Summary
In this paper, we review current practices for establishing the resolution in single-particle reconstructions. The classical Raleigh criterion for the resolution is not applicable in this case; and the resolution is commonly defined by a consistency test, whereby the data set is randomly split in half and the two resulting reconstructions are then compared. Such a procedure, however, may introduce statistical dependence between the two half-sets, which leads to a too optimistic resolution estimate. On the other hand, this overestimation is counteracted by the diminished statistical properties of a mere half of the data set. The “true” resolution of the whole data set can be estimated when the functional relationship between the data size and the resolution is known. We are able to estimate this functional by taking into account the B-factor and the geometry of data collection. Finally, the drawbacks of resolution estimation are entirely avoided by computing the correlation of neighboring voxels in the Fourier domain.
Keywords: Fourier shell correlation, Fourier neighbor correlation, spectral signal-to-noise ratio, B-factor
1. Introduction
The concept of resolution in optics is related to the minimal distance between two points in the image at which they can be still be distinguished from one another. According to the Rayleigh criterion, this happens when the central peak of the image of one point source falls exactly on the first zero of the image of the second point source. However, depending on experimental conditions (such as the signal-to-noise ratio) or a priori information regarding the imaged object, this limiting distance could be further reduced (see, e.g., (Di Francia, 1955; Shahram and Milanfar, 2006)). For example, if the image of a single point (the point spread function) is circularly symmetric and it is known that there are exactly two point sources, then their image will no longer be rotationally symmetric, and the distance between the point sources can be readily obtained by fitting. The definition of resolution is hence rather subjective and specific to the imaging modality and the underlying assumptions. This concept of resolution cannot be applied to high-resolution electron microscopy at any rate, since it does not lend itself to a suitable experiment (Frank, 2006).
In both crystallography and statistical optics, it is common to define resolution by the orders of Fourier components available for the Fourier synthesis of the signal part of the image. This so-called crystallographic resolution Rc and Raleigh's point-to-point resolution distance d for an instrument (which is diffraction limited to Rc) are related by d=0.61/Rc. In electron crystallography (Glaeser et al., 2007), the signal-related Fourier components of the image are concentrated on the points of a regular lattice, the reciprocal lattice, while the noise-related components form a continuous background. Thus, resolution can be determined by the radius of the highest diffraction orders that stand out from the background; e.g., by comparing the density of the peak with the mean density of the background surrounding the peak.
In single-particle averaging or reconstruction, on the other hand, due to the absence of a periodic order, there are no points of high concentration in the Fourier domain coming from the signal component. Therefore, a different approach to defining and determining the resolution is needed. Common practice today is to look for data consistency by splitting the data set randomly in half and compare the two resulting averages (or 3D reconstructions). The comparison is performed over rings (or shells, respectively) with increasing radius in Fourier space using a suitable measure of reproducibility. An alternative approach, which is gaining popularity, is to analyze the 3D density map reconstructed from the whole data set, by computing the cross-correlation between neighboring voxels in the Fourier domain. The two techniques can be related by a simple formula as we will show below.
The main factors limiting the theoretical resolution (which is beyond 0.1 Å for typical electron wavelengths) in high-resolution electron microscopy are inelastic scattering, specimen movement, and foremost imperfections of the electron lens (e.g., astigmatism, aberrations, aperture function, etc.). An additional factor is the low signal-to-noise ratio in the data, which is due to the low electron dose required to avoid radiation damage. Furthermore, in single-particle reconstruction, alignment of particles introduces an additional source of error.
In the next section, we describe the various approaches to resolution estimation known in the literature, which can be classified into three main categories: the half-set comparison methods, those that make multiple comparisons of the whole data set, and finally those that analyze the reconstructed 3D density map reconstructed from the total set. In contrast to methods for analyzing periodic structures, single-particle methods rely on the averaging over numerous correctly-aligned images, for the purpose of eliminating noise and bringing out common feature. Thus, the size of the data is an important determinant in achieving high resolution. Section 3 addresses the functional relationship between the resolution and the number of particles, which is inherent in the geometrical sampling in Fourier space. The concept of B-factor is also used, which models some of the aforementioned resolution-limiting factors. We believe our analysis is important in determining, among other relations, how many particles are needed to achieve a certain resolution or what is the resolution achievable for a given data size. In the last section, we provide conclusions and discussion. It should be noted that in this paper that, unless otherwise stated, “averaging” refers to both the usual averaging in 2D as well as the 3D reconstruction from projections.
2. Resolution estimation in single-particle reconstruction
In the development of single-particle techniques, measurement of resolution first surfaced in the context of 2D averages (Frank, 1972, 1975; Frank et al., 1970; Frank et al., 1981; Kessel et al., 1985; Saxton and Baumeister, 1982; Unser et al., 1987). The general principle is to find the limit, in Fourier space, out to which data are found to be consistent by some quantitative measure of similarity. Subsequently, definitions and recipes for measurement were readily generalized to the measurement of resolution in 3D reconstructions (Harauz and van Heel, 1986; Penczek, 2002; Sousa and Grigorieff, 2007; Unser et al., 2005). However, as we will see, the way in which the 3D reconstruction is synthesized from the projection data offers a novel opportunity for resolution determination, by multiple comparisons.
In the following, approaches to resolution measurement are initially introduced in the 2D formulation. Unless stated otherwise, these are immediately applicable to the 3D case, as well. We then proceed to discuss those methods that have no 2D equivalent since they are tied to the unique relationship between 2D experimental data and 3D reconstruction.
Approaches to defining resolution in single-particle reconstruction can be grouped into three main categories: one (Section 2.1) that is based on the comparison of two independent averages in the Fourier domain, another one (Section 2.2) that is based on multiple comparisons of all the images participating in the average, and finally, one category (Section 2.3) where solely the reconstructed 3D density map is analyzed (Sousa and Grigorieff, 2007). In the first group are the differential phase residual (Frank et al., 1981) and the Fourier ring correlation (Saxton and Baumeister, 1982; van Heel et al., 1982); while the spectral SNR (Unser et al., 1989; Unser et al., 1987) and the Q-factor (Kessel et al., 1985; van Heel and J., 1980) are part of the second group. We note, however, that all the listed criteria have in common that they ignore the falloff of the signal in Fourier space. That is, a resolution of 1/20 Å-1 might be found by a consistency test, even when the signal power is very low beyond 1/30 Å1. For example, in (van Heel and Stoffler-Meilicke, 1985) a resolution of 1/17 Å-1 found by the Fourier ring correlation has only a minimal signal power even beyond 1/25 Å-1. Thus, a resolution assessment ideally should be accompanied by an assessment of the range and falloff of the power spectrum.
2.1 The half-set comparison method
In the cross-resolution criteria, the aim is the reproducibility of a density map obtained by averaging, when based on two randomly drawn subsets of equal size. In order to avoid the inclusion of material surrounding the molecule, as its inclusion might lead to overly pessimistic estimate of the resolution, it is tempting to use a mask that narrowly defines the region of the molecule. In such attempts, however, it is advised to use a “soft” mask with a slow (e.g., a Gaussian) falloff at the edges, rather than a binary mask, since the latter introduces artificial correlation.
Let F1(k) and F2(k) be, respectively, the discrete Fourier transforms of the two averages, where k = (k1,k2) is the spatial frequency assuming all the values within the Nyquist range. The Fourier transforms are then compared, and a measure of discrepancy is computed. The discrepancy measure is in turn averaged (in the usual sense) over rings of width Δk and radius . The result is then plotted as a function of the ring radius, giving rise to a curve that characterizes the discrepancy between the two subset averages over the entire spatial frequency domain. Finally, a resolution figure is derived from this curve. The most popular discrepancy measures are the differential phase residual (Frank et al., 1981) and the Fourier ring (or shell, if in 3D) correlation (Saxton and Baumeister, 1982; van Heel et al., 1982).
In practice, special care must be taken to ensure statistical independence of the two half-sets. Both the reference-based alignment (Frank, 1975) and the reference-free alignment (Penczek et al., 1992) inevitably introduce statistical dependencies between the two half-set averages being compared and hence cause an overestimation of the resolution. This problem, which is primarily caused by the fitting of the noise into intermediate averages (also referred to as “over-fitting” in (Sousa and Grigorieff, 2007) during the alignment process, has been extensively studied in (Grigorieff, 2000; Penczek, 2002; Yang et al., 2003). It was suggested in (Grigorieff, 2000) to use two markedly different references at the outset, and to keep the two randomly selected image subsets separated throughout the procedure. It appears that this separation significantly reduces the resolution overestimation. On the other hand, it is unclear how different the initial references of the two subsets can be chosen without jeopardizing the performance of the projection alignment algorithm. Additional statistical limitations of the half-set comparison methods are discussed in Section 2.1.3.
2.1.1 Differential phase residual
The differential phase residual (DPR) measures the phase difference between the two Fourier transforms, weighted by the average Fourier amplitude. If Δϕ(k) is the phase difference between the two Fourier transforms for each discrete spatial frequency k, then the differential phase residual is defined as
| (1) |
The sums are computed over Fourier components falling within rings (or shells, in the 3D case) defined by spatial frequency radii k±Δk; k = |k| and plotted as a function of k. In principle, as in the case of the Fourier ring correlation to be introduced later, the entire curve is needed to characterize the degree of consistency between the two averages. However, it is convenient to use a single figure, k45, which is the spatial frequency for which Δϕ̄(k, Δk) = 45°. As a conceptual justification for the choice of this value, one can consider the effect of superimposing two sine waves differing by Δϕ̄. If Δϕ̄ is less than 45°, the waves tend to enforce each other, whereas for any Δϕ̄ > 45°, the maximum of the one wave already tends to fall in the vicinity of the zero of the other, and destructive interference starts to occur.
A characteristic of the DPR is that it is defined in the Fourier domain over successive rings (or shells) of radius k, rather than globally over the entire circle (or ball, if in 3D) of radius k. Since |F1(k)| falls off rapidly as Δϕ, the figure k45 obtained with the global measure would not be very meaningful: good agreement in the lower frequency range can make up for the poor agreement in the higher frequency range and hence produce an over-optimistic value for k45.
It was pointed out in (van Heel, 1987) that Eq. (1) is sensitive to the relative scaling of the two Fourier transforms. Therefore, to circumvent this undesired property one can, for a given frequency k, introduce an extra dimension s that multiplies |F1(k)|, let s vary, and choose Δϕ̄(k, Δk) that attains the minimum (Frank, 2006). The DPR relates to the measure often used in electron and X-ray crystallography to assess reproducibility and the preservation of symmetry.
2.1.2 Fourier ring/shell correlation
Like the DPR, the Fourier ring correlation (Saxton and Baumeister, 1982; van Heel et al., 1982) (or the Fourier shell correlation (FSC (Harauz and van Heel, 1986); if in 3D) is also computed over successive rings/shells of certain radius and width, but it directly compares the two Fourier transforms
| (2) |
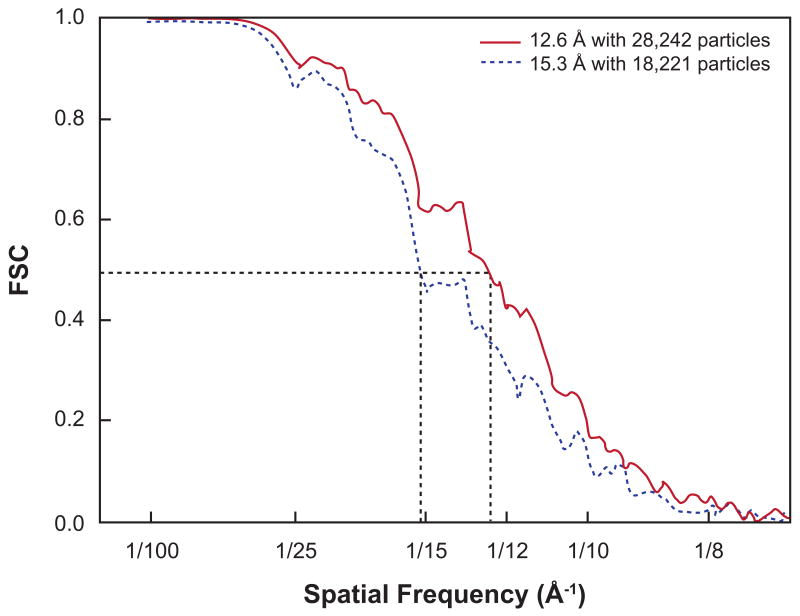
(The discussion in this paragraph applies to the FSC curve as well.) The FRC curve starts with a value of near one at low spatial frequencies, which indicates perfect correlation, then falls off more or less gradually, until it reaches region in which the curve becomes almost flat but with oscillations. The resolution is then typically derived according to either one of two criteria: (i) the measured FRC is compared with the standard deviation of the FRC for the pure noise case: , where N[k, Δk] denotes the number of samples in the Fourier ring with radius k and width Δk, or (ii) the measured FRC is compared with an empirical threshold value (see Figure 1). Under this notation, advocates of the first criterion have been using 2σ (van Heel and Stoffler-Meilicke, 1985), 3σ (Orlova et al., 1997), or 5σ (Radermacher, 1988; Radermacher et al., 2001); whereas a FRC=0.5 (Bottcher et al., 1997) is the most frequently used in the second criterion. It is argued in (Rosenthal and Henderson, 2003) that the threshold of 0.5 should be applied instead to the FRC between the best map obtainable from whole data set and the ideal noise-free structure; doing so leads to the FRC=0.143 criterion. When comparing the two criteria, the FRC= 3σ criterion tends to provide a much better numerical resolution than that from the FRC=0.5 criterion. A noteworthy comment on this matter can be found in the appendix section of (Malhotra et al., 1998), which puts forth reasons for a fixed-value FRC threshold. In contrast, in (van Heel and Schatz, 2005) it is argued strongly in favor of the criterion based on the σ.
Figure 1.
Example of Fourier shell correlation curves, with resolution criterion FSC=0.5 indicated. Reprinted from J. Mol. Biol., 382, J. Sengupta, J. Nilsson, R. Gursky, M. Kjeldgaard, P. Nissen, J. Frank, Visualization of the eEF2-80S Ribosome Transition-State Complex by Cryo-Electron Microscopy, 179-187, Copyright (2008), with permission from Elsevier.
DPR vs. FRC
The FRC is much more popular nowadays than the DPR. Experience has generally shown that the FRC=3σ criterion gives consistently a more optimistic answer than the DPR= 45°. This observation had been justified by a theoretical analysis in (Unser et al., 1987). Further numerical results reported in (Radermacher, 1988) showed that an FRC= 2σ cutoff is equivalent to SNR=0.2 and that the DPR= 45° cutoff is equivalent to SNR=1. Thus, the FRC cutoff and even the DPR cutoff with its fivefold increased SNR seem quite optimistic; on the other hand, for well-behaved data, the DPR curve is normally quite steep, so that even a small increase in the FRC cutoff will often lead to a rapid increase in the SNR.
It is also important to mention that a drawback of the FRC method (as already indicated by other researchers; see e.g. (Unser et al., 2005), which is also the case for the DPR method, is that both are invariant to isotropic filtering of the whole data set: the multiplication of the numerator and the denominator in Eq. (1) and Eq. (2) by a non-vanishing filter leaves the respective curves unchanged, which implies that the same resolution estimate is obtained even after, for example, a low-pass Gaussian filtration is applied to the data. The reader is also referred to (van Heel, 1987) for discussions on some arithmetic variants of these two measures as alternative means of defining and assessing the resolution.
2.1.3 Statistical limitations of the half-set criteria
The resolution estimation methods mentioned thus far (DPR and FRC) are all based on the comparison of two “independent” averages in the Fourier domain. In addition to a potential statistical dependence of the half-sets, which was discussed earlier, there are two other disadvantages with this type of method; one is the statistical fluctuation coming from the splitting of data set in half, and the other one is the systematic underestimation of the resolution, since the total set has better statistics than either half-set. The first drawback can be ignored if the number of particles N is large. As for the second disadvantage, one can resort to some numerical relationship between N and the estimated resolution, then perform an extrapolation from multiple resolution tests with increasing number of particles, up to N, as was done in (Morgan et al., 2000), in which the resolution dependence was assumed to be of the form log(N). Later, in Section 3.2, we discuss a more precise equation that relates the two quantities, based on both theoretical results and empirical observations.
2.2 Evaluation of the whole data set
In the second category, we point out the technique of the spectral signal-to-noise ratio, which is applicable to both 2D (Unser et al., 1989; Unser et al., 1987) and 3D (Penczek, 2002; Unser et al., 2005) and the Q-factor (Kessel et al., 1985; van Heel and J., 1980), which only makes sense in 2D.
2.2.1 Spectral signal-to-noise ratio
This resolution measure was first introduced in (Unser et al., 1987) (see also (Unser et al., 1989)), and it was later extended to 3D forms in (Penczek, 2002; Unser et al., 2005). It is based on a measurement of the SNR as a function of the frequency and has better statistical performance than the DPR and the FRC, since here the whole data set is used. It is remarked in (Unser et al., 1987) that the SSNR relates directly to the Fourier-based resolution criteria commonly used in crystallography and to the DPR and the FRC.
Considering first the 2D case: let be the Fourier transform of the n-th projection (n=1,…,N) at a pixel r(k) in a ring of radius k. There are nk pixels in this ring, so r = 1,…, nk. Assuming the following additive noise model: , where is the true but unknown signal in the r-th pixel, and is independent Gaussian noise in individual images, such that the expectation of is denoted by σr. The SSNR for the ring of radius k is then defined as
| (3) |
which is the ratio between the energy of the signal and that of the noise, scaled up by the size of the data set, in the ring of radius k. (Note that the dependency on k of the right hand side of Eq. (3) is implicit through the variable r.) An unbiased estimator of the SSNR in Eq. (3) is given by S(k)−1, if S(k) > 1, where S(k) is the spectral variance ratio
| (4) |
and Fr is an estimator of the signal component (also used in the numerator)
| (5) |
(The estimator is 0 if S(k) ≤1; it does not take into account the so called B-factor, which is addressed in Section 3.1.) Generally, the SSNR decreases with increasing spatial frequency, and the resolution is taken to be where SSNR(k)=2. Consideration of a numerical model has shown that this limit is roughly equivalent to DPR=45° (Unser et al., 1987). A desirable feature of this resolution measure is that a confidence interval has also been assessed (Unser et al., 1987). In relation to the FRC, it can be shown that
| (6) |
which is of the same kind of relation given earlier in (Frank and Al-Ali, 1975): the SNR there is related to the cross-correlation function in the same manner; we note, however, that the relationship of Eq. (6) is not mathematically exact but only approximate.
The concept of the SSNR for 3D reconstructions is defined as follows. Recall that in the 2D case, the signal component in Eq. (3) was estimated as the 2D average over the Fourier transform of the whole data set; i.e., Fr in Eq. (5). The equivalent operation in 3D would be that the signal component were estimated as the Fourier transform of a 3D reconstruction from the whole data set. We can write explicitly the 3D reconstruction in terms of the data if the Fourier nearest-neighbor interpolation reconstruction method is used (Penczek, 2002); i.e., Fr becomes
| (7) |
where is the Fourier transform of the n-th measured projection (n=1,…,N) and the summation covers only those Lr >0 measurements that pass through voxel r = r(k), situated in the shell of radius k. Because of the unevenness of the distribution of the central planes (a consequence of the geometrical sampling), the number of Fourier components in a voxel, Lr, will be variable and specific to each voxel. (In 2D, the number of Fourier components in all the pixels was simply N.) As for the noise component, recall that in 2D, differences between the data and the 2D average Fr needed to be calculated; see Eq. (4). In the 3D case, the noise component would be estimated by computing the differences (in the Fourier domain) between the data and the reprojections of the 3D reconstruction along the same directions as those in the data. (It is necessary to reproject the 3D reconstruction to compute the differences, since the reconstruction is 3D in nature and the data are in 2D.) Because of the choice of the reconstruction algorithm and by the virtue of the Fourier Slice Theorem, the Fourier transform of a reprojection of the 3D volume is precisely the right hand side of Eq. (7). Thus, the spectral variance ratio of Eq. (4) becomes
| (8) |
which is, after subtracting of 1, an estimator of the 3D SSNR
| (9) |
assuming that the Fourier nearest-neighbor interpolation is used. For the derivations of an estimator for an arbitrary reconstruction algorithm, see (Unser et al., 2005). The relationship in Eq. (6) is also valid in 3D.
2.2.2 Q-factor
Used only for 2D resolution estimation, the Q-factor (Kessel et al., 1985; van Heel and J., 1980) is simply the ratio between the length of the vector sum and the sum of the length of each vector
| (10) |
where Fn(k) is the Fourier transform of a 2D image from the data, which is viewed as a vector in the complex plane. Clearly, 0 ≤ Q ≤ 1. For pure noise, it can be shown that the expected (Einstein equation). The Q-factor is a reasonable indicator for the presence of a signal component in a given Fourier coefficient as realized in N images. A map of Q(k) readily shows weak signal at high spatial frequencies standing out from the background and thus enables the ultimate limit of resolution recoverable (potential resolution) to be established. A quantitative statement can be obtained by averaging this measure over rings in the Fourier domain, and plotting the result, say Q′(k), as a function of the radius k = |k|. The stipulation that Q′(k) should be at least can be used as resolution criterion. For some additional considerations regarding the statistics of the Q-factor, see (Grigorieff, 1998).
A variant of the Q-factor is called the S-factor (Sass et al., 1989), which is related to the structural content (or energy): . For pure noise, the expected S(k) is 1/N[k, Δk]; hence, a resolution criterion can be that S(k)= 3/N[k, Δk].
2.3 Evaluation of 3D reconstruction: Fourier neighbor correlation
The third and last category comprises the so-called Fourier neighbor correlation (FNC) (Sousa and Grigorieff, 2007), which computes the correlation between neighboring voxels in the Fourier domain. It is used for 3D reconstructions only; it can be related to the SSNR via a simple formula; and, unlike the SSNR, availability of the projection data is not required. The FNC is defined as
| (11) |
where, for a given voxel corresponding to frequency k, N(k) is a neighborhood of that voxel, consisting of the six face-adjacent voxels and F(k) is the Fourier transform of the volume. It can be shown (Sousa and Grigorieff, 2007) that
| (12) |
where FNCT(k, Δk) and FNCN(k, Δk) are the Fourier neighbor correlation of, respectively, the noise-free structure and a pure-noise volume (additive noise model is assumed). From Eq. (12), one can then derive the SSNR (and in turn the FSC via Eq. (6)), based solely on the reconstructed structure, without the need of the data; see Figure 2. An example of the use of the FNC for resolution assessment can be found in (Lau and Rubinstein, 2010).
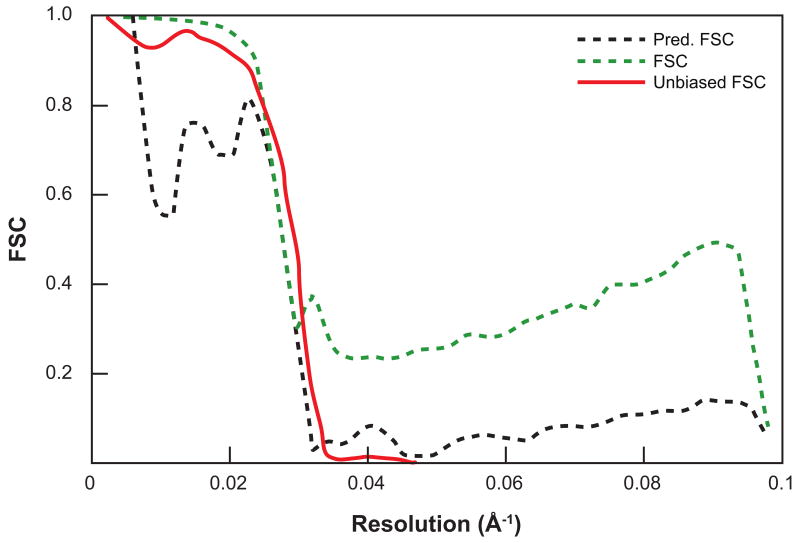
Figure 2.
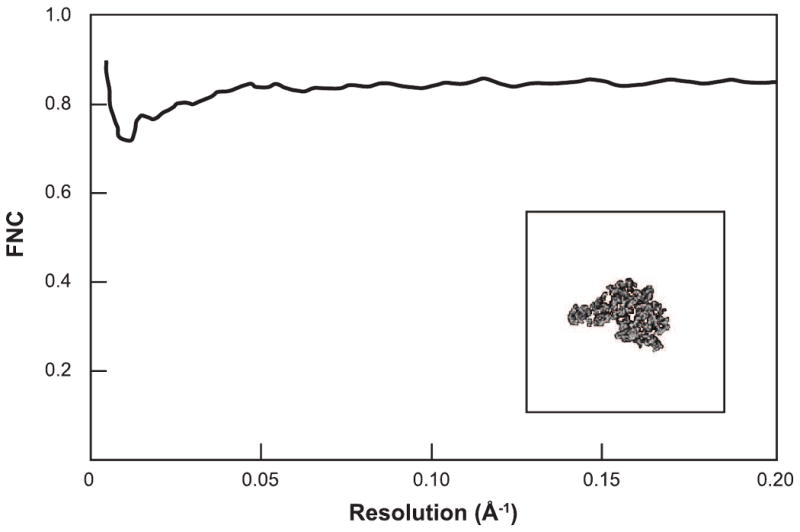
Example of Fourier neighbor correlation curve and its derived FSC curve. Left panel shows the FNC curve corresponding to an atomic model of the 50S large ribosomal subunit (Klein et al., 2001) (PDB code 1JJ2). The density was rendered with a pixel size of 2.44 Å and centered in a box of size 180 pixels along each side. Right panel displays the FNC-based FSC curve (Pred. FSC) and the usual FSC curve, after refinement and reconstruction from 30,000 simulated noisy projections (SNR=0.01) of the subunit. The unbiased FSC was produced by comparing the refined structure with the PDB model. The similarity between the Pred. FSC and the unbiased FSC shows that the Pred. FSC does not suffer from noise fitting as does the usual FSC. Reprinted from J. Struct. Biol., 157, D. Sousa, N. Grigorieff, Ab initio resolution measurement for single particle structures, 201-210, Copyright (2007), with permission from Elsevier.
2.4 Experimental evidence from resolved structures
There is now an emerging consensus that evidence for distinct structural features resolved is the best, irrefutable criterion for actual resolution achieved in a single-particle reconstruction. Examples for such indicators are the visibility of the major and minor groove in A-form RNA helices (∼22 Å and 12 Å, respectively); the appearance of alpha-helices as cylindrical rods (thickness ∼5-7 Å); the appearance of phosphorus atoms as bumps along an RNA helix with expected distance (∼5 Å); or the ability to see individual strands of beta-sheets (4.7 Å) (see (Chiu et al., 2005)). These indicators have been used by various authors to fortify claims of resolution achieved, and they may be used to gauge the utility of data analysis-based criteria of the kind reviewed here. Alternatively, when an atomic structure is available for at least one of the components of the structure under investigation, then comparison with a gallery of low-pass filtered versions will reveal which nominal resolution figure most adequately describes the structural content (see an example in (Frank, 2006)).
3 Resolution vs. number of particles
The resolution criteria most extensively used are perhaps those based on half-set comparison, and in particular the FSC=0.5 criterion. However, as already mentioned in Sections 2.1 and 2.1.3, they come with two shortcomings with opposite effects: one is that it is inherently based on the statistical properties of merely half the data set and thus leads to a pessimistic estimate of the resolution; the other is that before being split in half, the entire data set has been processed in the same way, thus sharing intermediate references in the entire angular refinement process. The halves of the data set are, therefore, not statistically independent, which makes the resolution estimate overly optimistic (see (Grigorieff, 2000)).
To “fully utilize” the entire data set, it has been proposed to estimate the “true” resolution of the full data set by extrapolation from multiple resolution tests with increasing numbers of particles, up to the total number in the data set. Legitimate extrapolation could be done if there were a known functional dependence between the resolution and the number of particles. A logarithmic type of dependence was in fact observed in (LeBarron et al., 2008; Liu et al., 2007; Morgan et al., 2000; Rosenthal and Henderson, 2003). In this section, we attempt to look at a theoretical basis for such a relationship. The same type of relationship was already discussed in (Henderson, 1995; Rosenthal and Henderson, 2003); however, we use a simple geometric consideration in our derivations, as well as an important resolution limiting factor, known as the B-factor.
3.1 B-factor approximation
In analogy with the temperature factor or the B-factor in X-ray crystallography, there is also a Gaussian falloff in the Fourier transform of the images, which limits the resolution in electron microscopy techniques. Specifically, resolution-limiting factors such as charging, specimen instability, and posterior image processing procedures (e.g., alignment and interpolation errors) can be all modeled together as a Gaussian envelope term that affects the Fourier transform of the images in a multiplicative way
| (13) |
where B is referred to as the B-factor. Some authors (e.g. (Glaeser and Downing, 1992; Thuman-Commike et al., 1999)) introduce the notion of the electron microscopy B-factor, which is simply one-fourth of B. Equation (13) suggests a way to correct for the B-factor; i.e., by multiplying Faffected(k) with . However, it should be noted that a multiplicative envelope term is already taken into account in the theory of partial coherence, in which the defocus spread term goes like exp (−ak2); while the spatial partial coherence term goes like exp(−bk6 −ck4 −dk2), where a, b, c, and d are constants. However, in practice, the effect of spatial partial coherence in modern FEG microscopes as used in biological structure determination is relatively small, so the envelope term is usually quite well approximated by a Gaussian envelope term.
Since the Fourier amplitude decay is dependent on instrument quality and performance, it is not surprising that the values quoted for B in the cryo-EM literature cover a wide range: 110 Å2 (Miyazawa et al., 2003), 400 Å2 (Conway and Steven, 1999), 500 Å2 (Bottcher et al., 1997), over 1,200 Å2 (Gabashvili et al., 2000), and even over 2,100 Å2 (Thuman-Commike et al., 1999). These values are usually derived by comparing the radial profiles of power spectra from electron micrographs with those obtained by low-angle X-ray solution scattering. However, it was found in (Thuman-Commike et al., 1999) that the subtraction of noise in the power spectrum results in substantially higher B values than without such a correction-since at higher spatial frequencies, the Fourier amplitude originating from the structure is usually overestimated due to the presence of the noise term. To avoid this complication, it would seem beneficial to use the power spectrum of the density map (in which noise is supposedly absent), rather than the power spectrum of the electron micrographs, for comparison with the X-ray scattering profile.
According to the theory of partial coherence, the falloff term should actually include terms of order higher than 2 (Frank, 1973). Hence, since the only reliable way to determine the parameter B is by measuring low-angle X-ray scattering data anyway, it is more straightforward to compare the actual Fourier amplitude of the cryo-EM map (after the usual CTF correction) and the scattering amplitude (if available). A resulting empirical function is thus obtained and used, rather than a B of Gaussian falloff (see, e.g., (Gabashvili et al., 2000)).
3.2 Relationship between the number of particles and resolution
We turn our attention to establishing a relationship between the resolution and the number of particles. A theoretical estimate based on electron scattering is in fact already given in (Henderson, 1995) and used in (Rosenthal and Henderson, 2003) on experimental data. Following (Rosenthal and Henderson, 2003) (caption of Figure 11), the number of particles required to achieve a resolution k can be estimated by
| (14) |
where Nasymm is the number of asymmetric units, 〈S〉2/〈N〉2 is the signal-to-noise ratio of amplitudes, Ne is the electron dose, and σe is the elastic cross-section of electron. Note that k in Eq. (14) is equivalent to d-1 in the original formulation.
As far as the dependency of Npart on k goes, one can deduce a similar type of functional relationship, by just using the geometry of data collection and the B-factor. Recall that in order to estimate the 3D SSNR (Eq. (9) and with uniform noise) the estimator for the true signal was Fr(k) and a B-factor was not considered. Taking into account the definition of the B-factor (Eq. 13), the estimator becomes
| (15) |
Let us call then Φ is a function of Npart and the radius k; this is because the number of Fourier components, Lr, in a voxel depends on these two quantities. In fact, for a fixed 3D angular distribution of the particle orientations, Lr is proportional to Npart; whereas the number of voxels intersecting a great circle of radius k is proportional to k; hence, Φ is proportional to , and Eq. (15) reads
| (16) |
where C is the constant of proportionality. Under the fixed-FSC criterion, let γ be the threshold; then the SSNR is δ = 2γ/(1−γ) (see Eq. (6); e.g., when FSC=γ=0.5, SSNR=δ =2); and therefore, setting SSNR(k)= δ in Eq. (16), we arrive at
| (17) |
which is a similar type of functional with respect to k as that in Eq. (14). Formula (17) is sensible in that more particles are needed when either the noise σ2 in the data increases, the Fourier amplitudes of the structure (in the absence of the B-factor) decreases, or the prescribed SSNR δ increases. Since the B-factor captures the falloff of the Fourier amplitude, for high frequency regime (which is of interest, when it comes to resolution determination), the dominant term of the functional is the exponential term . This is also the case in Eq. (14). Even though our formula does not provide a precise Npart for a specified resolution k (since the constant C is unknown), it can nevertheless be used to estimate Npart from multiple resolution tests. This is done for example to determine the resolution achievable if the “whole” data were used (see earlier discussions in the beginning of Section 3). Examples of this type of strategy can be found in (LeBarron et al., 2008; Liu et al., 2007).
4 Conclusions and discussion
Common practices today for defining and measuring resolution in single-particle methods check for “internal” consistency of the results directly or indirectly, rather than by the Raleigh criterion, nor by analyzing diffraction spots as permitted in crystallography. The criterion encountered most often for 3D reconstruction is perhaps the Fourier shell correlation FSC=0.5, although it comes with two shortcomings with opposite effects: one is that it is inherently based on the statistical properties of merely half the data set and thus leads to a pessimistic estimate of the resolution; the other is that before being split in half, the entire data set has been processed in the same way, sharing intermediate references in the entire angular refinement process. The halves of the data set are, therefore, not statistically independent, which makes the resolution estimate overly optimistic. The former drawback can be overcome by extrapolating from multiple resolution tests with increasing numbers of particles, up to the total number in the data set, using the functional dependence between the resolution and the number of particles. Such dependency was already established in (Henderson, 1995; Rosenthal and Henderson, 2003). However, based only on the B-factor and the geometry of data collection, we could deduce a similar formula, which from a practical point of view is equivalent to that given in (Henderson, 1995; Rosenthal and Henderson, 2003). As for the latter disadvantage, the undesired effect can be reduced by aligning the two half-sets separately. These two issues are not present in the more recent resolution assessment strategy known as the Fourier neighbor correlation, which also proves to be more resilient to noise-fitting, as compared to the FSC.
Highlights.
Review of current practices for establishing the resolution in single-particle reconstructions
Drawbacks of the methods of comparison between two half-sets
Functional relationship between the data size and the resolution revisited
Fourier neighbor correlation as a new way of establishing the resolution
Acknowledgments
The authors are grateful to Jie Fu for her contributing ideas on finding the functional dependence between the resolution and the number of particles. This work was supported by HHMI and NIH R37 GM29169 (to J.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- Di Francia GT. Resolving Power and Information. J Opt Soc Am. 1955;45:497–499. [Google Scholar]
- Frank J. Observation of the relative phases of electron microscopic phase contrast zones with the aid of the optical diffractometer. Optik. 1972;35:608–612. [Google Scholar]
- Frank J. The envelope of electron microscopic transfer functions for partially coherent illumination. Optik. 1973;38:519–536. [Google Scholar]
- Frank J. Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy. 1975;1:159–162. doi: 10.1016/s0304-3991(75)80020-9. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. New York: Oxford University Press; 2006. [Google Scholar]
- Frank J, Al-Ali L. Signal-to-noise ratio of electron micrographs obtained by cross correlation. Nature. 1975;256:376–379. doi: 10.1038/256376a0. [DOI] [PubMed] [Google Scholar]
- Frank J, Bussler P, Langer R, Hoppe W. Einige Erfahrungen mit der rechnerischen Analyse und Synthese von elektronenmikroskopischen Bildern hoher Aufloesung. Ber Bunsenges Phys Chem. 1970;74:1105–1115. [Google Scholar]
- Frank J, Verschoor A, Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Downing KH. Assessment of resolution in biological electron crystallography. Ultramicroscopy. 1992;47:256–265. doi: 10.1016/0304-3991(92)90201-t. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Downing KH, DeRosier D, Chiu W, Frank J, editors. Electron Crystallography of Biological Macromolecules. New York: Oxford University Press; 2007. [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Resolution measurement in structures derived from single particles. Acta Crystallogr D Biol Crystallogr. 2000;56:1270–1277. doi: 10.1107/s0907444900009549. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry 3-dimensional reconstruction. Optik. 1986;73 [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Kessel M, Radermacher M, Frank J. The structure of the stalk surface layer of a brine pond microorganism: correlation averaging applied to a double layered lattice structure. J Microsc. 1985;139:63–74. doi: 10.1111/j.1365-2818.1985.tb04662.x. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WC, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Proc Natl Acad Sci U S A. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBarron J, Grassucci RA, Shaikh TR, Baxter WT, Sengupta J, Frank J. Exploration of parameters in cryo-EM leading to an improved density map of the E. coli ribosome. J Struct Biol. 2008;164:24–32. doi: 10.1016/j.jsb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Penczek P, Agrawal RK, Gabashvili IS, Grassucci RA, Junemann R, Burkhardt N, Nierhaus KH, Frank J. Escherichia coli 70 S ribosome at 15 A resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Menetret JF, Radermacher M, Neuhof A, Akey IV, Rapoport TA, Akey CW. A comparison of the yeast and rabbit 80 S ribosome reveals the topology of the nascent chain exit tunnel, inter-subunit bridges and mammalian rRNA expansion segments. J Mol Biol. 2000;301:301–321. doi: 10.1006/jmbi.2000.3947. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Harris JR, Beckman E, Zemlin F, Markl J, van Heel M. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 A resolution by electron cryomicroscopy and angular reconstitution. J Mol Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Penczek PA. Three-dimensional spectral signal-to-noise ratio for a class of reconstruction algorithms. J Struct Biol. 2002;138:34–46. doi: 10.1016/s1047-8477(02)00033-3. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Wieczorek H, Gruber G. The structure of the V(1)-ATPase determined by three-dimensional electron microscopy of single particles. J Struct Biol. 2001;135:26–37. doi: 10.1006/jsbi.2001.4395. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Sass HJ, Buldt G, Beckmann E, Zemlin F, van Heel M, Zeitler E, Rosenbusch JP, Dorset DL, Massalski A. Densely packed beta-structure at the protein-lipid interface of porin is revealed by high-resolution cryo-electron microscopy. J Mol Biol. 1989;209:171–175. doi: 10.1016/0022-2836(89)90180-0. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Shahram M, Milanfar P. Statistical and Information-Theoretic Analysis of Resolution in Imaging. IEEE Trans Inf Theory. 2006;52:3411–3437. [Google Scholar]
- Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Thuman-Commike PA, Tsuruta H, Greene B, Prevelige PE, Jr, King J, Chiu W. Solution x-ray scattering-based estimation of electron cryomicroscopy imaging parameters for reconstruction of virus particles. Biophys J. 1999;76:2249–2261. doi: 10.1016/S0006-3495(99)77381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unser M, Sorzano CO, Thevenaz P, Jonic S, El-Bez C, De Carlo S, Conway JF, Trus BL. Spectral signal-to-noise ratio and resolution assessment of 3D reconstructions. J Struct Biol. 2005;149:243–255. doi: 10.1016/j.jsb.2004.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unser M, Trus BL, Frank J, Steven AC. The spectral signal-to-noise ratio resolution criterion: computational efficiency and statistical precision. Ultramicroscopy. 1989;30:429–433. doi: 10.1016/0304-3991(89)90074-0. [DOI] [PubMed] [Google Scholar]
- Unser M, Trus BL, Steven AC. A new resolution criterion based on spectral signal-tonoise ratios. Ultramicroscopy. 1987;23:39–51. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- van Heel M. Similarity measures between images. Ultramicroscopy. 1987;4:95–99. [Google Scholar]
- van Heel M, J H. The stretching of distorted images of two-dimensional crystals. In: Baumeister W, editor. Electron Microscopy at Molecular Dimensions. Berlin New York: Springer; 1980. pp. 256–260. [Google Scholar]
- van Heel M, Keegstra W, Schutter WG, van Bruggen EFJ. Arthropod hemocyanin studied by image analysis. Life Chem Rep Suppl. 1982;1:69–73. [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- van Heel M, Stoffler-Meilicke M. Characteristic views of E. coli and B. stearothermophilus 30S ribosomal subunits in the electron microscope. EMBO J. 1985;4:2389–2395. doi: 10.1002/j.1460-2075.1985.tb03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yu X, Galkin VE, Egelman EH. Issues of resolution and polymorphism in singleparticle reconstruction. J Struct Biol. 2003;144:162–171. doi: 10.1016/j.jsb.2003.09.016. [DOI] [PubMed] [Google Scholar]