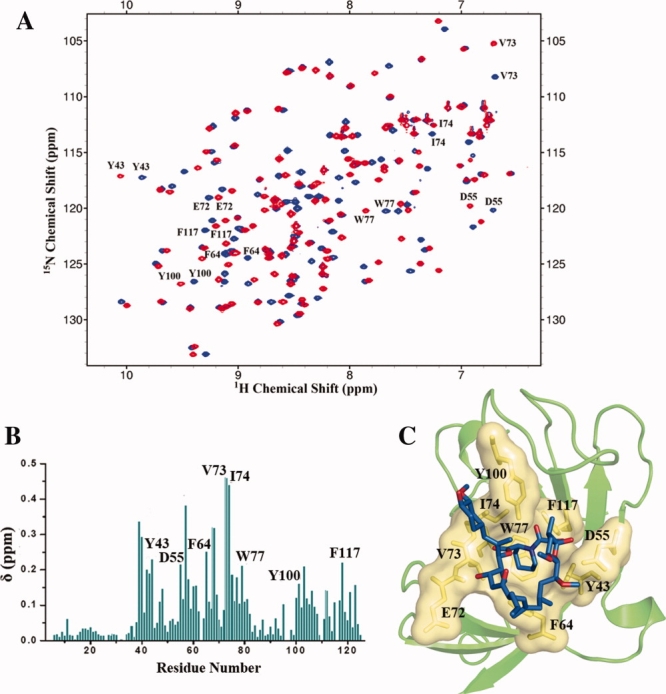
NMR chemical shift perturbation analysis of PvFKBD upon complex formation with FK506. (A) Overlay of 2D 1H-15N spectra of 15N- labeled PvFKBD in the absence (blue) and the presence (red) of non-labeled FK506 at a molar ratio (PvFKBD:FK506) of 1:2. (B) Averages of the 1H and 15N chemical shift changes on the two spectra in panel A, (δ) plotted against the residue number. (C) Mapping of the residues (yellow) located in and near binding pocket and involve in interaction with FK506 (shown as stick in blue colour) on the 3Dstructure of PvFKBD (Protein Data Bank entry 2KI3). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 6.
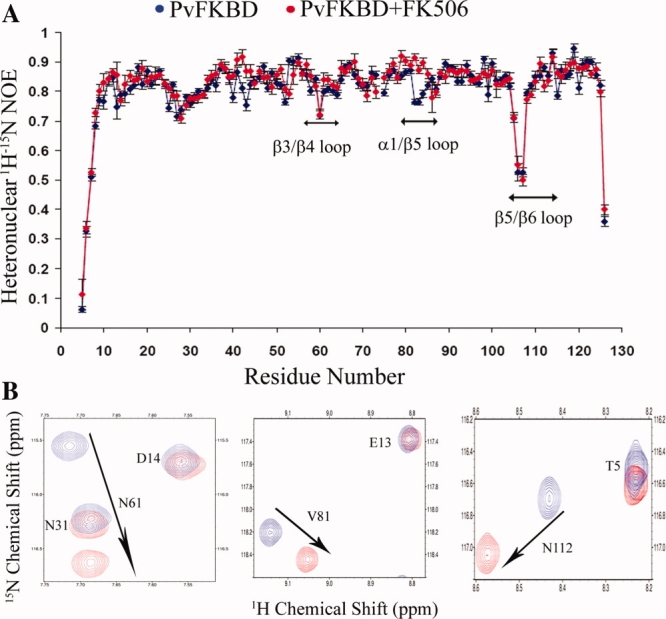
(A) Heteronuclear 1H-15N NOE for the backbone amides of PvFKBD. Backbone 1H-15N NOE values as functions of residue number for PvFKBD were measured. The NOE values for free PvFKBD and complexed with FK506 are colored blue and red, respectively. The regions showing backbone dynamics changes are labeled. (B) Sections of the regions β3/β4, α1/β5, and β5/β6 which show backbone dynamics changes upon FK506 binding. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Biochemical and structural comparison of PvFKBD and other FKBP family members
To assess the catalytic profiles of PvFKBP35 and other FKBP family members, the PPIase activity of PvFKBD was measured and compared with those of PfFKBD and HsFKBP12. Our results showed that HsFKBP12 exhibits highest catalytic efficiency followed by PvFKBD and PfFKBD (Table III). We then measured the inhibitory effect of FK506 on the PPIase activities of HsFKBP12, PfFKBD, and PvFKBD, showing corresponding IC50 values of 100 nM, 260 nM, and 160 nM, respectively (Table III). We also tested the inhibitory effect of PvFKBD/FK506 complex on the calcineurin activity and compared with those of HsFKBP12/FK506 and PfFKBD/FK506 complexes. Our results demonstrated that the FKBP family members complexed with FK506 inhibit calcineurin activity (Fig. 7) to a similar degree, suggesting that the FKBP members tested in this study share canonical profiles of the binary complex formation between the FKBPs and FK506 and subsequent suppression of the phosphatase activity of calcineurin upon ternary complex formation of FKBPs/FK506/calcineurin.
Table III.
PPIase Activity of HsFKBP12, PfFKBD, and PvFKBD and its Inhibition by FK506
| Protein | Kcat/Km (M−1S−1) × 105 | IC50 (nM) |
|---|---|---|
| HsFKBP12 | 3.1 | 100 |
| PfFKBD | 1.0 | 260 |
| PvFKBD | 1.5 | 160 |
Figure 7.
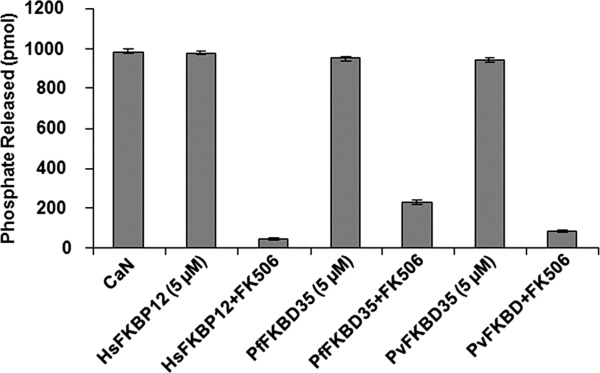
Inhibitory effect of PvFKBD on the phosphatase activity of calcineurin. Calcineurin-inhibitory activity by PvFKBD was examined in the absence or presence of FK506. For comparison, PfFKBD and HsFKBP12 were also used in the assay. The calcineurin activity was measured by observing the release of phosphate. In the assay, 5 μM of proteins and 10 μM of FK506 were used. The phosphatase activities represent averages of three independent experiments.
Some differences were observed between human and parasites chaperones in their ligand binding pockets. H87 and I90, which are present in the loop between β5 and β6 strands of HsFKBP12, are different from parasite FKBDs.12 These two hydrophobic residues are replaced with C105/C106 and S108/S109 in PvFKBD and PfFKBD, respectively. In HsFKBP12, H87 and I90 along with I91 form a hydrophobic pocket for methyl group present on the C11 position of FK506 and even form contacts with tetrahydropyran ring (Fig. 8). But in parasite's FKBDs such kind of interactions were not observed, suggesting that any small molecule or FK506 analogue which could target cysteine as well as serine residues and binds to binding pocket and devoid of immunosuppressive function would be specific for parasites proteins. Another approach would be introducing bulky/hydrophobic groups on C11 methyl group of FK506 and its derivatives may decrease the binding affinity to FKBP12 due to steric clashes with hydrophobic pocket formed by H87, I90, and I91.
Figure 8.
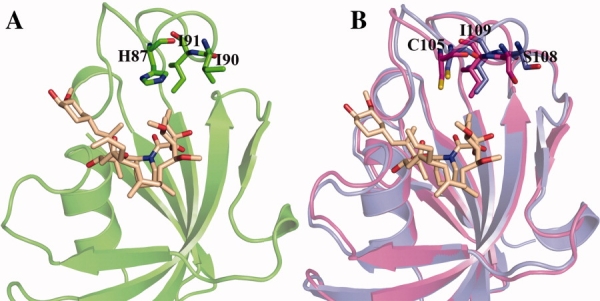
Structural differences in binding pockets of human and parasite FK506 binding domains. (A) The hydrophobic residues (H87, I90, and I91) on the loop between β5 and β6 which can interact with methyl group present on the C11 position of FK506 are shown. Human FKBP12 crystal structure with FK506 (PDBID: 1FKJ) was utilized to highlight the interaction pattern. (B) The variant amino acid residues in PvFKBD and PfFKBD (C105/106, S108/109, and I109/110, respectively) which replaces the H87, I90, and I91 of human FKBP12 are highlighted with sticks. Human FKBP12, PvFKBD, PfFKBD are displayed in green, blue, and magenta, respectively. FK506 ligand and the residues are indicated with sticks. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In conclusion, NMR solution structure of free PvFKBD and crystal structure in complex with FK506 reveal a canonical FKBD fold and ligand binding mode, which is similar to those of PfFKBP35. Despite the similarity, our structural data revealed minor differences between PvFKBD and HsFKBP12, suggesting that the high resolution crystallographic structural information gained in this study might provide insights into designing potentially non-immunosuppressive bifunctional antimalarial drugs against both P. falciparum and P. vivax by targeting either PvFKBP35 or PfFKBP35 alone.
Materials and Methods
Materials
FK506 was purchased from LC laboratories (Woburn, MA). Ni2+-nitriloacetic acid (NTA) was from Qiagen (Hilden, Germany). HiPrep 16/60 Sephacryl S-200 column was from GE Healthcare (Singapore). Isopropyl-thio-β-D-galactopyranoside (IPTG) was purchased from US Biological (Swampscott, MA). Protein molecular weight marker was from Bio-Rad Laboratories (Hercules, CA). Escherichia coli BL21 (DE3) cells and kanamycin were purchased from Invitrogen (Carlsbad, CA). The vector pSUMO was from Life Sensors (Malvern, PA). All other chemicals were purchased from Sigma–Aldrich (St. Louis, MO). Enzymes were from Roche (Lewes, East Susex, UK).
Expression plasmids, protein expression, and purification
The coding sequence of FK506 binding domain (M1 to E126) of PvFKBP35 was amplified from the genomic DNA of P. vivax (a kind gift from Dr. Zbynek Bozdech) by polymerase chain reaction. The following primers were used for cDNA amplification: forward primer contains BsaI restriction site (5′CGC CGC GGT CTC GAGGT ATG GAG CAG GAG ACC CTC GAG CAA GTG CAC 3′); reverse primer contains BamHI restriction site (5′CGC CGC GGA TCC TTA TTC TCT AAA GCT GAT TAG CTC TAT TTC 3′). After amplification, the amplified DNA fragment was digested with BsaI and BamHI to insert the product into pSUMO vector and generated pSUMO-FKBD vector with a hexahistidine tag and sumo fusion protein at the N-terminus. The resulting pSUMO-FKBD plasmid was transformed into E. coli BL21(DE3) cells, and transformed cells were grown in M9 medium containing 1 g/L of 15NH4Cl or 1 g/L of 15NH4Cl and 2 g/L of 13C-glucose at 37°C to obtain 15N and 15N/13C uniformly labeled proteins, respectively. When the cell culture reached Abs600 = 0.6–0.8, the protein expression was induced by adding IPTG at final concentration of 1 mM. The cells were harvested by centrifugation at 18,600 × g for 10 min. Pellet was resuspended in resuspension buffer (20 mM NaPO4, 500 mM NaCl, 10 mM Imidazole), and cells were lysed by sonication for 30 min on ice. The cell lysate was cleared by centrifugation at 48,400 × g for 20 min and purified on a Ni2+-NTA resin as described before.21 For further purification, the Ni2+-NTA elution fractions of 15N-labeled or 15N/13C labeled protein were loaded onto a Superdex-200 filtration column (GE Healthcare, Singapore). The NMR samples (0.5 mM) were prepared in a buffer containing 20 mM NaPO4 (pH 6.8), 50 mM NaCl, and 1 mM DTT, 0.01% NaN3.
Crystallization
Purified PvFKBD (10 mg/mL) in 10 mM phosphate buffer, 137 mM NaCl, 2.7 mM KCl, and FK506 was mixed at a 1:1 molar ratio and subjected to crystallization using a CyBio crystallization robot. Crystallization was performed using sitting-drop vapor diffusion in 96-well plates by mixing 0.2 μL protein solution with 0.2 μL precipitant solution against 0.1 mL precipitant solution at 277 K. Cubic-shaped crystals were obtained within two weeks from a reservoir solution comprising 100 mM BICINE pH 9.0, 2.4 M ammonium sulfate.
X-ray data collection and structure refinement
The crystal was transferred into a cryoprotectant composed of 25% (v/v) glycerol in addition to the components of the reservoir solution, mounted in a cryoloop (Hampton Research) and flash-frozen in a nitrogen-gas stream at 100K. X-ray diffraction data were collected using a MAR 345dtb image plate mounted on a Rigaku Micromax-007 HF X-ray generator at a wavelength of 1.54 Å. The resulting data set was processed, merged, and scaled using MOSFLM and CCP4 to a resolution of 1.66 Å (Table I).
Initial phases were determined by the molecular replacement method using Phaser19 with the coordinates of the previously solved structure of FK506 bound PfFKBD (PDB ID: 2VN1) as a search probe. Rigid body refinement was carried out followed by difference Fourier syntheses calculations. Inspection of the FO–FC and 2FO–FC maps of protein-ligand complexes clearly showed electron density corresponding to the bound ligand. Iterative cycles of model building and refinement were carried out using the programs Coot22 and Refmac23 of the CCP4 suite. The geometry of the final model was checked with PROCHECK24 and the figures were drawn using the program PyMOL.25 The atomic coordinates and structure factors have been deposited at the Protein Data Bank (PDB ID: 3IHZ).
NMR experiments and structure calculation
NMR spectra were acquired on a Bruker AV700 (equipped with a cryoprobe) spectrometer at 298 K. Spectra were processed with NMRPipe26 and analyzed using SPARKY (T.D Goddard and D.G. Kneller, UCSF, San Francisco, CA). Backbone assignments were performed using HNCA, HN (CO) CA, HNCO, HNCACB, CBCACONH spectra. Side chain assignments were performed by using HCCH-TOCSY, 13C-NOESY-HSQC, 15N-NOESY-HSQC. Aromatic ring resonances were assigned from 13C-NOESY-HSQC and 15N-NOESY-HSQC. The proton chemical shifts were referenced to the 2,2-dimethylsilapentane-sulfonic acid (DSS) externally. The 15N and 13C chemical shifts were referenced indirectly to DSS. The distance restraints were observed from 13C-NOESY-HSQC and 15N-NOESY-HSQC by manual and automated assignment of NOESY spectra by using CYANA 2.1.27 Dihedral angle restraints were calculated from chemical shifts using TALOS26and hydrogen bond restraints, which were obtained based on protein structure during structure calculations. Total 100 conformers were generated as initial structures by CYANA27 from 2434 NOE constraints. After calculation, initial conformers were sorted by target function values and the lowest 20 conformers were selected by their total energy values for display and structural analysis. The structure was generated with CYANA 2.1 and visualized by MOLMOL28 and PyMOL.25 NMR data have been deposited in the RCSB PDB with code 2KI3 and Biological Magnetic Resonance Data Bank with code 16260.
Heteronuclear single quantum correlation (HSQC) NMR spectroscopy and chemical shift perturbation analysis
Interaction between PvFKBD and FK506 was examined by 2D 1H-15N HSQC experiment at 298K on a Bruker Avance700. The 15N labeled PvFKBD (0.1 mM) was titrated with FK506 (0.2 mM), and the chemical shift perturbations were analyzed before and after the addition of FK506. Weighted average of the 1H and 15N chemical shift changes, δ1H and δ15N, respectively, were calculated with the function of the form, δ = [(δ1H) 2+ (δ15N/6.51)2]1/2.
NMR relaxation measurements
1H-15N heteronuclear nuclear Overhauser effect (NOE) experiment was carried out at 298 K on a Bruker Avance700 as described previously.29,30 The NOE values were measured using a 3 s interscan delay followed by either proton saturation for 3 s using a series of 120° 1H pulses or an additional 3 s delay at 298 K for free PvFKBD and the 1:2 PvFKBD-FK506 complex. NMR data were processed by using NMRPipe.26
PPIase assay
PPIase activity of recombinant proteins HsFKBP12, PfFKBD and PvFKBD and FK506 inhibition effect on this activity were determined using protease-coupled assays as described previously31 using tetra peptide substrate succinyl-Ala-Leu-Pro-Phe-p-nitroanilide (Peptide Institute, inc. Japan). 1 mL of reaction mixture contained 300 μg of α-chymotrypsin from Sigma (St. Louis, MO), 100 μM of substrate succinyl-Ala-Leu-Pro-Phe-p-nitroanilide and assay buffer [50 mM HEPES and 100 mM NaCl (pH 8.0)]. For checking FK506 inhibition on PPIase activity, stock solution of FK506 was prepared in methanol and solvent alone served as control. Reaction was performed at 10°C and change in absorbance was measured at 390 nm for 5 min in a Shimadzu UV-1601PC UV-vis spectrophotometer. First order rate constants for uncatalyzed (kun) and catalyzed reactions (kcat) were calculated in the absence and presence of protein, respectively, and then catalytic efficiency was calculated as described previously.31 IC50 values of FK506 for all proteins were calculated from the respective dose curves.
Calcineurin assay
The calcineurin assay was performed using commercial substrate RII (Asp-Val-Pro-Ile-Pro-Gly-Arg-Phe-Asp-Arg-Val-pSer-Val-Ala-Ala-Glu, MW. 2192) according to the protocol provided by the manufacturer (CalBiochem, USA). The reaction buffer for the assay contained 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 6 mM MgCl2, 0.5 mM CaCl2, 0.5 mM DTT, 0.025% NP-40, and 250 nM calmodulin. The reaction was performed at 37°C for 45 min in 50 μL of final volume and stopped by adding 100 μL Malachite green dye followed by measuring the absorbance at 620 nm. The release of phosphate was calculated according to the standard curve as described.32
Glossary
Abbreviations:
- CaM
Ca2+/calmodulin-binding module
- DTT
dithiothreitol
- HSQC
heteronuclear single quantum correlation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NTA
nitrilotriacetic acid
- PfFKBD
Plasmodium falciparum FK506 binding domain
- PPIase
peptidylprolyl cis-trans isomerase
- PvFKBD
Plasmodium vivax FK506 binding domain
- TPR
tetratricopeptide repeats.
References
- 1. World Health Organization (2005) World Malaria Report.
- 2.Pain A, Hertz-Fowler C. Plasmodium genomics: latest milestone. Nat Rev Microbiol. 2009;7:180–181. doi: 10.1038/nrmicro2096. [DOI] [PubMed] [Google Scholar]
- 3.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009;80:194–198. [PubMed] [Google Scholar]
- 5.Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. doi: 10.1016/s1286-4579(01)01524-6. [DOI] [PubMed] [Google Scholar]
- 6.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, Gastinel LN, Habuka N, Chen X, Maldonado F, Barker JE, Bacquet R, Villafranca JE. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 7.Galat A. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity—targets—functions. Curr Top Med Chem. 2003;3:1315–1347. doi: 10.2174/1568026033451862. [DOI] [PubMed] [Google Scholar]
- 8.Feske S, Okamura H, Hogan PG, Rao A. Ca2+/calcineurin signalling in cells of the immune system. Biochem Biophys Res Commun. 2003;311:1117–1132. doi: 10.1016/j.bbrc.2003.09.174. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 10.Bell A, Wernli B, Franklin RM. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin A, FK506, and rapamycin. Biochem Pharmacol. 1994;48:495–503. doi: 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- 11.Kang CB, Ye H, Yoon HR, Yoon HS. Solution structure of FK506 binding domain (FKBD) of Plasmodium falciparum FK506 binding protein 35 (PfFKBP35) Proteins. 2008;70:300–302. doi: 10.1002/prot.21623. [DOI] [PubMed] [Google Scholar]
- 12.Kotaka M, Ye H, Alag R, Hu G, Bozdech Z, Preiser PR, Yoon HS, Lescar J. Crystal structure of the FK506 binding domain of Plasmodium falciparum FKBP35 in complex with FK506. Biochemistry. 2008;47:5951–5961. doi: 10.1021/bi800004u. [DOI] [PubMed] [Google Scholar]
- 13.Kumar R, Adams B, Musiyenko A, Shulyayeva O, Barik S. The FK506-binding protein of the malaria parasite, Plasmodium falciparum, is a FK506-sensitive chaperone with FK506-independent calcineurin-inhibitory activity. Mol Biochem Parasitol. 2005;141:163–173. doi: 10.1016/j.molbiopara.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Monaghan P, Bell A. A Plasmodium falciparum FK506-binding protein (FKBP) with peptidyl-prolyl cis-trans isomerase and chaperone activities. Mol Biochem Parasitol. 2005;139:185–195. doi: 10.1016/j.molbiopara.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Monaghan P, Fardis M, Revill WP, Bell A. Antimalarial effects of macrolactones related to FK520 (ascomycin) are independent of the immunosuppressive properties of the compounds. J Infect Dis. 2005;191:1342–1349. doi: 10.1086/428454. [DOI] [PubMed] [Google Scholar]
- 16.Alag R, Bharatham N, Dong A, Hills T, Harikishore A, Widjaja AA, Shochat SG, Hui R, Yoon HS. Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide. Protein Sci. 2009;18:2115–2124. doi: 10.1002/pro.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiwad M, Edlich F, Kilka S, Erdmann F, Jarczowski F, Dorn M, Moutty MC, Fischer G. Comparative analysis of calcineurin inhibition by complexes of immunosuppressive drugs with human FK506 binding proteins. Biochemistry. 2006;45:15776–15784. doi: 10.1021/bi061616p. [DOI] [PubMed] [Google Scholar]
- 18.Kang CB, Hong Y, Dhe-Paganon S, Yoon HS. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- 19.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 20.Michnick SW, Rosen MK, Wandless TJ, Karplus M, Schreiber SL. Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science. 1991;252:836–839. doi: 10.1126/science.1709301. [DOI] [PubMed] [Google Scholar]
- 21.Kang CB, Tai J, Chia J, Yoon HS. The flexible loop of Bcl-2 is required for molecular interaction with immunosuppressant FK-506 binding protein 38 (FKBP38) FEBS Lett. 2005;579:1469–1476. doi: 10.1016/j.febslet.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 25.Delano WL. Palo Alto, CA: DeLano Scientific; 2002. The PyMOL molecular graphics system. Available at http://www.pymol.org. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 28.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 29.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 30.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 31.Harrison RK, Stein RL. Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK-506 binding proteevidence for the existence of a family of distinct enzymes. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. [DOI] [PubMed] [Google Scholar]
- 32.Mondragon A, Griffith EC, Sun L, Xiong F, Armstrong C, Liu JO. Overexpression and purification of human calcineurin alpha from Escherichia coli and assessment of catalytic functions of residues surrounding the binuclear metal center. Biochemistry. 1997;36:4934–4942. doi: 10.1021/bi9631935. [DOI] [PubMed] [Google Scholar]


