Abstract
Background
The Abelson helper integration-1 (AHI1) gene is required for both cerebellar and cortical development in humans. While the accelerated evolution of AHI1 in the human lineage indicates a role in cognitive (dys)function, a linkage scan in large pedigrees identified AHI1 as a positional candidate for schizophrenia. To further investigate the contribution of AHI1 to the susceptibility of schizophrenia, we evaluated the effect of AHI1 variation on the vulnerability to psychosis in two samples from Spain and Germany.
Methodology/Principal Findings
29 single-nucleotide polymorphisms (SNPs) located in a genomic region including the AHI1 gene were genotyped in two samples from Spain (280 patients with psychotic disorders; 348 controls) and Germany (247 patients with schizophrenic disorders; 360 controls). Allelic, genotypic and haplotype frequencies were compared between cases and controls in both samples separately, as well as in the combined sample. The effect of genotype on several psychopathological measures (BPRS, KGV, PANSS) assessed in a Spanish subsample was also evaluated. We found several significant associations in the Spanish sample. Particularly, rs7750586 and rs911507, both located upstream of the AHI1 coding region, were found to be associated with schizophrenia in the analysis of genotypic (p = 0.0033, and 0.031, respectively) and allelic frequencies (p = 0.001 in both cases). Moreover, several other risk and protective haplotypes were detected (0.006<p<0.036). Joint analysis also supported the association of rs7750586 and rs911507 with the risk for schizophrenia. The analysis of clinical measures also revealed an effect on symptom severity (minimum P value = 0.0037).
Conclusions/Significance
Our data support, in agreement with previous reports, an effect of AHI1 variation on the susceptibility to schizophrenia in central and southern European populations.
Introduction
Because of its early onset, severity and chronic course schizophrenia is one of the most devastating neuropsychiatric disorders and represents a major public health concern [1]. Although the pathogenetic mechanisms leading to this disorder are largely unclear, it is well accepted that schizophrenia is the result of a complex interplay between genetic variants and environmental factors of different nature [2]–[4]. Until now, the search for susceptibility genes has given promising results, although some inconsistencies have also arisen [5]–[7]. Interestingly, several linkage studies have located putative schizophrenia as well as bipolar disorder susceptibility loci on the long arm of chromosome 6, and particularly in the 6q15–23.2 region [8]. Different genome-wide association studies (GWAS) also support association between Single Nucleotide Polymorphisms (SNPs) located in 6q21–6q25 and psychotic disorders [9]–[12]. In agreement with these findings, an autosomal scan of Arab-Israeli families [13], [14] supported linkage to schizophrenia to 6q23. This linkage peak contains a neurodevelopmental gene, the Abelson helper integration site 1 gene (AHI1). This gene is required for both cerebellar and cortical development in humans and several null-type and missense mutations in AHI1 have been shown to cause Joubert syndrome, a rare autosomal recessive neurodevelopmental disorder, characterized by cognitive and behavioural disturbances among other clinical symptoms [15], [16].
The human AHI1 gene contains 31 exons and generates at least three alternative isoforms. AHI1 protein, also known as Jouberin [16], contains seven Trp-Asp (WD) repeats, a Src homology 3 (SH3) domain and a coiled-coil domain. Jouberin is conserved in different mammalian species, particularly in the WD40 and SH3 domains [15]. The presence of both WD40 and SH3 domains in signaling and adaptor molecules suggests that AHI1 may play an important role in signal transduction in normal cells, as an adaptor protein recruiting other signaling molecules and modulating and integrating their action [16], [17]. However, little is known about how AHI1 is involved in the development of the central nervous system (CNS). AHI1 mRNA is highly expressed in both the developing and the mature brain [15], [16]. Interestingly, it has been found [18] that murine Ahi1 protein forms a stable complex with huntingtin-associated protein 1 (Hap1). This complex appears to be critical for neonatal development through its function in intracellular trafficking, neurogenesis and neuronal differentiation.
Taken together, AHI1 is an attractive candidate for a schizophrenia susceptibility gene and three association studies have been performed with promising results. In the first study in an inbred Arab-Israeli family sample and in an outbred nuclear family sample, Amann-Zalcenstein et al. [19] were able to identify seven markers strongly associated with schizophrenia. The association was found in a 500 kb linkage disequilibrium (LD) block on chromosome 6 harbouring AHI1. Subsequently, this association has been replicated in an independent European cohort from Iceland [20], in another large European sample [21], as well as in an enlarged sample from the original study [22]. In the latter, the significant SNPs were localized in a substructure of the large original LD block, which contains AHI1 and the C6orf217 gene, and may contain regulatory elements for both the AHI1 and the neighboring phosphodiesterase 7B gene (PDE7B). Furthermore, there is also evidence of an association between an AHI1 haplotype and autistic disorder (ASD) in a region of the gene that had been previously associated with schizophrenia [23].
Here we report the results from a case-control association study performed on two different samples from Germany and Spain. Our main aim was to test the hypothesis that AHI1 variants were associated with schizophrenia as well as dimensional measures of disease severity.
Results
Spanish sample
SNP18 (rs4896156) deviated from Hardy-Weinberg Equilibrium (HWE) in the control sample (P = 3.89.10−6), clearly suggesting a low genotyping quality for this polymorphism. For this reason, this SNP was discarded.
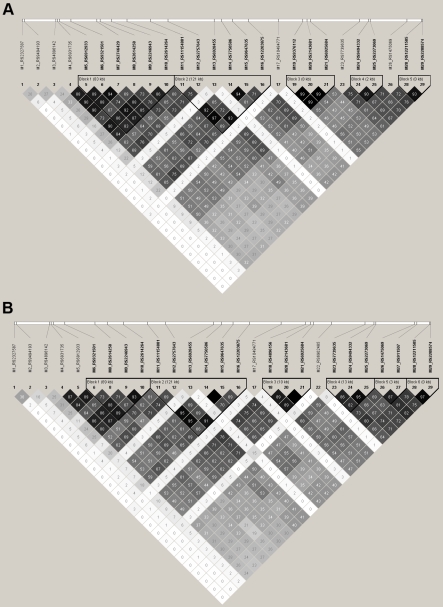
Figure 1a shows the pairwise linkage disequilibrium values observed in the control sample. The LD pattern from the schizophrenic sample (information not shown) was very similar. r2 values ranged between 0.0 and 0.993. According to Gabriel et al. [24] criteria, five LD blocks were detected. SNPs 1, 2, 3, 13 and 17 presented the lowest LD values (both r2 and D') when compared with the rest of the SNPs.
Figure 1. Linkage disequilibrium (LD) pattern of AHI1 gene.
Figures represent pairwise r2 values observed in control subjects from German (a) and Spanish (b) origin. Values are represented in a grayscale ranging from white (no LD) to black (high LD).
Single SNP association analysis revealed that SNP14 (rs7750586) was associated with psychosis, since both the G allele and the G/G genotype were more frequent in the group of patients with schizophrenia (P = 0.001 for the genotypic comparisons (recessive model); see table 1 for the allelic comparisons). The G allele of SNP27 (rs911507) also was associated with schizophrenia (table 1) and the same result was observed in the analysis of genotypic frequencies (P = 0.001, recessive model). Nevertheless, the significant values did not withstand the correction for multiple testing in the analysis of the allelic frequencies, although the P value corresponding to the analysis of genotypic frequencies still remained significant (P = 0.02 for both SNPs). We also performed a haplotype analysis, as well as an analysis of two-, three- and four marker haplotype sliding windows. As a result, several significant risk and protective haplotypes were detected (see table 1). The haplotypes encompassed SNPs 1, 2, 3 and 4 (located in the 3′ region of the analyzed genomic area), as well as markers 13, 14, 15 and 16 (located in intron 2 and in the 5′ putative regulatory region of AHI1 gene). Interestingly, 5 of these significant haplotypes remained significant after a 1000-permutation run.
Table 1. Single alleles and haplotypes of the AHI1 region associated with schizophrenia in the present study.
| GERMAN SAMPLE (N = 247 patients+360 controls) | SPANISH SAMPLE (N = 280 patients+348 controls) | COMBINED SAMPLE (N = 558 patients+708 controls) | ||||||||||
| Single SNP or haplotypea | Cont freq | SCZ freq | OR (95% CI) | P | Cont freq | SCZ freq | OR (95% CI) | P | Cont freq | SCZ freq | OR (95% CI) | P |
| SNP14 (rs7750586) | 0.353 (G) | 0.323 (G) | 0.88 (0.67–1.14) | 0.315 | 0.286 (G) | 0.380 (G) | 1.53 (1.15–2.04) | 0.003 (0.086) | 0.325 | 0.360 | 1.52 (1.13–2.06) | 0.006 |
| SNP27 (rs911507) | 0.342 (G) | 0.356 (G) | 1.07 (0.82–1.39) | 0.619 | 0.370 | 0.441 | 1.34 (1.03–1.76) | 0.031 (0.81) | 0.331 | 0.386 | 1.34 (1.02–1.77) | 0.038 b |
| SNP1-SNP4 (TAGG) | 0.108 | 0.081 | 0.43 (0.031–5.87) | 0.277 | 0.0868 | 0.025 | 0.69 (0.19–3.91) | 0.006 (global:0.004) | 0.099 | 0.055 | 0.71 (0.19–2.54) | 0.011 (global:0.126) |
| SNP1- SNP4 (TATA) | 0.222 | 0.223 | 0.57 (0.04–7.43) | 0.853 | 0.212 | 0.305 | 3.41 (0.73–16) | 0.007 (ns after perm. run) (global:0.004) | 0.217 | 0.264 | 1.55 (0.45–5.29) | 0.027 (global:0.126) |
| SNP13-SNP15 (TAA) | 0.588 | 0.600 | 0.85 (0.51–1.44) | 0.663 | 0.628 | 0.555 | 0.91 (0.51–1.61) | 0.036 (global:0.044) | 0.603 | 0.578 | 0.89 (0.59–1.31) | 0.324 (global:0.458) |
| SNP13-SNP15 (TGC) | 0.348 | 0.328 | 0.79 (0.46–1.35) | 0.469 | 0.293 | 0.380 | 1.33 (0.74–2.41) | 0.009 (global:0.044) | 0.325 | 0.353 | 1.00 (0.67–1.49) | 0.255 (global:0.458) |
| SNP14-SNP16 (AAA) | 0.572 | 0.588 | 1 (reference) | 0.613 | 0.609 | 0.529 | 1 (ref) | 0.025 (ns after perm. run) (global:0.046) | 0.587 | 0.560 | 1 (ref) | 0.276 (global:0.489) |
| SNP14-SNP16 (GCG) | 0.345 | 0.328 | 0.92 (0.70–1.22) | 0.584 | 0.288 | 0.369 | 1.48 (1.08–2.00) | 0.014 (global:0.046) | 0.321 | 0.348 | 1.13 (0.92–1.39) | 0.237 (global:0.489) |
Significant values are marked in bold. The corrected P-values are indicated in brackets.
To avoid redundant information, other significant haplotypes, which are variations of other larger significant haplotypes from this table, have not been included.
SNP27 had significantly different genotypic distributions in the German and the Spanish sample (heterogeneity P value = 0.016 uncorrected).
Abbreviations: Cont, control; SCZ, schizophrenia; ns, not significant; perm, permutation; ref, reference haplotype.
We next studied whether any of the SNPs associated to risk for psychosis in the single-SNP and haplotype analysis (SNP1, SNP2, SNP3, SNP4, SNP13, SNP14, SNP15, SNP16, and SNP27) had any impact on several clinical scores, namely Brief Psychiatric Rating Scale (BPRS), Psychiatric Assessment scale or KGV Symptom scale, and finally the total, general, positive and negative scores from the Positive and Negative Syndrome scale (PANSS). Significant results are displayed in table 2 with SNP4, SNP14 and SNP15 appearing to modulate the severity of psychotic symptoms, evaluated through PANSS, BPRS and KGV scales.
Table 2. AHI1 SNPs significantly associated with clinical traits in the Spanish sample.
| SNP | Genotypic distribution | Average response (SE) | Difference (95% CI) | Model | Pa |
| PANSS total score | |||||
| SNP4 (rs6931735) | A/A–G/A = 61 | 61.98 (2.06) | 0.00 | Recessive | 0.05 (0.3) |
| G/G = 19 | 70.68 (4.27) | 8.70 (0.12–17.29) | |||
| SNP14 (rs7750586) | A/A–A/G = 59 | 60.25 (1.66) | 0.00 | Recessive | 0.017 (0.136) |
| G/G = 18 | 70.06 (4.85) | 9.80 (1.94–17.66) | |||
| SNP15 (rs9647635) | A/A–C/A = 60 | 63.58 (2.09) | 0.00 | Recessive | 0.023 (0.161) |
| C/C = 13 | 75.62 (5.63) | 12.03 (1.86–22.20) | |||
| PANSS General Score | |||||
| SNP4 (rs6931735) | A/A–G/A = 61 | 31.54 (1.05) | 0.00 | Recessive | 0.042 (0.252) |
| G/G = 19 | 36.26 (2.37) | 4.72 (0.23–9.21) | |||
| SNP14 (rs7750586) | A/A–A/G = 59 | 30.78 (0.84) | 0.00 | Recessive | 0.016 (0.112) |
| G/G = 18 | 36.17 (2.86) | 5.39 (1.11–9.66) | |||
| SNP15 (rs9647635) | A/A–C/A = 60 | 32.28 (1.02) | 0.00 | Recessive | 0.0037 (0.029) |
| C/C = 13 | 40.23 (3.25) | 7.95 (2.76–13.13) | |||
| BPRS | |||||
| SNP14 (rs7750586) | A/A–A/G = 59 | 44.8 (1.22) | 0.00 | Recessive | 0.045 (0.315) |
| G/G = 19 | 50.5 (3.15) | 5.70 (0.21–11.19) | |||
| KGV scale | |||||
| SNP15 (rs9647635) | A/A–C/A = 157 | 9.35 (0.41) | 0.00 | Recessive | 0.049 (0.392) |
| C/C = 30 | 11.43 (1.05) | 2.08 (0.02–4.14) | |||
Significant P values (P<0.05) are indicated in bold.
aThe corrected P value is indicated in brackets.
Abbreviations: SE, standard error; CI, confidence interval.
German sample
The genotyping for SNP7 and SNP19 did not pass the quality control in the sample of healthy subjects and were omitted from further analysis. LD structure for the control sample can be seen in figure 1b. These values were very similar for the affected subjects (data not shown) and ranged between 0.0 and 0.87. 6 blocks according to Gabriel et al. [24] criteria were detected. By contrast, the 3′ region of the AHI1 gene (SNP1 to SNP3) featured lower pairwise LD values.
Regarding the single SNP association analysis (table 1), none of the 29 AHI1 SNPs selected for the study differed in their allelic and genotypic frequencies between schizophrenic patients and controls (P<0.01). The haplotype analysis also yielded negative results.
Combined sample
The German and the Spanish samples were combined to perform a heterogeneity test and an association analysis of the genotypic frequencies (Table 1) through Cochran-Mantel-Haenszel (CMH) tests, as well as a haplotype analysis through a retrospective likelihood algorithm combined with a χ2 test. Heterogeneity tests revealed a significantly different genotype distribution of SNPs 1, 11, 12, 17, 22 and 27 in both the Spanish and the German sample (data not shown). Overall, we detected differences between patients and controls for two SNPs (SNP14 and SNP27), which were most likely due to a specific association in the Spanish sample.
Discussion
Here we present the results for a case-control association analysis that explores the impact of variation at AHI1 locus on the vulnerability towards schizophrenia in two European samples of Caucasian origin from Southern Germany and East Spain. For this analysis, we focused on 29 tag SNPs located in the region corresponding to the linkage peak detected by Amann-Zalcenstein et al. [19]. Among these SNPs, we included those 7 SNPs which have been widely studied and associated with schizophrenia in different populations [19]–[21]. Several alleles and haplotypes were significantly associated with the risk for schizophrenia in the Spanish and, less pronounced, the combined sample. Some protective haplotypes were also found.
There are several reasons to consider AHI1 a candidate gene for psychotic disorders. First, AHI1 expression has been detected in brain areas particularly related to the pathophysiology of psychotic symptoms, including hippocampus, cerebral cortex, mesolimbic pathways, cerebellum, inferior colliculus and dorsal cochlear nucleus [15], [25]–[30]. Another interesting issue is the accelerated evolution, probably due to positive selection, of AHI1 along the primate lineage leading to humans [15], [31]. Given that AHI1 is critical for a proper neurodevelopment, accelerated evolution of AHI1 in the human lineage may indicate the effect of directional selection. These observations suggest the involvement of this gene in the acquisition of higher cognitive and mental abilities in humans, a phenomenon which can be linked to the appearance of mental disorders such as schizophrenia [32]–[34]. Remarkably, it is not uncommon that genes causing monogenic rare diseases can also contribute to common complex diseases [35], [36]. Therefore, it cannot be excluded that more subtle variation in AHI1 than those causing Joubert syndrome may have a role in the vulnerability to neuropsychiatric disorders.
While the study presented here was designed and performed, three case-control association studies reported the involvement of AHI1 in the susceptibility to schizophrenia [19], [20] and autistic disorder [23]. According to these antecedents, we performed the present study, which intends to extend the study of AHI1 gene to other populations in an attempt to find schizophrenia-associating markers in the Caucasian European context.
In summary, support for a role of AHI1 in psychotic disorder was obtained in the sample from Valencia (East Spain), in which two SNPs located upstream from AHI1 gene (SNP14-rs7750586 and SNP27-rs911507) were significantly associated with schizophrenia, with a moderate effect size (Odds Ratios (ORs) were 1.53 and 1.34, respectively). However, with regard to rs7750586, the risk allele associated with schizophrenia in our study (G allele) was different than in the two previous studies [19]–[20]. Moreover, we also found protective as well as risk haplotypes spanning SNP1 to SNP4, and SNP13 to SNP16, i.e. including the associated risk marker. This marker also impacted dimensional severity scores of schizophrenia, lending further support to the categorical case-control finding. Remarkably, many of the significant SNPs are located in putative regulatory regions. The significant haplotypes including SNP1 to SNP4 are of special interest because they span a region encoding the SH3 domain, one of the most important regions of Jouberin and particularly conserved in mammals [15], [16].
With regard to the negative results in the German sample, it should be noted that this sample was enriched for chronic, non-remitting schizophrenia, so that one might hypothesize that AHI1 is associated with less severe, remitting forms of this disorder, a hypothesis which could easily be tested in further studies. In addition, we may also consider that the existence of differences in the clinical phenotyping of patients (DSM-IV criteria were used in the Spanish sample, whereas ICD-10 was the reference in the German dataset) may explain the differences observed between both samples in the case-control association analysis.
Interestingly, we also found some SNPs associating with schizophrenia in the combined German-Spanish sample. The higher size of this sample is of special interest because it increases the statistical power to detect association with the risk for schizophrenia (table 2). Indeed, sample size is one of the most important issues in the study of the genetics of complex diseases. However, the significant association observed in the joint dataset reduced significance of the results from the Spanish sample, therefore suggesting that the positive finding in the combined sample is mainly due to the SNP distribution in the Spanish sample. The differences among samples observed in the heterogeneity tests for some SNPs can also make it difficult to perform and interpret a study with a joint group of individuals from different origin. Indeed, it cannot be discarded that the differences observed between the Spanish and the German sample could be related to differences in the genetic structure of both populations. Although European populations share an important part of their genetic structure, they also present clear differences which should be taken into account when a case-control association analysis is designed [37]–[39]. These differences in the genetic structure of Spanish and German populations can involve other genetic interacting factors, which, in turn, may modulate the effect of the polymorphism of interest on the vulnerability to a complex disease. In addition, the differences in the directionality of the findings between the Spanish sample and previous reports also point to different issues. One paradigmatic example of this situation is the dysbindin gene, where every one of its five major haplotypes has been associated with schizophrenia [40]. This fact can be related, on one hand, to differences among populations and, on the other hand, to different study designs. In our study, to avoid an effect due to population stratification [41], [42], affected and control subjects were carefully matched on ethnicity, gender and age, and for the German sample the absence of population substructure has already been shown [43]. Finally, as it has been previously commented, we cannot discard that differences in the LD pattern may mask the existence of common causal variants among populations.
Finally, as a complement to our case-control association study, we evaluated the impact on several clinical variables (PANSS, BPRS and KGV scales) of those AHI1 SNPs associated with schizophrenia in this study. Although we found a slight effect of AHI1 gene on symptom severity, additional studies on more specific variables (particularly of cognitive nature) and with more individuals would be necessary to confirm this finding.
In conclusion, the findings from the present study lend support to the notion that the AHI1 locus increases the genetic risk for psychoses at least in some populations. However, additional analyses with larger samples are necessary. Indeed, while this manuscript was in preparation, two additional analyses were published, one of them including a large number of different European samples [21], as well as another study that focused on a dense SNP map of the AHI1 region [22]. Both studies also supported the hypothesis that AHI1 is involved in the pathogenesis of schizophrenia. Therefore, our study, together with previous studies in different Caucasian populations, supports the relationship between genetic variation in AHI1 and the risk for schizophrenia.
Materials and Methods
Ethics Statement
All subjects gave their written informed consent to participate in this study, which was approved by the Ethical Committees of the Medical Faculty, University of Valencia and the University of Würzburg.
Clinical samples
The Spanish sample consisted of 280 unrelated psychotic patients and 348 control subjects. Patients came from the psychiatric in-patient and out-patient units of the Mental Health Service 4 of the Clinical Hospital, University of Valencia, Spain. Age ranged from 18 to 77 years. The retrospective clinical data collected from each patient were compared with the information provided from previous clinical reports and family members. All patients met DSM-IV criteria for different psychoses, mainly schizophrenia (76%). These diagnoses were confirmed for every patient by a consensus meeting with the treating psychiatrist and one of the psychiatrists of our research group. Patients also had a minimum one-year evolution of the illness and were on antipsychotic treatment at evaluation time. Exclusion criteria for this study included incoherence of speech and/or the incapacity for basic comprehension of the questions. Symptom severity was assessed in some patients through BPRS, KGV and PANSS scales (80, 200 and 80 individuals, respectively). The 348 healthy unrelated subjects had no history or familial background of psychiatric disorders. To avoid sample stratification, these subjects had similar demographic characteristics (Caucasian ethnic group, similar age) to the psychotic group. They were between 18 and 91 years old and were also of Spanish origin. Drug abuse was also considered among the exclusion criteria.
The German sample was enrolled at the Department of Psychiatry, Psychosomatics and Psychotherapy, University of Würzburg. A total of 247 unrelated patients meeting ICD-10 criteria for schizophrenia (mean age, 42 years) took part in the study. They were at least once in-patients at the Department of Psychiatry and none of the patients remitted completely during the course of disease, i.e. the sample was enriched for chronic and severe schizophrenia. Diagnoses were made by an extensive, semi structured interview performed by an experienced psychiatrist. Clinical data obtained from the interview was contrasted with the information obtained from family members and previous clinical assessments. Symptom severity was assessed in some individuals through BPRS. Patients did not show other disorders (neurological disorders, epilepsy, mental retardation) which could be the underlying cause of the psychiatric disorder. Substance-induced psychosis and affective disorders were also considered as exclusion criteria. A total of 360 control subjects were also included in the study. They were healthy blood donors from the same catchment area as the patient group (Lower Franconia, Bavaria). To avoid ethnic stratification, they were also from Caucasian origin. Although these individuals were not assessed for psychiatric disorders, all of them were free of medication and the aim of the study was explained to them, so it is unlikely that the subjects from the control group are suffering from severe psychiatric disorders. Only those patients and controls who gave their written informed consent were accepted for the study.
Genotyping
Genomic DNA was isolated from the peripheral blood of patients and controls according to standard procedures. Thirty single-nucleotide polymorphisms (SNPs) were initially selected from previous studies. However, one of them (rs6935033) was discarded during the design of the genotyping assay. Finally, twenty-nine SNPs were included in the study (see table 3 and figure 2 for more information about the markers). These SNPs covered the entire AHI1 genomic region, as well as a 25 kb region located 5′ of AHI1. Upstream SNPs of AHI1 were analyzed because the highest association signals from the original study from Amann-Zalcenstein et al. [19] were located in this region. SNP genotyping was performed in both samples through the iPLEX assay on the MassARRAY platform (Sequenom, San Diego, CA, USA), which allows high throughput genotyping through multiplex reactions. Exclusion criteria during quality control of the genotyping procedure were the following: HWE P-value below 0.05 in the control sample, genotyping rate below 70% and minor allele frequency (MAF) below 0.04. According to these criteria, one SNP (SNP18) was discarded in the Spanish sample (HWE P-value = 0.0389 in the control sample, genotyping rate 53,1%), as well as two SNPs (SNP7 and SNP19) were discarded in the German sample (genotyping rate 0% in the control sample). Moreover, in the Spanish sample, 25 controls and 37 patients were discarded due to the low genotyping rate, while two controls and three patients were discarded in the German sample. The high number of discarded samples in the Spanish dataset may be due to the DNA extraction procedures, given that a subset of Spanish DNA samples was extracted by the phenol-chloroform methodology.
Table 3. List of SNPs included in the present study.
| rs code (or other name) | Position (dbSNP Build 127) | Marker order | Distance from SNP1 | Allele change | CEU MAF | Location/Function |
| rs2327587 | 135622646 | SNP1 | 0 | C/T | 0.408 (A) | 3′ near gene |
| rs9494193 | 135625316 | SNP2 | 2670 | A/C | 0.433 (C) | 3′ near gene |
| rs4896142 | 135664512 | SNP3 | 41866 | G/T | 0.383 (G) | Intron 25 |
| rs6931735 | 135666504 | SNP4 | 43858 | A/G | 0.408 (G) | Intron 25 |
| rs6912933 | 135669227 | SNP5 | 46581 | A/G | 0.425 (G) | Intron 25 |
| rs9321501 | 135683110 | SNP6 | 60464 | A/C | 0.397 (C) | Intron 24 |
| rs2746429 | 135714977 | SNP7 | 92331 | C/T | 0.395 (C) | Intron 23 |
| rs2614258 | 135718895 | SNP8 | 96249 | A/G | 0.350 (A) | Intron 23 |
| rs2246943 | 135733209 | SNP9 | 110563 | A/T | 0.400 (A) | Intron 22 |
| rs2614264 | 135752387 | SNP10 | 129741 | A/G | 0.400 (G) | Intron 22 |
| rs11154801 | 135781048 | SNP11 | 158402 | A/C | 0.317 (A) | Intron 19 |
| rs2757643 | 135802926 | SNP12 | 180280 | A/C | 0.381 (A) | Intron 13 |
| rs6928455 | 135859828 | SNP13 | 237182 | C/T | 0.051 (C) | Intron 2 |
| rs7750586 | 135869366 | SNP14 | 246720 | A/G | 0.325 (G) | 5′ near gene |
| rs9647635 | 135882749 | SNP15 | 260103 | A/C | 0.322 (C) | 5′ near gene |
| rs12203875 | 135902107 | SNP16 | 279461 | A/G | 0.383 (G) | 5′ near gene |
| rs10484771 | 135939628 | SNP17 | 316982 | C/T | 0.058 (T) | 5′ near gene |
| rs4896156 | 135968556 | SNP18 | 345910 | A/G | 0.322 (G) | 5′ near gene |
| rs9376112 | 135969693 | SNP19 | 347047 | A/C | 0.308 (C) | 5′ near gene |
| rs2143681 | 135974698 | SNP20 | 352052 | A/G | 0.314 (A) | 5′ near gene |
| rs6925684 | 135978571 | SNP21 | 355925 | A/G | 0.312 (A) | 5′ near gene |
| rs6902485 | 135999485 | SNP22 | 376839 | A/G | 0.042 (G) | 5′ near gene |
| rs7739635 | 136039471 | SNP23 | 416825 | C/T | 0.322 (T) | 5′ near gene |
| rs9494332 | 136050301 | SNP24 | 427655 | A/G | 0.280 (G) | 5′ near gene |
| rs2273069 | 136052491 | SNP25 | 429845 | A/G | 0.310 (G) in Asians | 5′ near gene |
| rs1475069 | 136097927 | SNP26 | 475281 | A/C | 0.283 (C) | 5′ near gene |
| rs911507 | 136101116 | SNP27 | 478470 | A/G | 0.230 (G) | 5′ near gene |
| rs12211505 | 136116514 | SNP28 | 493868 | C/T | 0.250 (C) | 5′ near gene |
| rs2208574 | 136117310 | SNP29 | 494664 | A/G | 0.258 (A) | 5′ near gene |
Abbreviations: CEU, CEPH collection - DNA samples of Utah residents with ancestry from northern and western Europe; MAF, minor allele frequency.
Figure 2. Genomic region and SNPs analyzed in this study.
The position of each SNP is indicated with an orange arrow.
Statistical analysis
Prior to any association analysis, it is important to make a priori calculation to know the statistical power of the study. This power varies attending to different parameters, such as the sample size, the risk allele frequency, the incidence of the disease and the risk (odds ratio) assumed for each polymorphism, among others. For this study, QUANTO software version 1.2.3 was used to calculate the statistical power to find associations between genetic polymorphisms and the risk for schizophrenia. We assumed an odds ratio (OR) between 1.3 and 2. We also set the incidence of schizophrenia at 1% and the inheritance model as additive. The estimations for our different samples are summarized in table 4. Particularly, this power ranged between 0.149 and 0.999 depending on the sample size, the risk associated to each allele and the allele frequencies. Thus, for those polymorphisms with a low MAF, the statistical power to detect association with the disease is expected to be lower, especially if the risk is low. In our particular case, 21 of the 29 SNPs presented a MAF equal to or higher that 0.3, and only 3 SNPs had a MAF lower than 0.1. Therefore, we consider the statistical power of the study to be adequate.
Table 4. Estimation of the statistical power associated to the different case-control and family-based studies.
| Type of study | Sample | N | Frequency of the risk allelea | Statistical Power | ||
| OR = 1.3 | OR = 1.6 | OR = 1.9 | ||||
| Case-control | Spanish | 243 patients | 0.04 | 0.149 | 0.307 | 0.678 |
| 323 controls | 0.09 | 0.261 | 0.554 | 0.933 | ||
| 0.14 | 0.355 | 0.709 | 0.984 | |||
| 0.19 | 0.429 | 0.801 | 0.995 | |||
| 0.24 | 0.487 | 0.855 | 0.998 | |||
| 0.29 | 0.529 | 0.887 | 0.999 | |||
| 0.34 | 0.558 | 0.906 | 0.999 | |||
| 0.39 | 0.577 | 0.916 | 0.999 | |||
| 0.44 | 0.586 | 0.919 | 0.999 | |||
| Case-control | German | 243 patients | 0.04 | 0.153 | 0.318 | 0.698 |
| 358 controls | 0.09 | 0.271 | 0.573 | 0.943 | ||
| 0.14 | 0.368 | 0.728 | 0.987 | |||
| 0.19 | 0.445 | 0.818 | 0.997 | |||
| 0.24 | 0.503 | 0.870 | 0.999 | |||
| 0.29 | 0.546 | 0.899 | 0.999 | |||
| 0.34 | 0.576 | 0.917 | 0.999 | |||
| 0.39 | 0.594 | 0.926 | 0.999 | |||
| 0.44 | 0.603 | 0.929 | 0.999 | |||
| Case-control | Combined | 486 patients | 0.04 | 0.252 | 0.540 | 0.930 |
| 681 controls | 0.09 | 0.464 | 0.843 | 0.978 | ||
| 0.14 | 0.613 | 0.945 | 0.989 | |||
| 0.19 | 0.713 | 0.978 | 0.999 | |||
| 0.24 | 0.779 | 0.989 | 0.999 | |||
| 0.29 | 0.821 | 0.994 | 0.999 | |||
| 0.34 | 0.847 | 0.996 | 0.999 | |||
| 0.39 | 0.862 | 0.997 | 0.999 | |||
| 0.44 | 0.869 | 0.997 | 0.999 | |||
aThe risk allele was considered to be the minor allele.
Abbreviation: OR, Odds Ratio.
Genotypes were assessed for Hardy-Weinberg Equilibrium (HWE) in both patient and control samples by applying a χ2 test implemented with Haploview Program version 4 [44]. As the deviation from HWE in the control sample could be indicating a genotyping error, those SNPs which deviated from HWE in the control sample (P<0.1) were discarded from the analysis.
Differences in the allelic frequencies between patients and controls were evaluated with a χ2 test via Unphased program version 3.0.12 [45], [46]. Moreover, the web-based application SNPStats [47] was used to compare, by means of logistic regression, the genotypic frequencies between groups. Different inheritance models (codominant, dominant, recessive, and additive) were tested. ORs and 95% confidence intervals as indicators of the risk associated to each genotype were also calculated. Finally, if any SNP was found to be associated with the risk for schizophrenia, a linear regression analysis with SNPstats was used to evaluate the effect of those SNPs on several clinical scores (BPRS, PANSS and KGV). Bonferroni sequential test for multiple comparisons [48] was applied to correct all the reported p-values.
Regarding the haplotype analysis, haploblock frequencies were compared between patients and controls with Haploview Program version 4 [44]. Moreover, frequencies of sliding windows of two, three and four-marker haplotypes were estimated through a retrospective likelihood algorithm and compared between patients and controls with the Unphased software version 3.0.12 [45], [46]. It should be noted that haplotypes with a frequency below 0.03 were considered as rare and therefore discarded from the statistical analysis. A 1000-permutation run was performed in all haplotype analyses to better estimate the significance of the positive associations.
We also performed a joint analysis by combining the affected individuals and healthy subjects from Spain and Germany. The linkage disequilibrium patterns between SNPs were determined to assess the validity of such type of analysis. Moreover, these analyses were only stratified according to the region of origin of each individual (Germany or Spain) since there were no differences in other possible stratification variables, for example ethnicity. PLINK software version 1.0.2 [49] was used to apply two different CMH tests, which are based on an “average” OR that controls for the potential confounding due to the cluster variable. Two variants of the CMH tests were performed: a) a CMH test that performs the case-control association test but controlling the effect of the cluster variable (in our particular case, the cluster variable was the country of origin); b) a CMH test that is used as a measure of the heterogeneity between samples, which assesses whether a particular SNP varies between clusters. Finally, we also performed a haplotype analysis following the same procedures described above for the Spanish and the German sample separately.
Acknowledgments
We thank all the patients who have participated in this study. We also thank A. Tolosa, I. Toirac and JL Ivorra for their valuable help in sample collection.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the Spanish Ministry of Health: Instituto de Salud Carlos III, FIS02/0018 (to OR, JS, and CN), Red de Genotipación y Psiquiatría Genética-ISCIII G03/184 (to OR, JS, MDM and CN), Red de Enfermedades Mentales (REM-TAP Network) and CIBER de Salud Mental (CIBERSAM) (to OR, JS, MDM and CN). Furthermore, this work was supported by the Deutsche Forschungsgemeinschaft (Grant RE1632/1-5 to AR, Clinical Research Unit - KFO 125 to AR and KPL; SFB 581 to KPL, SFB TRR 58 to AR and KPL), Bundesministerium für Bildung und Forschung (IZKF Würzburg, 01KS9603, to KPL; IZKF N-27-N, to AR) and the European Comission (NEWMOOD LSHM-CT-2003-503474, to KPL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jablensky A, Sartorius N, Ernberg G, Anker M, Korten A, et al. Schizophrenia: manifestations, incidence and course in different cultures. A World Health Organization ten-country study. Psychol Med Monogr. 1992;(Suppl 20):1–97. doi: 10.1017/s0264180100000904. [DOI] [PubMed] [Google Scholar]
- 2.Jablensky A. The epidemiological horizon. In: Hirsch SR, Weinberger DR, editors. Schizophrenia. Oxford: Blackwell Science Ltd; 2003. [Google Scholar]
- 3.Shih RA, Belmonte PL, Zandi PP. A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int Rev Psychiatry. 2004;16:260–283. doi: 10.1080/09540260400014401. [DOI] [PubMed] [Google Scholar]
- 4.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 5.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 6.Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20:687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 7.Burmeister M, McInnis MG, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 8.Kohn Y, Lerer B. Excitement and confusion on chromosome 6q: the challenges of neuropsychiatric genetics in microcosm. Mol Psychiatry. 2005;10:1062–1073. doi: 10.1038/sj.mp.4001738. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, et al. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2009;14:796–803. doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerer B, Segman RH, Hamdan A, Kanyas K, Karni O, et al. Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol Psychiatry. 2003;8:488–498. doi: 10.1038/sj.mp.4001322. [DOI] [PubMed] [Google Scholar]
- 14.Levi A, Kohn Y, Kanyas K, Amann D, Pae CU, et al. Fine mapping of a schizophrenia susceptibility locus at chromosome 6q23: increased evidence for linkage and reduced linkage interval. Eur J Hum Genet. 2005;13:763–771. doi: 10.1038/sj.ejhg.5201406. [DOI] [PubMed] [Google Scholar]
- 15.Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 16.Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, et al. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louie CM, Gleeson JG. Genetic basis of Joubert syndrome and related disorders of cerebellar development. Hum Mol Genet. 2005;14 Spec No. 2:R235–242. doi: 10.1093/hmg/ddi264. [DOI] [PubMed] [Google Scholar]
- 18.Sheng G, Xu X, Lin YF, Wang CE, Rong J, et al. Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J Clin Invest. 2008;118:2785–2795. doi: 10.1172/JCI35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur J Hum Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- 20.Ingason A, Sigmundsson T, Steinberg S, Sigurdsson E, Haraldsson M, et al. Support for involvement of the AHI1 locus in schizophrenia. Eur J Hum Genet. 2007;15:988–991. doi: 10.1038/sj.ejhg.5201848. [DOI] [PubMed] [Google Scholar]
- 21.Ingason A, Giegling I, Cichon S, Hansen T, Rasmussen HB, et al. A large replication study and meta-analysis in European samples provides further support for association of AHI1 markers with schizophrenia. Hum Mol Genet. 2010;19:1379–1386. doi: 10.1093/hmg/ddq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torri F, Akelai A, Lupoli S, Sironi M, Amann-Zalcenstein D, et al. Fine mapping of AHI1 as a schizophrenia susceptibility gene: from association to evolutionary evidence. FASEB J. 2010 doi: 10.1096/fj.09-152611. 2010. In press. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez Retuerto AI, Cantor RM, Gleeson JG, Ustaszewska A, Schackwitz WS, et al. Association of common variants in the Joubert syndrome gene (AHI1) with autism. Hum Mol Genet. 2008;17:3887–3896. doi: 10.1093/hmg/ddn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Doering JE, Kane K, Hsiao YC, Yao C, Shi B, et al. Species differences in the expression of Ahi1, a protein implicated in the neurodevelopmental disorder Joubert syndrome, with preferential accumulation to stigmoid bodies. J Comp Neurol. 2008;511:238–256. doi: 10.1002/cne.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreasen NC, Paradiso S, O'Leary DS. “Cognitive dysmetria” as an integrative theory of schizophrenia: a dysfunction in cortical-subcortical-cerebellar circuitry? Schizophr Bull. 1998;24:203–218. doi: 10.1093/oxfordjournals.schbul.a033321. [DOI] [PubMed] [Google Scholar]
- 27.Glahn DC, Ragland JD, Abramoff A, Barrett J, Laird AR, et al. Beyond hypofrontality: a quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum Brain Mapp. 2005;25:60–69. doi: 10.1002/hbm.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirayasu Y. Brain imaging in schizophrenia. Neuropathology. 2007;27:601–603. doi: 10.1111/j.1440-1789.2007.00827.x. [DOI] [PubMed] [Google Scholar]
- 29.White T, Cullen K, Rohrer LM, Karatekin C, Luciana M, et al. Limbic structures and networks in children and adolescents with schizophrenia. Schizophr Bull. 2008;34:18–29. doi: 10.1093/schbul/sbm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shergill SS, Brammer MJ, Williams SC, Murray RM, McGuire PK. Mapping auditory hallucinations in schizophrenia using functional magnetic resonance imaging. Arch Gen Psychiatry. 2000;57:1033–1038. doi: 10.1001/archpsyc.57.11.1033. [DOI] [PubMed] [Google Scholar]
- 31.Yu F, Keinan A, Chen H, Ferland RJ, Hill RS, et al. Detecting natural selection by empirical comparison to random regions of the genome. Hum Mol Genet. 2009;18:4853–4867. doi: 10.1093/hmg/ddp457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crow TJ. Is schizophrenia the price that Homo sapiens pays for language? Schizophr Res. 1997;28:127–141. doi: 10.1016/s0920-9964(97)00110-2. [DOI] [PubMed] [Google Scholar]
- 33.Horrobin DF. Schizophrenia: the illness that made us human. Med. Hypotheses. 1998;50:269–288. doi: 10.1016/s0306-9877(98)90000-7. [DOI] [PubMed] [Google Scholar]
- 34.Dean B. Is schizophrenia the price of human central nervous system complexity? Aust N Z J Psychiatry. 2009;43:13–24. doi: 10.1080/00048670802534416. [DOI] [PubMed] [Google Scholar]
- 35.Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15 Spec No 1:R67–74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 36.Selch S, Strobel A, Haderlein J, Meyer J, Jacob CP, et al. MLC1 polymorphisms are specifically associated with periodic catatonia, a subgroup of chronic schizophrenia. Biol Psychiatry. 2007;61:1211–1214. doi: 10.1016/j.biopsych.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Marchini J, Cardon L, Phillips M, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 38.Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 40.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, et al. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet. 2006;79:903–909. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langefeld CD, Fingerlin TE. Association methods in human genetics. Methods Mol Biol. 2007;404:431–460. doi: 10.1007/978-1-59745-530-5_21. [DOI] [PubMed] [Google Scholar]
- 42.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reif A, Herterich S, Strobel A, Ehlis AC, Saur D, et al. A neuronal nitric oxide synthase (NOS-I) haplotype associated with schizophrenia modifies prefrontal cortex function. Mol Psychiatry. 2006;11:286–300. doi: 10.1038/sj.mp.4001779. [DOI] [PubMed] [Google Scholar]
- 44.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 45.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 46.Dudbridge F. Technical Report 2006/5, MRC Biostatistics Unit, Cambridge, UK; 2006. UNPHASED user guide. [Google Scholar]
- 47.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 48.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 49.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]