Ellis JP, Culviner PH, Cavagnero S. (2009) Protein Sci 18:2003–2015. The ribosome-bound nascent chain denoted as PIR37 should be listed as PIR33, throughout the manuscript. In addition, some order parameters reported in Table 2 need to be corrected to the values shown in bold in the updated Table 2 below. The corrections listed here do not affect any of the reported cone semi-angles, rotational correlation times, result interpretation nor conclusions from this study.
Table 2.
Order Parameters Describing the Spatial Confinement and Dynamic Disorder Associated with the apoMb RNC Local Motions. Data on the Natively Unfolded Protein PIR Are Also Shown, as a Comparison
| Timescales of motion |
||||||||
|---|---|---|---|---|---|---|---|---|
| Fast (sub-ns) |
Intermediate (ns) |
|||||||
| Polypeptide chain | SFa,b |
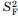 a a
|
τc,F (ns)a | SIa,b |
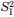 a a
|
τc,I (ns)a | r(0) | Reduced χ2 of fitsc |
| ApoMb16d,f | 0.85 ± 0.01 | 0.72 ± 0.02 | 0.3 ± 0.1 | 0.37 | 1.86 | |||
| ApoMb24d | 0.88 ± 0.01 | 0.78 ± 0.01 | 0.14 ± 0.02 | 0.37 | 0.44 | |||
| ApoMb35d,f | 0.86 ± 0.01 | 0.74 ± 0.01 | 0.4 ± 0.1 | 0.37 | 1.90 | |||
| ApoMb89d,f | 0.89 ± 0.01 | 0.80 ± 0.01 | 0.4 ± 0.1 | 0.94 ± 0.01 | 0.88 ± 0.02 | 5 ± 1 | 0.37 | 0.43 |
| ApoMb124d,f | 0.88 ± 0.02 | 0.78 ± 0.04 | 0.3 ± 0.1 | 0.94 ± 0.01 | 0.88 ± 0.03 | 7 ± 1 | 0.37 | 0.41 |
| ApoMb153d,f | 0.88 ± 0.01 | 0.77 ± 0.03 | 0.3 ± 0.1 | 0.91 ± 0.01 | 0.84 ± 0.01 | 5 ± 1 | 0.37 | 0.26 |
| Chaperone-free apoMb153 RNCse,f,h | 0.88 ± 0.02 | 0.77 ± 0.02 | 0.1 ± 0.01 | 0.93 ± 0.02 | 0.87 ± 0.03 | 2.2 ± 0.2 | 0.37 | 0.40 |
| Ribosome-released apoMb153d,f,i | 0.82 ± 0.01 | 0.67 ± 0.01 | 0.8 ± 0.1 | 0 | 0 | 41 ± 2 | 0.29 ± 0.02 | 0.74 |
| PIR19d,g | 0.91 ± 0.01 | 0.83 ± 0.01 | 0.15 ± 0.02 | 0.37 | 0.73 | |||
| PIR22d,g | 0.89 ± 0.01 | 0.80 ± 0.01 | 0.14 ± 0.03 | 0.37 | 0.46 | |||
| PIR33d,g | 0.85 ± 0.01 | 0.73 ± 0.01 | 0.12 ± 0.02 | 0.37 | 3.6 | |||
| PIR90d,f,g | 0.79 ± 0.01 | 0.63 ± 0.02 | 0.29 ± 0.03 | 0.37 | 4.4 | |||
| Chaperone-free PIR90 RNCse,g,h | 0.79 ± 0.01 | 0.63 ± 0.03 | 0.14 ± 0.01 | 0.37 | 2.8 | |||
| Ribosome-released PIR90d,g,i | 0.40 ± 0.02 | 0.17 ± 0.02 | 0.40 ± 0.03 | 0 | 0 | 13 ± 2 | 0.23 ± 0.01 | 0.77 |
Uncertainties are expressed as ± one standard error (2–7 repeats). In case the standard error was smaller than the propagated fitting error of the fluorescence fractions for any of the experimental repeats, the propagated error is reported.
The order parameter is calculated as the square root of the order parameter squared.
Defined as in (Jameson and Hazlett, 1999).
RNCs generated in wild-type cell-free strain.
These RNCs are generated in Δtig cell-free strains.
The rotational correlation times and pre-exponential factors for these chain lengths are reproduced from (Ellis et al. 2008).
All data for PIR are regarded here as reference information and are reproduced from Ellis & Cavagnero, submitted (2009).
Chaperone-free is intended to mean “devoid of any RNC-bound chaperones.”
For these species, the intermediate timescale (I) dynamics denotes global motions.


