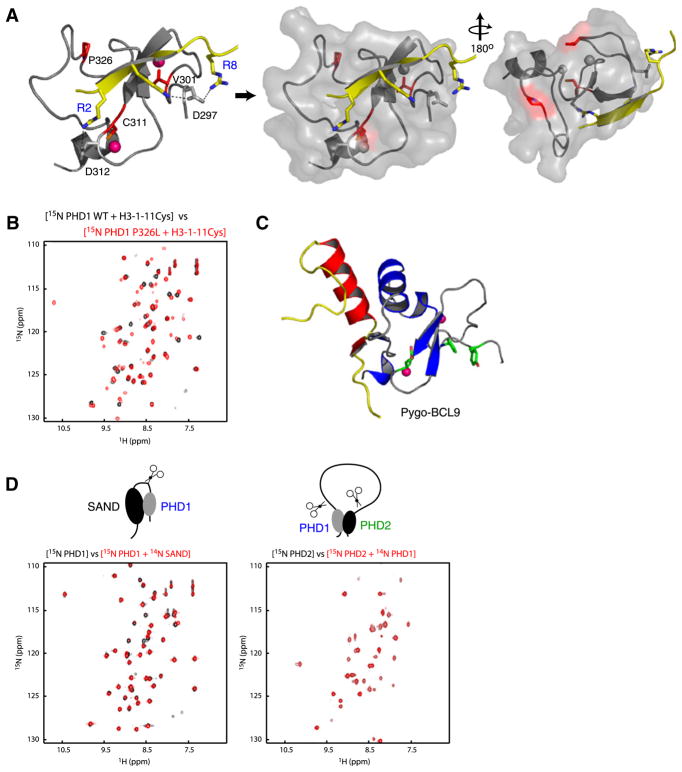
Figure 3. APECED Mutants.
(A) Location of the mutant residues (red) on the protein (gray)-peptide (yellow) complex structure in ribbons (left) and transparent surface representation (middle, right). The pink Zn atoms in left panel are shown in gray in surface representation for clarity.
(B) Comparison of two-dimensional 1H-15N HSQC spectra of AIRE-PHD1 wild-type with that of P326L mutant in presence of a peptide derived from N-terminal H3 residues 1–11 (black, wild-type; red, P326L mutant).
(C) Nonhistone interacting surface of Pygo-PHD (gray) in complex with the HD1 domain of BCL9 (yellow/red) (left). The aromatic cage residues of Pygo-PHD involved in H3K4me2 are green.
(D) Two-dimensional 1H-15N HSQC spectra assessing AIRE’s interdomain interaction at 1:1 molar ratio with 0.2 mM respective labeled proteins. The 15N-labeled PHD1 and PHD2 are labeled blue and green, respectively.