Summary
The unfolded protein response (UPR) is linked to metabolic dysfunction, yet it is not known how ER disruption might influence metabolic pathways. Using a multilayered genetic approach, we find that mice with genetic ablations of either ER stress sensing pathways (ATF6α, eIF2α, IRE1α), or of ER quality control (p58IPK), share a common dysregulated response to ER stress that includes the development of microvesicular steatosis. The rescue of ER protein processing capacity by the combined action of UPR pathways during stress prevents the suppression of a subset of metabolic transcription factors that regulate lipid homeostasis. This suppression occurs in part by unresolved ER stress perpetuating expression of the transcriptional repressor CHOP. As a consequence, metabolic gene expression networks are directly responsive to ER homeostasis. These results reveal an unanticipated direct link between ER homeostasis and the transcriptional regulation of metabolism and suggest mechanisms by which ER stress might underlie microvesicular steatosis.
Introduction
Cellular protein folding homeostasis is protected when the depletion of chaperone reserve leads to the activation of proximal signaling molecules, which ultimately results in alterations in gene expression to alleviate stress. In the endoplasmic reticulum (ER), three principal pathways are activated in response to ER stress and comprise the unfolded protein response (UPR): PERK, IRE1, and ATF6. PERK and IRE1 are ER-resident transmembrane kinases that lead to translational inhibition through eIF2α phosphorylation, and production of XBP1 transcription factor by an unconventional splicing mechanism, respectively. ATF6 is a transmembrane transcription factor liberated by stress-regulated intramembrane proteolysis. Each pathway culminates in transcriptional regulation of gene expression and contributes to the overall maintenance of homeostasis in the ER during stress (Ron and Walter, 2007; Schröder, 2008; Wek and Cavener, 2007; Wu and Kaufman, 2006). Gene expression profiling has demonstrated that numerous cellular processes beyond protein folding in the ER are regulated by UPR activation (Harding et al., 2003; Shen et al., 2005; Travers et al., 2000). However, how these processes are activated and temporally regulated by ongoing stress, and how such regulation alters cellular functions only tangentially related to secretory pathway function to improve the chances for adaptation, is not understood.
ER stress has been associated with metabolic dysfunction caused by dietary demand (Lee et al., 2008; Oyadomari et al., 2008; Özcan et al., 2004; Özcan et al., 2006). How the various pathways of the UPR protect cells from stress in vivo and how these pathways intersect with other cellular functions, such as metabolism, in different circumstances is not clear. Thus there is a need to examine the functions of the individual UPR signaling pathways in parallel and in vivo. The liver presents an ideal model system for studying these connections because it is a key organ for both protein secretion (principally in the form of lipoproteins and other serum factors) and metabolism (Postic et al., 2004). The coupling of these processes involves families of transcriptional activators and coregulators. Among these are members of the C/EBP, PPAR, PGC-1, and SREBP families, that reprogram liver metabolic mRNA expression as environmental cues dictate (Ferre and Foufelle, 2007; Lee et al., 2003; Lekstrom-Himes and Xanthopoulos, 1998; Lin et al., 2005). Liver function is also sensitive to environmental or genetic perturbation, and a number of these perturbations, including both alcoholic and nonalcoholic steatohepatitis, viral hepatitis, hyperhomocysteinemia, acute exposure to hepatotoxins, and high carbohydrate or high fat diets, have been suggested to lead to hepatic ER stress (Jaeschke et al., 2002; Lee et al., 2008; Nguyen et al., 2008; Oyadomari et al., 2008). A common thread amongst these insults is disruption of lipid homeostasis (i.e., fatty acid metabolism) in the liver. Elements of the UPR, including the transcription factor XBP1 and the translational regulator eIF2α, have been proposed to directly regulate lipid metabolic pathways (Lee et al., 2008; Oyadomari et al., 2008), although it is not clear whether these interactions are influenced by ER stress, whether they are pathway specific, or whether they apply equally to the many diverse causes of hepatic dyslipidemia.
ATF6α protects ER function by augmenting the upregulation of ER protein processing factors such as chaperones and ER-associated protein degradation (ERAD) machinery during stress (Adachi et al., 2008; Wu et al., 2007; Yamamoto et al., 2007). ATF6α deletion sensitizes cells and animals to persistent ER stress. In vivo, this failure to recover from ER stress results in fatty liver, uncovering a potential connection between ER stress and lipid metabolism (Wu et al., 2007). In this study, we have used a genetic approach to identify the proximal mechanistic connections between activation of the UPR by ER stress and metabolic control. This approach has revealed that the three arms of the UPR cooperate to maintain ER function, and that failure to do so leads to unresolved ER stress and suppression of several key regulators of metabolic gene expression independent of any specific UPR pathway. These results suggest that an intact UPR guards liver function by improving ER protein processing to maintain lipid homeostasis.
Results and Discussion
Unresolved ER stress perturbs hepatic lipid homeostasis
Animals nullizygous for the Atf6α gene display no overt phenotype under unchallenged conditions, being viable, fertile, and grossly normal. However, when injected intraperitoneally with an ordinarily sub-lethal dose of the ER-stress inducing agent tunicamycin (TM), Atf6α-null animals succumb and show evidence of persistent ER stress in the liver and kidneys (Wu et al., 2007). Consistent with our previous findings, TM-challenged Atf6α-null animals displayed prolonged upregulation of the apoptotic protein CHOP and its target GADD34, at a time when the expression of these genes is attenuated in wild-type animals (Figure 1A). While adaptation to or recovery from ER stress is accompanied by rapid degradation of Chop mRNA and protein (Rutkowski et al., 2006), persistent CHOP expression correlates with unresolved stress. The most likely cause of this persistent stress is a failure of Atf6α-null animals to fully upregulate ER protein folding and processing machinery (Wu et al., 2007; Yamamoto et al., 2007), which renders them less able to recover from acute challenge.
Figure 1.
ATF6α deletion reveals an intersection between ER stress and metabolic homeostasis. (A) Livers from wild-type, heterozygous, or Atf6α-null mice injected with vehicle or 1 mg/kg b.w. TM were probed by immunoblot for expression of tubulin (loading control) BiP, CHOP, GADD34, and the phosphorylated form of eIF2α. Efficacy of the TM was reflected in inhibition of TRAPα glycosylation, for which the glycosylated (closed arrow) and unglycosylated (open arrow) species are indicated. (B) Wild-type and Atf6α−/− mice were injected with 1 mg/kg TM and livers were visualized in situ at the indicated times post-injection. (C) Wild-type and Atf6α−/− mice were injected with TM. Cryosections (6 μM) of liver isolated 48 hours post injection were stained by Hematoxylin & Eosin (H&E) and visualized at 400× magnification. (D) Wild-type and Atf6α−/− mice were injected with vehicle or TM (1 mg/kg). Livers were surgically removed 48hrs post injection, fixed in 2.5% glutaraldehyde, then prepared for transmission electron microscopic analysis. Endoplasmic reticulum (ER), nuclei (N), mitochondria (M) and lipid droplets (LD) are indicated. White scale bar equals 500 nm. (E) Protein lysates from liver, isolated 8 or 48 hrs post-injection, were probed by immunoblot as labeled. RNA was prepared from the same liver tissue samples and assayed by RT-PCR that simultaneously detects both spliced (sp) and unspliced (us) Xbp1 mRNA. (F) Mice were injected with 1 mg/kg TM for 48 hours. Mice were fasted for 6 hours and then injected with insulin (2 U/kg) 20 minutes prior to sacrifice. Immunoblotting detected phosphorylated or total AKT.
Also consistent with our previous results (Wu et al., 2007), wild-type or heterozygous animals challenged with TM showed initial liver perturbation at the molecular and morphological levels, but recovered thereafter, while Atf6α−/− animals failed to recover. By 48 hours after injection, livers of Atf6α−/− animals were much lighter in color than wild-type counterparts and produced a distinct fat cap upon homogenization and sedimentation (Figure 1B and data not shown). Hematoxylin and eosin (H&E) failed to stain large swaths of Atf6α−/− liver tissue after TM injection, Oil Red O staining confirmed the presence of fat deposits, and substantially more intracellular triglyceride was found in Atf6α-null animals than in wild-type (Figures 1C, S1 and data not shown). Ultrastructural analysis revealed a uniform accumulation of cytosolic lipid droplets in knockout livers indicative of microvesicular steatosis (Figure 1D). In addition, while TM induced breakdown of the lamellar structure of the ER into smaller vesicles in wild-type animals, Atf6α-null animals appeared to show an even greater loss of structural integrity of the ER (Figure 1D). Confirming the accumulation of lipid, the cytosolic lipid droplet protein marker adipose differentiation related protein (ADRP) (Martin and Parton, 2006) was upregulated by ER stress in all genotypes, but more so in Atf6α-null animals (Figure 1E). This accumulation of lipid occurred concomitant with the persistent presence of XBP1 protein derived from spliced mRNA, that reflects ongoing stress (Figures 1E and S2) Thus, lipid accumulation in the livers of Atf6α-null animals is an active process that accompanies persistent UPR signaling. Similar fat accumulation and ADRP upregulation were seen in the kidneys of TM challenged Atf6α-null animals (Figure S3). ER stress and fat deposition were not observed in other tissues, consistent with liver and kidneys being the primary target organs of TM (Hetz et al., 2006; Marciniak et al., 2004; Zinszner et al., 1998). Because Atf6α deletion produced such a dramatic phenotype upon challenge but did not otherwise result in lethality [in contrast to constitutive ablation of IRE1α or eIF2α signaling (Harding et al., 2001; Reimold et al., 2000; Scheuner et al., 2001; Zhang et al., 2005)] we used this genetic model to further explore how ER stress influences lipid metabolism.
Injection of wild-type animals with a higher dose of TM (2 mg/kg BW rather than 1 mg/kg) led to animal mortality along a similar time frame as the lower dose in Atf6α-null animals (~3 days after injection, data not shown). However, this higher dose only modestly increased lipid deposition in wild-type animals (Figures 1C and 1E). This result suggests that the divergent phenotype of Atf6α-null animals is not simply a consequence of increased toxicity of TM in these animals. Several additional lines of evidence support this hypothesis. First, Atf6α−/− animals were capable of upregulating cytochrome P450 isoforms, that detoxify compounds in the liver, in response to TM or other challenge (Supplementary Figure S4 and data not shown). Second, TM injection led to only very modest increases in plasma alanine transaminase (ALT) levels in Atf6α-null animals compared to wild-type, and only at very late time points after injection, arguing against gross hepatic necrosis or apoptosis (Figure S5). Third, hepatocellular apoptosis, judged by cleavage of the nuclear PARP protein, was not significantly increased in Atf6α-null animals compared with wild-type (Figure S5).
While steatosis is the most obvious consequence of TM injection in Atf6α-null mice, other metabolic pathways are progressively perturbed as well. One consequence of fat accumulation in the liver is the development of insulin resistance (Postic and Girard, 2008). Challenge of wild-type animals with TM rendered them resistant to signaling by endogenous insulin produced by refeeding after fasting, as assessed by phosphorylation of AKT at Ser473, consistent with the idea that ER stress produces insulin resistance (Özcan et al., 2004) (Figure S6). However, on top of this stress-dependent resistance to endogenous insulin, we observed resistance to exogenous insulin in Atf6α-null animals, but not wild-type animals, by 48 hours after TM injection (Figure 1F). Thus, while both wild-type and Atf6α-null animals became resistant to endogenous insulin during ER stress, Atf6α-null animals eventually became profoundly resistant to even exogenous insulin, concomitant with lipid dysregulation. Atf6α-null animals also displayed hypoglycemia and reduced glycogen storage (data not shown) consistent with gross metabolic disruption in the liver. Combined with the lipid deposition phenotype, these results suggest that TM treatment perturbs metabolic homeostasis and that this phenotype becomes progressively more severe selectively in Atf6α-null animals.
ER stress suppresses expression of genes involved in maintaining energy and lipid homeostasis
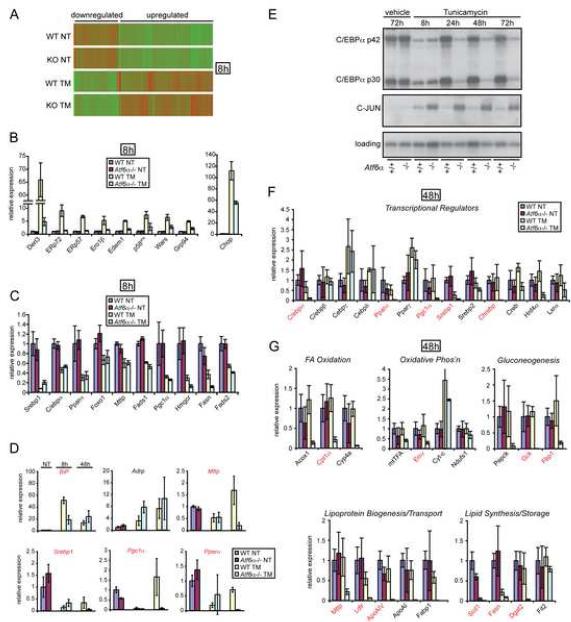
Because ATF6α is a transcription factor, we reasoned that the connection between Atf6α deletion and disruption of lipid homeostasis might be most readily identified by global transcriptional analysis. Microarray profiling 8 hours after TM challenge revealed that, as in mouse embryonic fibroblasts (MEFs) (Adachi et al., 2008; Wu et al., 2007), Atf6α deletion had little effect on basal gene expression (Figure 2A and Suupplemental Data). However, when animals were challenged with TM for 8 hours, a subset of ER stress-regulated genes was altered in their expression by Atf6α deletion (Figure 2A). The majority of these were genes that were upregulated by stress in wild-type liver, and either to a lesser or greater extent in Atf6α-null animals. As in MEFs, the expression of ER chaperones and co-chaperones (Erp72, Erp57, p58IPK, Grp94), as well as genes involved in ERAD (Derl3, Edem1), was attenuated in Atf6α-null livers (Figure 2B). In addition, early upregulation of the gene encoding the pro-apoptotic transcription factor CHOP depended partially upon ATF6α.
Figure 2.
Persistent ER stress in vivo suppresses expression of a subset of metabolic genes. (A) Cluster analysis of gene expression and transcriptional profiling analysis of liver RNAs were performed as described in the Experimental Procedures. Graphic representation of the average expression levels of 4262 differentially-expressed genes is shown. Each vertical bar represents a single gene. Green coloration indicates lower expression, and red indicates higher expression. (B, C) The expression levels of two subsets of mRNAs from microarray analysis are shown, normalized against vehicle-injected wild-type samples. Note that the differences in expression levels of these genes in the absence of ER stress were generally not significant. Error bars represent means ±S.D.M. from three animals in each group. For (B), all genes shown were significantly (p<0.05) less-induced by TM in Atf6α-null animals than wild-type. For (C), all genes shown were significantly (p<0.05) downregulated by stress in both genotypes. The ATF4-dependent tryptophanyl tRNA synthetase (Wars) is here Atf6α-dependent likely because of the failure of Atf6α-null mice to stimulate eIF2α phosphorylation in response to TM challenge. (D) Real-time RT-PCR analysis was used to assess the expression of selected mRNAs in wild-type or Atf6α-null livers 8 or 48 hours after TM injection. Error bars are means ±S.D.M. from 2 (vehicle) or 3 (TM) animals. Expression values were normalized against β-actin levels and are shown relative to the expression level in wild-type unchallenged animals. (E) Protein lysates from livers of wild-type or Atf6α−/− mice injected with vehicle or TM (1 mg/kg) were probed by immunoblot for either C/EBPα (which exists in a 42kd long form and 30kd short form) or C-JUN. (F, G) The expression of metabolic genes subdivided into various categories was determined 48 hours after TM injection by real-time RT-PCR. For (D, F, G, and H). Red lettering indicates genes significantly (p<0.05) different in expression comparing TM-challenged Atf6α-null animals to unchallenged.
Notably, there was no obvious connection between Atf6α deletion and expression of genes involved in hepatic lipid metabolism, consistent with the phenotypic separation between genotypes developing progressively and manifesting more so at later time points rather than only 8 hours after challenge. However, we observed that a number of key mediators of lipid homeostasis were significantly downregulated by TM independent of genotype (Figure 2C and Table S1). These included genes encoding the transcription factors and cofactors SREBP1, C/EBPα, PPARα, FOXO1, and PGC1α; and components of lipid metabolism including Microsomal Triglyceride Transfer Protein (MTTP), Fatty acid desaturases and synthetases (FADS1 and FADS2; and FASN), and HMG CoA reductase. This regulation was selective only for a subset of metabolic genes, in that the expression of many others was unaltered after 8 hours of ER stress (Table S1).
Deletion of Atf6α does not immediately sensitize cells to stress, but instead renders them less able to recover from and adapt to ongoing stress (Wu et al., 2007). Indeed, the initial response of the liver to TM injection in Atf6α-null mice was very similar to that of wild-type mice in terms of activation of ER stress pathways, and it was only at later time points that knockout animals showed persistent ER stress when wild-type animals had recovered (Figure 1A). Thus, we speculated that Atf6α deletion might sensitize animals to ER stress not by loss of direct regulation of metabolic gene expression by ATF6α, but instead by indirectly leading to ongoing stress in the liver because of failure to properly improve the protein folding environment in the ER. In support of this hypothesis, quantitative RT-PCR demonstrated that expression of the genes Mttp, Srebp1, Pgc1α, and Pparα was suppressed by ER stress in both wild-type and Atf6α-null animals 8 hours after TM injection, but to a much greater extent in Atf6α-null animals 48 hours after injection (Figure 2D). Expression of C/ebpα mRNA and protein followed a similar trend, being rapidly suppressed by TM in both genotypes, but remaining suppressed in Atf6α-null animals over the course of the experiment (Figure 2E and data not shown). C/EBPα protein is synthesized in both long (p42) and short (p30) forms by virtue of translational regulation (Calkhoven et al., 2000) and, consistent with the decrease in C/ebpα mRNA, both forms were suppressed in Atf6α-null animals. We also observed upregulation of c-jun at both the mRNA and protein levels (Figure 2E and data not shown). This is notable because elevated hepatic expression of c-jun accompanies C/ebpα deletion (Flodby et al., 1996).
We considered four possible mechanisms to account for fatty liver in TM-challenged Atf6α-null mice: (1) Enhanced de novo synthesis of fatty acids in the liver; (2) Increased mobilization and uptake of fatty acids from adipose tissue to the liver; (3) Diminished fatty acid oxidation by the liver; and (4) Decreased lipoprotein secretion. Because the genotype-specific changes in gene expression upon ER stress became evident at later time points after TM challenge, we used quantitative RT-PCR to probe the expression of several categories of genes involved in lipid and carbohydrate homeostasis 48 hours after challenge, when wild-type animals have largely recovered. The transcriptional regulators C/ebpα (regulates gluconeogenesis and lipogenesis), Pparα (fatty acid oxidation and gluconeogenesis), Pgc1α (fatty acid oxidation and gluconeogenesis), Srebp1 (lipogenesis), and ChREBP (a coregulator of lipogenesis with SREBP1) all remained significantly downregulated by persistent ER stress in Atf6α-null livers (Figure 2F). In contrast, other regulators including other C/ebp family members, Pparγ, Creb, and Lxrα, were not significantly affected by ER stress in these animals. Consistent with these results, we sampled the expression of key targets of these transcriptional regulators, finding that genes involved in fatty acid oxidation, gluconeogenesis, and lipogenesis were all suppressed in Atf6α-null animals (Figure 2G). In addition, genes involved in lipoprotein synthesis and transport were similarly suppressed, suggestive of a defect in activity of the transcription factor SREBP2. Immunoblot confirmed that both uncleaved and cleaved forms of SREBP1 and SREBP2 were downregulated in Atf6α-null animals, suggesting that the primary regulation of their activity after TM injection is transcriptional (data not shown). Since key proteins involved in lipogenesis (SCD1, FASN, DGAT2) are downregulated in Atf6α-null animals and a key protein that regulates the formation of new lipid droplets [FIT2—(Kadereit et al., 2008)] is not affected, de novo lipogenesis in the liver is an unlikely source of cytosolic lipid droplet formation. Likewise, Fabp1 suppression implies limited mobilization of fatty acids from adipose tissue. Taken together, these data suggest that lipid accumulates in the livers of Atf6α-null animals because of a defect in fatty acid oxidation, possibly augmented by impaired lipoprotein secretion. In support of this idea, microvesicular steatosis can accompany genetic defects in both fatty acid oxidation (Rao and Reddy, 2001) and lipoprotein secretion (Raabe et al., 1999).
ER stress sensing pathways cooperate to maintain homeostasis and restore metabolic gene expression
We next turned our attention to the mechanism by which ER stress leads to the suppression of these transcription factors. The rapid downregulation of genes encoding metabolic transcription factors by ER stress independent of genotype does not support the idea that ATF6α directly regulates these genes. Rather, there more likely exists a difference between wild-type and Atf6α-null mice that is exacerbated as the latter fail to recover from TM challenge, that is regulated by ATF6α indirectly. One noticeable difference between the responses of wild-type and Atf6α-null livers to TM is that Xbp1 mRNA splicing and production of XBP1 protein from the spliced product persisted in the latter (Figures 1E and S2). Thus, we considered the possibility that suppression of metabolic gene expression might depend upon signaling through the IRE1/XBP1 axis. Ire1α- and Xbp1-null animals die during embryogenesis because of liver defects (Reimold et al., 2000; Zhang et al., 2005). To circumvent this problem, we created animals with a floxed Ire1α allele and mated them with mice expressing the CRE recombinase under the control of the albumin promoter, thus generating a liver-specific deletion of Ire1α. Confirming efficient Ire1α deletion, these animals showed very little splicing of Xbp1 mRNA in the liver in response to TM injection (Figure S7).
Rather than being dependent upon IRE1α, the suppression of metabolic gene expression was exacerbated in liver-specific Ire1α-null animals in a manner very similar to Atf6α-null animals. Both forms of C/EBPα protein were downregulated in Ire1α-null livers 30 hours after TM injection to a much greater extent than in wild-type animals, while expression of C-JUN was upregulated (Figure 3A). Liver-specific Ire1α-null animals developed fatty liver upon injection (data not shown), and displayed upregulation of Adrp along with suppression of transcription factors involved in lipid metabolism (Figure 3B). As with Atf6α-null animals, Ire1α-deleted livers showed persistent upregulation of the proapoptotic CHOP protein after CHOP upregulation had been largely attenuated in wild-type animals (Figures 3A and 3B). Thus, Atf6α-null and Ire1α-null livers display similar sensitivity to TM and perturbation of metabolic gene expression. While XBP1 has been proposed to regulate basal expression of lipogeneic genes (Lee et al., 2008), TM-induced steatosis occurs in the presence of either spliced or unspliced XBP1 (Figures 1E, 3A, S2, and S7B), suggesting that XBP1 does not directly regulate ER stress-induced microvesicular steatosis.
Figure 3.
Each arm of the canonical UPR contributes to protection from metabolic dysregulation. (A) Wild-type mice and mice with a hepatocyte-specific deletion of Ire1α were injected with vehicle or TM (2 mg/kg). Protein lysates from liver isolated 30 hours post-injection were probed by immunoblot as indicated. The long (open arrowhead) and short (closed arrowhead) forms of C/EBPα are indicated. Note that the apparent difference in inhibition of TRAPα glycosylation is likely a consequence of this protein itself being a UPR target at least partially regulated by IRE1α (Nagasawa et al., 2007), rather than any difference in the pharmacological efficacy of TM between genotypes. (B) RNA was prepared from wild-type and Ire1α−/− liver tissue samples from (A) and analyzed by real-time RT-PCR. n = 3 animals per group. (C) eIF2α S51A heterozygous or homozygous knockin mutant mice, rescued by a constitutively expressed wild-type eIF2α floxed transgene, were mated with mice expressing CRE recombinase under control of the albumin promoter to delete the transgene in hepatocytes. “eIF2α genomic” denotes whether the mice were heterozygous or homozygous for the S51A genomic allele, and “Δtg” indicates whether or not the transgene was deleted by CRE expression. Protein lysates from liver isolated 30 hours post-injection (2 mg/kg) were probed by immunoblot as labeled. Here inhibition of glycosylation was followed using the Transferrin Receptor (TfR) protein. Asterisk indicates non-specific background bands. (D) Expression of metabolic mRNAs from the animals shown in (C) was quantitated by real-time RT-PCR as in (B). n = 2-4 animals per group. While the difference in upregulation of Adrp mRNA was not statistically significant comparing SA and AA mice after TM challenge, more robust upregulation of ADRP protein was evident (data not shown). Red lettering indicates different expression levels comparing Ire1α−/− or AA TM-challenged animals to unchallenged, p<0.1.
The similar phenotypes in these two genetic models led us to ask whether ablating the third, PERK/eIF2α-dependent arm of the UPR would produce a similar outcome. To test this hypothesis, a constitutively expressed eIF2α transgene flanked by LoxP sites was introduced into mice harboring the nonphosphorylatable S51A mutation in genomic eIF2α. This transgene rescued the otherwise neonatal lethal eIF2α S51A homozygous mutant, and either these homozygous (AA) or heterozygous (SA) animals carrying the transgene were bred to mice expressing CRE under control of the albumin promoter (see Supplemental Experimental Procedures and Figure S8). No phosphorylation of eIF2α was observed in homozygous S51A animals with a deleted transgene upon TM injection (Figure 3C, lanes 13-16) confirming efficient eIF2α transgene deletion. As with ATF6α or IRE1α deletion, we also observed suppression of C/EBPα protein expression, upregulation of C-JUN and ADRP protein, and fatty liver in these animals, yet to a lesser extent in mice heterozygous for the genomic S51A locus (Figures 3C, compare lanes 10-12 with lanes 13-16; also data not shown) Similar results were obtained at the mRNA level (Figure 3D). Thus, while increased eIF2α dephosphorylation protects against steatosis induced by severe dietary stress (Oyadomari et al., 2008), eIF2α phosphorylation is clearly necessary to prevent fat accumulation in response to a direct challenge of ER stress.
Our data demonstrate that ongoing ER stress, irrespective of the UPR pathway ablated, perpetuates suppression of metabolic gene expression. These results suggest that the UPR protects lipid homeostasis not by selective regulation of gene expression mediated by the ATF6α, IRE1α, or PERK pathways, but instead by the three pathways of the UPR contributing to overall maintenance of ER function. In this view, the suppression of metabolic gene expression should be directly responsive to the ER stress burden in real-time, and should correlate with molecular markers of ER dysfunction. This hypothesis is supported by the following observations: First, the suppression of C/EBPα expression and upregulation of C-JUN can be recapitulated in cultured FaO rat hepatoma cells in response to either TM treatment or induction of ER stress by the ER calcium depleting agent thapsigargin (TG) (Figure 4A). This observation confirms that the effect is not TM-specific, but is likely to be a general consequence of ER stress. Second, at least one secretory protein was retained intracellularly in Atf6α-null TM-challenged mice. While the gene encoding serum amyloid protein (SAP) was transcriptionally induced by TM treatment to comparable extents in both wild-type and Atf6α-null animals (Figure S9), its secretion was essentially blocked in the latter, and it was instead detected readily in the cellular homogenate (Figures 4B and 4C). In addition, the concentration of plasma cholesterol in the form of ApoB-containing lipoproteins (VLDL and LDL), was diminished by TM in both genotypes 24 hours after challenge, but to a greater extent in Atf6α-null than wild-type animals 48 hours after challenge (Figures 4D and 4E). Together these data point to suboptimal function of the secretory pathway during challenge in the livers of TM-treated UPR-compromised animals.
Figure 4.
Disruption of ER protein processing suppresses metabolic gene expression. (A) FaO rat hepatoma cells were treated in duplicate with 500 nM TG or 5 μg/ml TM for 8 or 24 hrs, followed by cell lysis and immunoblot for GRP94, BiP, C/EBPα, and C-JUN. In hepatocytes, slower-migrating phospho-C-JUN, induced by ER stress, is also observed. (B, C) Wild-type and Atf6α−/− mice were injected with vehicle or TM (1 mg/kg bodyweight). Protein lysates from liver isolated at the indicated time points after injection were probed by immunoblot for intracellular SAP (B); Plasma level of SAP was assayed by ELISA (C). (D) Total cholesterol and triglyceride levels were measured by enzymatic assay from plasma samples of mice injected with vehicle or TM for 24 hours, with a 6 hour fast preceding sacrifice. (E) Cholesterol and triglyceride represented either as total content, or in the form of LDL and VLDL particles purified by differential precipitation, was measured as in (D). In this case samples were collected 48 hours after TM injection rather than 24 hours. Note that the absolute values are somewhat different comparing panels (D) and (E), likely due to experimental variation. However, in panel (D) there is no significant difference in either cholesterol or triglyceride levels between genotypes after TM injection, while cholesterol levels recover to a greater extent in wild-type animals than Atf6α-null animals in panel (E). The fact that TM has a much greater effect on cholesterol-rich lipoprotein particles than triglyceride-rich particles suggests that TM inhibits liver lipoprotein production but not intestinal production. * = p<0.05
If the suppression of metabolic gene expression exacerbated by genetic deficiency of UPR components is in fact caused by disruption of protein folding and processing in the ER rather than loss of specific UPR-mediated signaling cascades, then direct perturbation of ER protein folding independent of the UPR should produce a similar phenotype. To test this prediction, we used TM to challenge animals nullizygous for the ER-resident DnaJ protein and BiP cochaperone p58IPK (Ladiges et al., 2005). p58-null cells and animals are more sensitive to ER stress than wild-type counterparts, most likely due to the role for p58IPK in enhancing BiP-dependent protein folding (Rutkowski et al., 2007). Strikingly, p58-null animals developed fatty liver after TM injection (Figure 5A) and suppressed expression of C/ebpα (Figures 5B and 5C). As with Atf6α-null and Ire1α-null animals, p58-null animals also showed persistent CHOP upregulation after challenge (Figures 5B and 5C). Thus, the suppression of metabolic genes such as C/ebpα is a direct consequence of ER stress and not dependent upon any single pathway of UPR signaling.
Figure 5.
Disrupted ER protein folding leads to steatosis despite an intact UPR. (A, B, C) Wild-type and p58IPK−/− mice were injected with vehicle or TM (1 mg/kg) and sacrificed 48 hrs post-injection. (A) Livers from TM challenged mice were visualized in situ. (B) Protein lysates from livers were probed by immunoblot as indicated. (C) RNA was analyzed by real-time RT-PCR. Red lettering represents p<0.05 comparing p58−/− treated to untreated.
Compromised ER protein folding suppresses metabolic gene expression partially through CHOP
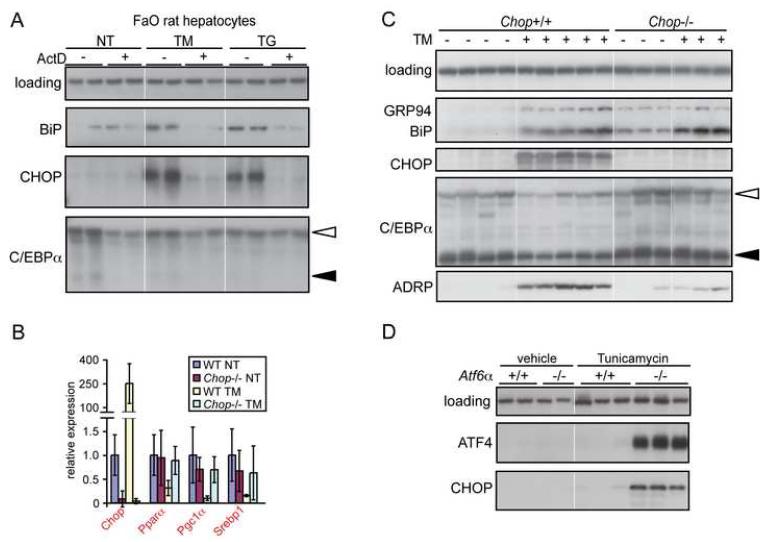
The rapid downregulation of C/ebpα mRNA and others during ER stress could be explained by either induced degradation of these mRNAs, or by inhibition of new synthesis. To discriminate between these possibilities, we treated FaO hepatoma cells with TM or TG in the presence or absence of the transcription inhibitor Actinomycin D (ActD). In the absence of stress, ActD alone dramatically reduced C/EBPα expression, and it essentially nullified the effect of TM or TG on C/EBPα (Figure 6A). Therefore, C/ebpα mRNA is basally degraded very rapidly in hepatocytes, and its stress-mediated downregulation is most likely due to inhibition of its transcription.
Figure 6.
CHOP suppresses expression of metabolic transcription factors. (A) FaO rat hepatoma cells were treated in duplicate with 5 μg/ml ActD and either no stress, 500 nM TG, or 5 μg/ml TM for 4 hours. Protein lysates were then harvested and probed by immunoblot as indicated. Note that ActD treatment completely blocks the upregulation of BiP and CHOP as expected. (B, C) Wild-type and Chop−/− mice were injected with vehicle or TM (1 mg/kg) and sacrificed 24 hours after injection. (B) RNA was analyzed by real-time RT-PCR as Figure 2D. n = 4-10 animals per group. Red lettering represents p<0.05 comparing Chop−/− treated to wild-type treated. (C) Protein lysates from liver were probed by immunoblot as indicated. (D) Nuclear lysates from TM-injected wild-type or Atf6α-null animals (48 hours) were probed for immunoblot as indicated.
One commonality amongst the Atf6α, Ire1α, and p58 genetic models presented here is persistent upregulation and nuclear localization of CHOP as a consequence of ongoing stress, while CHOP is rapidly, but only transiently, upregulated in response to TM challenge in wild-type mice (Figures 1A, S10, and S11). CHOP is a member of the C/EBP family of transcription factors, and has been proposed to be a dominant-negative regulator of their function (Ron and Habener, 1992). We considered the possibility that CHOP upregulation during stress is at least partially responsible for the suppression of metabolic gene expression. Upon challenge of Chop-null animals with TM, expression of the transcriptional regulators C/ebpα, Pparα, Pgc1α, and Srebp1 was less suppressed than in wild-type animals (Figure 6B). This was also observed upon analysis of C/EBPα protein (Figure 6C). Chop deletion also partially protected against lipid accumulation, seen in reduced upregulation of ADRP (Figure 6C). Thus, CHOP forms at least a component of the mechanistic linkage between persistent ER stress and the suppression of transcription factors like C/EBPα.
CHOP was persistently upregulated by stress in our genetic models, from early time-points through late, despite lack of an apparent stimulation of eIF2α phosphorylation (which is presumably a consequence of persistent upregulation of GADD34 downstream of CHOP (Marciniak et al., 2004)) [(Wu et al., 2007) and Figure 1A]. Conversely, in wild-type animals CHOP upregulation is rapidly attenuated despite persistent stimulation of eIF2α phosphorylation. Surprisingly, we found that ATF4 was also upregulated by ER stress in Atf6α−/− animals at both early and late time-points (Figure 6D and data not shown). These data suggest that the upregulation of CHOP, which then influences lipid metabolism, occurs through a dissociation of ATF4 protein levels from the phosphorylation status of eIF2α, by a mechanism that is the subject of ongoing investigation.
Hepatic fat accumulation accompanies diverse ER stresses
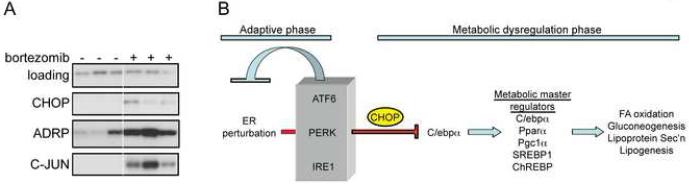
The diversity of physiological causes of hepatic microvesicular steatosis led us to ask whether other less severe forms of ER stress could also lead to fat accumulation and suppression of C/EBPα expression. Remarkably, the proteasome inhibitor bortezomib, which is used clincally to treat multiple myeloma, led to lipid accumulation, as seen by ADRP upregulation (Figure 7A). Published reports suggest possible hepatoxicity of bortezomib (Hernandez-Espinosa et al., 2008; Rosinol et al., 2005). In addition, ADRP and C-JUN upregulation, and suppression of lipogenic genes, were also seen as a consequence of overexpression of the misfolding-prone Factor VIII clotting protein (Figure S12). Thus, lipid accumulation and suppression of C/EBPα are conserved features of the hepatic response to ER stress in general.
Figure 7.
Diverse ER stresses lead to lipid accumulation. (A) Normal mice were injected intravenously with the proteasome inhibitor Velcade (bortezomib) at a dose of 1mg/kg and sacrificed 8 hours after injection. Lysates were probed for upregulation of CHOP, ADRP, and C-JUN. (B) A schematic diagram depicting the joint role of the ATF6, PERK, and IRE1 pathways in maintaining lipid homeostasis. Failure of the UPR to adequately protect the ER results in ongoing production of CHOP, which suppresses metabolic genes via C/EBPα and ultimately leads to disruption of fatty acid oxidation, lipoprotein secretion, gluconeogenesis, and other metabolic processes.
Conclusions and Perpsectives
Our data demonstrate that protection of ER homeostasis by the combined action of the three UPR pathways maintains metabolic function in part by suppressing CHOP expression and the attendant downregulation of genes involved in lipid and energy balance. This work is the first to our knowledge to use a comprehensive genetic approach to understand how the pathways of the vertebrate UPR contribute to the homeostasis of an organ in vivo. Our data allow us to propose a model to describe the proximal events that connect ER stress to lipid homeostasis (Figure 7B). Exposure to ER stress activates all three ER-resident sensors, IRE1α, ATF6α, and PERK. Activation of these sensors induces expression both of genes that facilitate adaptation and/or recovery, such as ER chaperones, and also of anti-adaptive genes including Chop. One of the consequences of Chop upregulation is suppression of genes involved in lipid homeostasis, likely mediated through dominant-negative inhibition of C/EBP family members by CHOP.
We conclude that negative regulation of C/ebpα by CHOP is of key importance in metabolic dysregulation by ER stress in part because the phenotypes we observe—fatty liver, hypoglycemia, and depletion of hepatic glycogen—are also observed upon postnatal deletion of C/ebpα, as are many similar changes in gene expression profiles including expression of metabolic transcriptional regulators (Yang et al., 2005). Indeed, potential binding sites for C/ebpα exist in the promoters of Pparα and Srebp1 (Villacorta et al., 2007). It seems most plausible that ER stress will lead to suppression of C/ebpα partially through CHOP, and consequent suppression of other master regulators of metabolic gene expression such as Srebp1, Pparα, and Pgc1α. Thus, we speculate that unresolved ER stress recapitulates a knockdown of C/EBPα. Whether C/EBPα suppression alone is sufficient to drive these changes, and whether CHOP alone is sufficient to negatively regulate C/EBPα expression, are the subject of ongoing investigation, and we think it is likely that other as yet unknown mechanisms contribute as well. Further, which individual metabolic transcription factors actually directly control fatty acid oxidation under these circumstances is not clear. It is possible that steatosis might be driven entirely by downregulation of PGC1α and/or PPARα, both of which ordinarily control fatty acid oxidation. Thus, it will be important to elucidate the hierarchy of genetic regulation downstream of CHOP and C/EBPα.
Our results suggest that an initial perturbation—even a transient one—will initiate changes in gene regulation that impact lipid metabolism. Assuming the adaptive response is sufficient to overcome the perturbation, the UPR is then largely deactivated and production of proteins involved in lipid metabolism is restored due to rapid degradation of CHOP (Rutkowski et al., 2006). However, if the stress is too severe (elicited in this case by genetic compromise of either UPR pathways or ER quality control) then CHOP-directed suppression of gene expression persists, leading to profound metabolic disruption. Our data support the idea that each of the UPR pathways contributes to adaptation and/or recovery, and that the overlap of UPR targets ensures that the strongest predictor of recovery versus ongoing stress is the expression of adaptive (e.g., ER chaperones) versus anti-adaptive (e.g., CHOP) downstream gene products (Rutkowski et al., 2006).
An important question for future work is what components of the ER stress-lipid homeostasis nexus facilitate adaptation and so are likely to be observed in the course of adaptation to chronic disruptions of liver function, versus those are purely consequences of severe stress and represent metabolic dysfunction. Indeed, while steatosis accompanies organismal susceptibility to ER stress in Atf6α-null models, the cause of death of these animals is not yet known, and inhibition of fatty acid oxidation and lipoprotein secretion might even be protective, at least in certain physiological circumstances. The topic of the role of components of the UPR in regulating lipid homeostasis has been the subject of very recent attention. Both XBP1 and eIF2α have been shown to participate in basal and/or diet-induced regulation of lipid metabolism (Lee et al., 2008; Oyadomari et al., 2008). We have observed in a model cultured cell system that cells adapted to chronic ER stress paradoxically become more resistant to further challenge than naïve cells (Rutkowski et al., 2006). Thus the extent to which dietary stresses might allow for gradual adaptation of UPR-deficient cells, while focal stresses might not, could significantly impact the resultant phenotype. Indeed, the observation that C/EBPα expression apparently promotes steatosis in the dietary context (Oyadomari et al., 2008) while its downregulation promotes steatosis in response to ER stress (this work) is reminiscent of the observation that constitutive embryonic deletion of C/EBPα prevents hepatic lipid accumulation (Flodby et al., 1996; Wang et al., 1995) while deletion in the adult exacerbates it (Yang et al., 2005). Whether our study and the recent XBP1 and eIF2α papers together represent three faces of the same basic signaling pathway or alternatively whether they illuminate the mechanisms of different forms of hepatic dysfunction remains to be investigated.
Our work demonstrates a direct connection between ER dysfunction and the regulation of lipid metabolism through the protective role of the UPR. These findings raise the possibility that ER stress is a contributing factor to the development of hepatic steatosis.
Experimental Procedures
General notes
Many of the techniques described here, including RNA and protein analysis, utilized previously published reagents and standard experimental techniques [(Rutkowski et al., 2006; Wu et al., 2007) and references therein].
Animal experiments
All protocols for animal use were reviewed and approved by the University Committee on Use and Care of Animals at the University of Michigan. Animals were fed standard rodent chow and housed in a controlled environment with 12 hr light and dark cycles. Littermate controls were used for all experiments where possible, and age and gender were matched for each experiment, though neither of these variables was observed to have any effect on the phenotype. Plasma from blood samples was collected by cardiac puncture, using a BD Microtainer with lithium herapin. Plasma levels of SAP were determined using the murine SAP ELISA kit (Immunology Consultants Laboratory, Inc.). TM injections were as described (Zinszner et al., 1998). Insulin injection was intraperitoneally with 2 U/kg BW. Plasma and liver cholesterol and triglyceride levels were determined as described (Iqbal et al., 2008a; Iqbal et al., 2008b). Velcade (bortezomib, from Millenium) was administered intravenously at a dose of 1 mg/kg and mice were sacrificed 8 hours after treatment.
Array analysis
Mice were injected intraperitoneally with 2 mg/kg body weight of TM, or vehicle, and livers were isolated 8 hours after injection. Total RNA was isolated using RNeasy (Qiagen) and analyzed by the University of Michigan Comprehensive Cancer Center (UMCCC) Affymetrix and cDNA Microarray Core Facility (Ann Arbor, Michigan) exactly as described (Wu et al., 2007).
Protein and RNA analysis
Briefly, tissue was homogenized using an electronic homogenizer in RIPA buffer containing protease inhibitors or in Trizol RNA reagent (Invitrogen), and centrifuged at 15,000 rpm for 10 minutes in a microfuge at 4°C. RNA was isolated using TRIzol RNA reagent or RNeasy (Qiagen), according to the manufacturers’ protocols. Additional real-time primer sequences are described in the Supplemental Material (Table S2).
Histological analysis of tissues
Livers were prepared for electron microscopy (EM) by fixing liver tissue using 2.5% glutaraldehyde in 0.1M phosphate salt buffer (pH 7.4). Samples were processed and imaged as described (Rutkowski et al., 2006). For H&E staining, liver tissues were frozen in OCT® in liquid nitrogen-cooled 2-methylbutane. The sections were cut to 6 microns. Livers were visualized in situ using a Leica MZ16FA stereomicroscope.
Cell culture and analysis
FaO rat hepatoma cells were from ATCC and cultured in DMEM medium with 10% fetal bovine serum. Treatment of cells with TM or TG was as described (Rutkowski et al., 2006).
Generation of mouse models
Creation of Atf6α-null, p58IPK-null, and Chop-null mice has been described elsewhere (Ladiges et al., 2005; Wu et al., 2007; Zinszner et al., 1998). eIF2αAA:tg/+:Alb-cre mice were created by introduction of a transgene encoding constitutively expressed wild-type eIF2α flanked by LoxP sites into the eIF2α S51A knockin background (SHB and RJK, submitted). These mice were then bred to mice expressing CRE recombinase under control of the albumin promoter for liver-specific transgene deletion. Similarly, liver-specific Ire1α knockout mice were created by ES-targeted homologous recombination of Exons 16 and 17 flanked by LoxP sites. Mating to albumin-CRE expressing mice allowed for liver-specific deletion of these exons, resulting in loss of the IRE1α kinase domain. Additional information for eIF2αAA:tg/+:Alb-cre mice can also be found in the Supplemental Materials. Ire1α deletion is described in the Supplemental Materials section of (Sakaki et al., 2008).
Supplementary Material
Acknowledgments
Portions of this work were supported by NIH grants DK042394, HL052173, and HL057346 to RJK. RJK is an Investigator of the Howard Hughes Medical Institute. Additional funding sources were UI Department of Anatomy and Cell Biology startup funds to DTR, NIH grant P60 DK020572 to the Michigan Diabetes Research and Training Center Cell and Molecular Biology Core facility, a grant from the Hemophilia of Georgia Foundation to MUC, and NIH grant DK46700 to MMH. We are particularly grateful to D. Ron for Chop−/− mice. We also thank J. Goldstein for SREBP1 and SREBP2 antibodies, D. Silver and J. Estall for insightful advice, D. Ginsburg and J. Engelhardt for equipment use, A. Goodman and B. Spiegelman for critically evaluating the manuscript, and members of the Kaufman lab for stimulating discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 Is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Espinosa D, Minano A, Martinez C, Ordonez A, Perez-Ceballos E, de Arriba F, Mota RA, Ferrer F, Gonzalez M, Vicente V, Corral J. Inhibition of proteasome by bortezomib causes intracellular aggregation of hepatic serpins and increases the latent circulating form of antithrombin. Lab Invest. 2008;88:306–317. doi: 10.1038/labinvest.3700717. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. [Google Scholar]
- Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, Ron D, Tabas I, Hussain MM. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008a;7:445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Rudel LL, Hussain MM. Microsomal triglyceride transfer protein enhances cellular cholesteryl esterification by relieving product inhibition. J Biol Chem. 2008b doi: 10.1074/jbc.M800398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ. Mechanisms of Hepatotoxicity. Toxicological Sciences. 2002;65 doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, Macauley A, Goodman AG, Leboeuf RC, Katze MG. Pancreatic {beta}-Cell Failure and Diabetes in Mice With a Deletion Mutation of the Endoplasmic Reticulum Molecular Chaperone Gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Nagasawa K, Higashi T, Hosokawa N, Kaufman RJ, Nagata K. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep. 2007;8:483–489. doi: 10.1038/sj.embor.7400933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Leray V, Diez M, Serisier S, Le Bloc’h J, Siliart B, Dumon H. Liver lipid metabolism. Journal of Animal Physiology and Animal Nutrition. 2008;92:272–283. doi: 10.1111/j.1439-0396.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Özcan U, Yilmaz E, Özcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Dentin R, Girard J. Role of the liver in the control of carbohydrate and lipid homeostasis. Diabetes Metab. 2004;30:398–408. doi: 10.1016/s1262-3636(07)70133-7. [DOI] [PubMed] [Google Scholar]
- Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe M, Veniant MM, Sullivan MA, Zlot CH, Bjorkegren J, Nielsen LB, Wong JS, Hamilton RL, Young SG. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Invest. 1999;103:1287–1298. doi: 10.1172/JCI6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Reddy JK. Peroxisomal beta-oxidation and steatohepatitis. Semin Liver Dis. 2001;21:43–55. doi: 10.1055/s-2001-12928. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosinol L, Montoto S, Cibeira MT, Blade J. Bortezomib-induced severe hepatitis in multiple myeloma: a case report. Arch Intern Med. 2005;165:464–465. doi: 10.1001/archinte.165.4.464. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Akha A. A. Sadighi, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki K, Wu J, Kaufman RJ. Protein Kinase C{theta} Is Required for Autophagy in Response to Stress in the Endoplasmic Reticulum. J Biol Chem. 2008;283:15370–15380. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Villacorta L, Garcia-Barrio MT, Chen YE. Transcriptional regulation of peroxisome proliferator-activated receptors and liver X receptors. Curr Atheroscler Rep. 2007;9:230–237. doi: 10.1007/s11883-007-0024-5. [DOI] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yang J, Croniger CM, Lekstrom-Himes J, Zhang P, Fenyus M, Tenen DG, Darlington GJ, Hanson RW. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J Biol Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.