Abstract
Ever since their existence, there has been an everlasting arms race between viruses and their host cells. Host cells have developed numerous strategies to silence viral gene expression whereas viruses always find their ways to overcome these obstacles. Recent studies show that viruses have also evolved to take full advantage of existing cellular chromatin components to activate or repress its own genes when needed. While in most cases viruses encode certain proteins to recruit or inhibit cellular factors through physical interactions, growing examples show that viral encoded enzymes affect host chromatin structure through post-translationally modifying histones or other cellular proteins important for chromatin function. The most well studied example is vSET encoded by paramecium bursaria chlorella virus 1. vSET specifically methylates histone H3 at lysine 27, causing genome-wide silencing of Polycomb target genes upon infection, thus mimicking the function of Polycomb repressive complex 2 (PRC2) in eukaryotes. Other examples include BGLF4 from Epstein-Barr virus that affects both condensin and topoisomerase II activity and Us3 from Herpes Simplex virus 1 that inhibits HDAC1 function through phosphorylation.
Introduction
DNA molecules in eukaryotes, unlike those in prokaryotes or archeae, do not exist in free forms but rather in the forms of nucleosomes, which consists of approximate 147 base pairs of DNA wrapped around a histone octamer consisting of two copies of each H2A, H2B, H3 and H4 [1]. Nucleosomes can be further packaged to form higher order structures such as chromatin fibers and chromosomes. The structural disordered histone tails, which protrude from the nucleosome core structure, can be processed by various enzymes to obtain diverse range of covalent modifications including methylation, acetylation, phosphorylation and ubiquitination, which are important in regulation of gene transcriptional activation or repression [2]. While DNA methylation is usually associated with repressed gene transcription [3], histone modification can lead to different outcomes depending on the type of modification and the position of the histone residue that is modified. For example, histone H3 lysine 4 (H3K4) methylation generally correlates to transcriptional activation [4], whereas H3K9 or H3K27 methylation corresponds to transcriptional repression [5, 6]. Recently, there has also been significant development in the field demonstrating the role of non-coding RNA in epigenetic gene regulation [7].
Many viruses integrate viral DNA into host cells, where they have to overcome the obstacle of chromatin formation. Viruses have evolved to take advantage of chromatin structure to increase viral genome stability, and more importantly to activate or repress gene transcription when needed. Viruses use a plethora of approaches to utilize and modulate the cellular chromatin-modifying activities to ensure their survival and propagation [8, 9]. For instance, VP16 of herpes simplex virus type I (HSV-1) has been shown to recruit general transcription factors and co-activators to IE promoters and to be responsible for dramatically reducing histone association with these promoters [10]. Human immunodeficiency virus type I (HIV-1) uses its trans-activator protein Tat to recruit the cellular SWI/SNF chromatin remodeling complex to HIV-1 long terminal repeat (LTR) [11]. The same viral protein has also been demonstrated to bind TAFII250 component of the general transcription factor TFIID, thus inhibiting its histone lysine acetyl transferase (KAT) activity. This leads to the repression of MHC class I transcription and accounts for one of the mechanisms through which HIV evades host immune response [12]. Human cytomegalovirus (CMV) IE1 and IE2 act as promiscuous transcription activators through their inhibition of HDAC3 by binding [13].
Most viral proteins affect the chromatin structures of cellular or their own genes by recruiting cellular chromatin-modifying proteins or by inhibiting their enzymatic activities, both of which are achieved by direct physical interactions. However, more cases are emerging where viral encoded proteins use their enzymatic activity to modify histones or other cellular proteins involved in chromatin modification. In this review, we will focus our discussion on a SET domain containing histone lysine methyltransferase (vSET) from paramecium bursaria chlorella virus 1 (PBCV-1) that represses host cell gene transcription through H3K27 methylation. vSET methyltransferase activity has been demonstrated both in vitro and in vivo, as has its gene silencing activity in vivo [14-16], and it represents a family of vSET-like lysine methyltransferases (KMT) that are encoded in probably all chlorella viruses. Finally, we will talk about several other viral proteins that could affect gene transcription through means beyond simply binding to cellular components.
vSET from PBCV-1
SET domain family proteins, which are originally identified in Drosophila suppressor of variegation (Su(var)3-9), enhancer of zeste (E(z)) and trithorax, are known to catalyze site- and state-specific methylation of lysine residues in histone tails that is important in epigenetic regulation of gene transcription [17]. One extensively studied SET domain KMT is EZH2 (also known as KMT7) [18] of Polycomb repressive complex 2 (PRC2). PRC2 tri-methylates hisonte H3 lysine 27 (H3K27) [6] and creates binding sites for the chromodomain of the CBX protein of Polycomb repressive complex 1 (PRC1) [6, 19] (Fig. 1). Polycomb group complexes are crucial regulators of genomic programming and differentiation. They achieve gene silencing through an array of approaches, which include chromatin compaction [20], histone H2A ubiquitination [21], and blocking transcription elongation [22]. The methylation mark on H3K27 can be removed by histone lysine demethylases (KDMs) [23] and further converted to acetyl group by lysine acetyl transferases (KATs) to result in the recruitment of chromatin remodeling complex and activation of gene transcription.
Fig. 1.
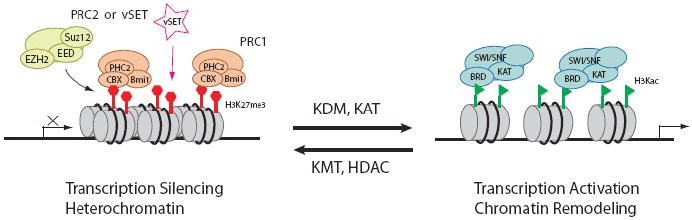
vSET mimicks the function of Polycomb repressive complexes (PRCs) for gene transcriptional silencing. vSET or Ezh2 of PRC2 methylates histone H3 at K27 to create binding sites for the chromodomain of CBX protein of PRC1, which ultimately results in heterochromatin formation and gene silencing. The process can be reversed by the action of lysine demethylases (KDM) and acetyltransferases (KAT) converting H3K27me3, a gene repression state, to lysine-acetylated H3 (H3Kac), which recruits the bromodomain-containing proteins of chromatin remodeling complexes and transcriptional proteins, leading to euchromatin formation and transcription activation.
It has been shown that a viral SET domain protein, vSET from PBCV-1 virus (119 aa, the smallest known SET protein), is also capable of methylating H3K27 in vitro [14]. Recent data further demonstrated its gene repression activity in vivo by mimicking the role of PRC2 [15] (Fig. 1). This is the first systematic study of a viral encoded protein that regulates host transcriptional machinery through direct modifications of chromatin.
vSET is a histone lysine methyltransferase specific for H3K27
In vitro methyltransferase assay using histone substrates showed that vSET is able to transfer 14C-labeled methyl group from S-adenosyl methionine (SAM) to histone H3 but not the other histones [14]. Methyl transfer also occurred when synthetic H3 peptide containing N-terminal residues 15-30 was used as the substrate. However, little activity was seen when H3 peptide with di-methylated K27 or phosphorylated S28 was used. GST-H3 peptide fusion substrate with mutations at individual lysine sites within the H3 peptide sequence showed unequivocally that vSET is very specific for K27 of histone H3 [15].
Three-dimensional structure of vSET
The structure of vSET was solved by NMR in its free state [14], and in a ternary complex with S-(5′-adenosyl)-L-homocysteine and histone H3 peptide containing mono-methylated K27 [16]. The structure of vSET is a two-fold symmetric dimer (Fig. 2A), which is consistent with gel filtration and sedimentation equilibrium analyses. Each protomer consists of two domains. Domain I has an antiparallel β-barrel. Domain II, an insert in the domain I sequence, contains a three-stranded open-faced sandwich. The primary dimer interface forms between the domain II of each protomer. A unique feature of the SET domain is the knot-like structure involving the C-terminal sequence. The peptide substrate and the methyl cofactor bind on two opposite sides of the vSET surface, with a small pore connecting the reactive entities. Residues around this pore likely help align the reactants to facilitate the methyltransferase reaction in the absence of general base catalysis. In comparison to vSET, the mammalian SET domain proteins have extra sequences at both termini (Fig. 2B) [16, 24, 25]. They also function only as monomers, and usually require the presence of other subunits for optimal methyltransferase activity. It is intriguing to see whether dimer formation of vSET is essential for its methyltransferase activity. This would require identification of key residue(s) at dimer interface, mutation of which would convert dimeric vSET to monomers. One could also measure the activity of a heterodimer that consists of one wild type monomer and one inactive mutant monomer. This would tell us whether each active site in a dimer works independently or cooperatively.
Fig. 2.
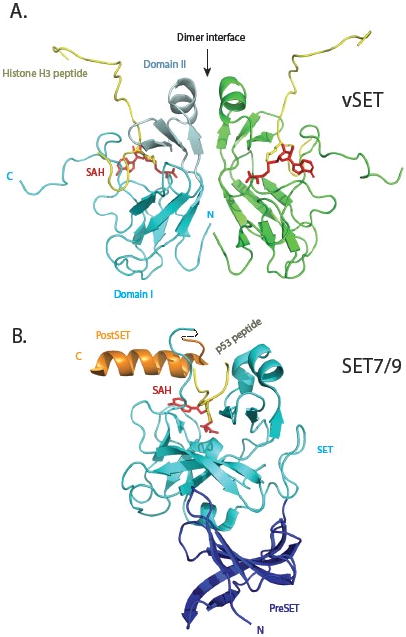
Comparison of three-dimensional structures of vSET and mammalian SET7/9 lysine methyltransferases. (A) Solution structure of vSET bound to cofactor SAH and histone H3 peptide containing mono-methylated lysine 27. The two protomers of the C2-symmetric dimer are shown in cyan and green. Domain I and domain II of one protomer are depicted in regular and pale cyan, respectively. The histone peptides are shown in gold, and SAH molecules in red. The N- and C-termini of the cyan protomer are labeled. (B) Crystal structure of SET7/9 in complex with p53 peptide and SAH. The preSET, SET and postSET domains are colored blue, cyan and orange, respectively. The peptide and cofactor are shown in the same colors as those in A.
vSET represses transcription of host cellular genes
PBCV-1 infection of green algae Chlorella causes massive inhibition of total RNA synthesis in host cells [15]. This is achieved by a two-tiered attack. Small amount of vSET is packaged in the virion and gets released right after infection to cause quick initial inhibition of host transcription, which is followed by the expression of more vSET protein to maintain a sustained and higher level repression. Western blot analysis showed that di- and tri-methylation of H3K27 increased after infection. Due to difficulty in manipulating chlorella cells, mammalian cells were used as a model system to study the in vivo function of vSET, since H3K27 methylation as an epigenetic mark for gene silencing is highly conserved among all eukaryotic species. A nuclear localization signal was discovered in vSET by fluorescence microscopy imaging analysis. While wild type GFP-vSET localized to the nucleus, a triple mutant (K85A/R86A/R88A) remained in the cytoplasm. Since EZH2 of PRC2 can cause H3K27 methylation, EZH2 knockdown Hela cells were generated to study the effect of vSET. vSET treatment of nuclear extract from these cells restored H3K27 di- and tri-methylation. Similar results were obtained when vSET was expressed in situ.
The effect of vSET methylation on gene transcription was also studied in a luciferase-based reporter assay. The luciferase promoter contained Gal4 binding sites. When vSET was expressed as a fusion protein with Gal4 DNA binding domain, 95% inhibition of luciferase expression was observed. Remarkably, when a single mutation was introduced at Y105 (a residue close to the active site pore, and its mutation to Phe or Ala resulted in almost a complete loss of H3K27 methylation by vSET), there was no repression of gene transcription. The high effectiveness of the fusion protein towards the engineered expression system demonstrates the feasibility of creating fusion protein of vSET with some other type of DNA binding module that would inhibit any chosen endogenous promoter with high specificity and efficiency.
To examine the biological consequence of H3K27 methylation by vSET, Polycomb group protein occupation at the H3K27 methylation site was monitored in the absence or presence of vSET expression in EZH2 knockdown Hela cells [15]. The expression level of PRC1 complex protein CBX8 was not changed, but its occupancy at the H3K27me2 and H3K27me3 sites was enhanced tremendously upon vSET induction. So vSET not only creates the same epigenetic mark as PRC2 does, it also causes the same recruitment of PRC1 and subsequent repression of gene transcription (Fig. 1).
The effect of vSET methylation of H3K27 on the transcription of Polycomb target genes was also studied by a quantitative RT-PCR assay. As expected, EZH2 knockdown by siRNA increased the transcription of all five Polycomb target genes that were checked (HOXA7, HOXA9, HOXB9, HOXD8 and Hey1). Upon vSET expression, however, the transcription levels of these genes returned to those of normal Hela cells.
To look at the effect of vSET on one Polycomb target gene promoter in more detail, HOXA7 promoter was coupled to a luciferase gene and its transcription monitored by luciferase expression [15]. Induction of vSET repressed luciferase expression in both EZH2 siRNA-treated and untreated cells. Chromatin immunoprecipitation assay of the endogenous HOXA7 promoter confirmed that vSET caused di- and tri-methylation of H3K27 and resulted in the enrichment of CBX8 of PRC1. Similar results were obtained for HIV Tat-dependent LTR promoter.
vSET also played an important role in cell cycle. Flow cytometric analysis indicated that vSET caused cell accumulation at the G2/M phase in transiently transfected NIH-3T3 cells [15]. Similar results were obtained when Hela cells were stably transfected with vSET in a tetracycline-controlled expression vector.
The prevalence of SET domain proteins
vSET homologues exist in other chlorella viruses as well. When a vSET gene probe was used to hybridize genomic DNA from 36 other chlorella viruses, 34 of them showed a positive result [15]. SET proteins from a number of chlorella viruses were shown to methylate H3K27 with the same stringency as vSET. In fact, a SET domain protein has also been found in an acetate-utilizing archaeal methanogen, Methanosarcina mazei strain Gö1 [26]. Gö1-SET exhibits selective methyltransferase activity towards one of the major archaeal DNA interacting proteins MC1-α at lysine 37. SET domain proteins are even present in bacteria. The chlamydial-SET physically interacts with chlamydial histone-like proteins Hc1 and Hc2, and functions as a histone methyltransferase to methylate mouse histone H3 and Hc1 [27]. SET domain proteins have survived the test of evolution and emerged as very important tools for all three kingdoms of life.
Other viral encoded enzymes that affect chromatin function
Over the past decade, more and more examples of viral encoded enzymes involved in chromatin function other than vSET have been reported, such as BGLF4 from Epstein-Barr virus (EBV) [28] and UBCv1 from African swine fever virus (ASFV) [29] (Fig. 3). They can be divided into three groups based on their cellular targets. Group 1 enzymes target histone tails directly for methylation, ubiquitination or phosphorylation. Group 2 works on cellular proteins, which themselves modify histone tails. Group 3 consists of viral enzymes that affect the activity of proteins involved in other aspects of chromatin structure and function. The biological functions of these viral enzymes in host chromatin modifications and recruitment of host transcriptional machineries for viral gene transcription and replication are described below.
Fig. 3.
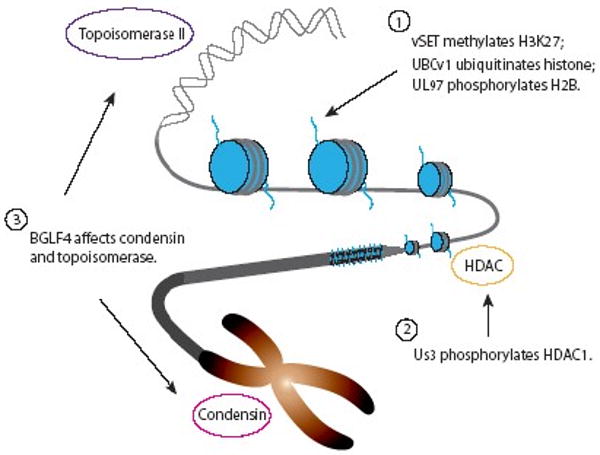
Viral encoded enzymes that interfere host chromatin functions. Examples of viral enzymes are shown that affect diverse range of infected host chromatin structure and function, ranging from covalent modifications of histone tails (Group 1), phosphorylation of HDAC (Group 2), to alteration of the activity of Topoisomerase II and condensin (Group 3).
Viral enzymes that target histone tails
UBCv1 from African Swine Fever Virus (ASFV)
The ARID (AT-rich interaction domain) family of proteins plays a pivotal role in the regulation of development and tissue-specific gene expression [30]. Some representative members in human are p270 of the SWI/SNF complexes, retinoblastoma binding proteins, and jumonji. SMCp, a pig ARID family protein, has been shown to be a possible in vivo target of UBCv1, a ubiquitin conjugating enzyme encoded by ASFV [29]. UBCv1 is also capable of phosphorylating histone in vitro [31]. UBCv1 is likely another example of viral encoded enzymes that influence chromatin function. SMCp was found to interact with UBCv1 by yeast two-hybrid system [29]. This interaction between these two proteins was confirmed by in vitro binding assay that utilized GST fusion protein of SMCp ARID DNA binding domain and radioactive labeled UBCv1 protein. Both SMCp and UBCv1 were shown to localize to the nucleus of ASFV infected cells using indirect immnofluorescence. An acidic C-terminal extension in UBCv1 played an important role in its localization based on deletion study. The same extension also suggests that UBCv1 might conjugate ubiquitin to its substrate in an E3 independent manner [32]. The role of UBCv1 may be to target SMCp for destruction during viral infection.
Several other viral enzymes that cause histone modification
There are several other examples of viral kinases that are capable of phosphorylating histone in vitro. However, the lack of any in vivo data makes impossible any definitive conclusion. Human cytomegalovirus UL97 has been shown to phosphorylate histone H2B using in vitro kinase assay with radioactive ATP [33]. Mass spectroscopic analyses revealed several potential phosphorylation sites. Phosphorylation seems to depend on the presence of a positively charged residue at the P+5 position. Other viral encoded kinases that can use histone as a substrate include HSV-2 UL13 [34] and human herpesvirus 6 U69 [35].
Viral enzymes that target cellular histone-modifying proteins
Instead of modifying histone tails directly, some viruses encode enzymes that target existing cellular factors that are involved in the process. Us3 from Herpes Simplex Virus 1 (HSV1) is such an example.
In cells affected with low ratios of HSV-1 per cell, VP16 transactivates α genes, the first set to be expressed after infection. Two out of the six α proteins, ICP4 and ICP0, are required for the expression of β genes. ICP0 has been shown to dissociate HDAC1 from the CoREST/REST/HDAC1 complex through a possible mimicry of part of the N-terminal sequence of CoREST, thus blocking the silencing of viral post-α genes by HDAC1 [36]. Another HSV-1 encoded protein Us3 kinase was later found to block histone deacetylation through an entirely different mechanism [37]. Us3 kinase induces HDAC1 phosphorylation and alters its amount and localization.
The expression of reporter genes in both restrictive (SK-N-SH) and permissive (U2OS) cells was examined in the presence or absence of an HDAC inhibitor sodium butyrate and compared to co-transduction with the Us3 gene. The reporter genes were expressed at very low levels in the absence of either sodium butyrate or Us3. Addition of sodium butyrate increased the expression levels as expected, and co-transduction of Us3 accumulated the reporter gene products to comparable levels as those in the sodium butyrate treatment cases. Consistent with its role on HDAC, transduction of Us3 several hours before the transduction of the reporter gene resulted in a much higher expression of target gene. HDAC1 was post-translationally modified in cells transduced with Us3 based on the appearance of slower migrating HDAC1 bands in SDS gel. The kinase activity of Us3 is essential for its function because transduction of an active site mutant caused neither posttranslational modification of HDAC1 nor accumulation of the reporter gene. Transduction of Us3 in SK-N-SH cells caused almost complete elimination of HDAC1 expression. The same transduction in U2OS cells also decreased HDAC1 level significantly and resulted in a speckled distribution instead of rather uniform distribution in native cells. By inhibiting HDAC1 through both ICP0 and Us3 with different mechanisms HSV wins the competition against host cells whose mission is to silence viral DNA.
Viral enzymes that affect chromatin function through other mechanisms
Besides their roles in controlling chromatin transcription levels, viral enzymes are also known for their functions in higher order chromatin structure formation. BGLF4 kinase, the only identified serine/threonine kinase in EBV, is conserved in all herpesviruses. It is indispensable for virion maturation and packaging due to its activity on viral DNA polymerase accessory factors [38, 39] and cellular translation factor [40]. Recent study further demonstrated that BGLF4 kinase alone can induce cellular DNA condensation through a concerted event of condensin phosphorylation and topoisomerase II (Topo II) activation [28].
Lee et al. observed cellular chromatin condensation and interchromosomal space enlargement upon EBV reactivation by either chemical treatment or expression of lytic transactivator [28]. This was attributed to BGLF4 kinase as its expression in many different cell types caused a prophase-like individualized condensation pattern for chromatin. BGLF4 kinase also induced other mitosis-like events such as nuclear lamina disassembly and reorganization of the cytoskeleton. However, these were not coordinated with centrosome separation. Consistent with this, BGLF4 induced premature chromosome condensation independently of G2/M accumulation and this event did not require the activity of Cdc2 kinase, which is responsible for normal cell cycle progression. Immunoprecipitation experiment showed that BGLF4 interacted with hCAP-D2 regulatory subunit and hCAP-E core subunit of human condensin I complex. Western blotting using phosphospecific antibody further demonstrated that BGLF4 through Cdc2 mimicry phosphorylated condensin regulatory subunits hCAP-G and hCAP-D2 at the Cdc2 consensus motifs. BGLF4 also stimulated Topo II activity as its expression caused the enhancement of Topo II-DNA complex in the presence of Topo II inhibitor, and elevated Topo II activity as revealed by in vitro decatenation assay. So BGLF4 induces chromosome condensation through the activation of condensin and Topo II, both of which are indispensable factors for this process [41]. This function seems to be conserved in all gammaherpesviruses as a similar protein from HSV-1 (UL13 kinase) showed the same ability. Lee et al. postulated that a compact chromosome structure might provide enough nuclear space for the viral compartments in EBV-replicating cells [28].
Concluding Remarks
Growing evidence argues that viruses have evolved to replicate themselves in infected host cells by using viral proteins that help recruit cellular proteins and hijack host transcriptional proteins for viral life cycle, and also by encoding viral enzymes that are capable of making host chromatin modifications thereby directly tapping into host histone-mediated gene transcriptional machinery for viral replication. The latter strategy may benefit a virus more than the former for several reasons. Firstly, by avoiding the participation of any cellular component in a particular step of chromatin regulation, the virus eliminates one option that the host could use to fight back, i.e. up- or down-regulation of the very cellular component. Secondly, because of its catalytic activity, only a small amount of a viral enzyme is needed to achieve a major effect in biology. Thirdly, there may be cases where physical molecular interaction is simply not enough to enhance or inhibit certain cellular function.
Any of the above reasons could explain why PBCV-1 utilizes vSET to accomplish massive repression of host gene transcription. The major difference between vSET and mammalian SET methyltransferases is that the former functions as a dimer while the latter work as monomers (Fig. 2). Dimer formation might be important for creating an optimal balance between flexibility and rigidity at the active site, as our ongoing study suggests that vSET monomers obtained through mutagenesis are inactive towards methyltransferase reaction. The similar balance has been achieved by mammalian SET methyltransferases through the addition of extra domains to the SET domain and/or recruitment of other protein partners to form complexes. The fact that vSET exists as a dimer could also have other functional consequences. For instance, it is conceivable that both active sites could bind histone H3 tails in chromatin and catalyze the reaction. The active site that finishes the reaction first could find another substrate in the vicinity before the dimeric enzyme dissociates from the chromatin. The vSET dimer may function by “walking” along chromatin to methylate histone H3K27, which is far more efficient than a monomeric SET protein that would undergo constant association and dissociation. Motor proteins such as myosins and kinesins are perfect examples that function via such a walking mechanism [42]. It remains to be seen whether vSET uses the same mechanism to accomplish its host gene silencing activity in the infected cells.
Acknowledgments
We wish to thank M. Walsh and S. Mujtaba for helpful discussion. The related work in the Zhou lab was supported by the grants from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 4.Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A. 2002;99 4:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakayama J, Rice J, Strahl B, Allis C, Grewal S. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones R, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 7.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman PM. Chromatin regulation of virus infection. Trends Microbiol. 2006;14:132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman PM. Chromatin organization and virus gene expression. J Cell Physiol. 2008;216:295–302. doi: 10.1002/jcp.21421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78:9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman JD, Brown JA, Howcroft TK, Hwang J, Chawla A, Roche PA, Schiltz L, Nakatani Y, Singer DS. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci U S A. 1998;95:11601–11606. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A. 2004;101:17234–17239. doi: 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzur K, Farooq A, Zeng L, Plotnikova O, Koch A, Sachchidanand, Zhou M. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat Struct Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 15.Mujtaba S, Manzur KL, Gurnon JR, Kang M, Van Etten JL, Zhou MM. Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol. 2008;10:1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian C, Wang X, Manzur K, Sachchidanand, Farooq A, Zeng L, Wang R, Zhou M. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol. 2006;359:86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Qian C, Zhou M. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein E, Duncan E, Masui O, Gil J, Heard E, Allis C. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 21.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao B, Jing C, Wilson J, Walker P, Vasisht N, Kelly G, Howell S, Taylor I, Blackburn G, Gamblin S. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 25.Chuikov S, Kurash J, Wilson J, Xiao B, Justin N, Ivanov G, McKinney K, Tempst P, Prives C, Gamblin S, Barlev N, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 26.Manzur K, Zhou M. An archaeal SET domain protein exhibits distinct lysine methyltransferase activity towards DNA-associated protein MC1-alpha. FEBS Lett. 2005;579:3859–3865. doi: 10.1016/j.febslet.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Murata M, Azuma Y, Miura K, Rahman MA, Matsutani M, Aoyama M, Suzuki H, Sugi K, Shirai M. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology. 2007;153:585–592. doi: 10.1099/mic.0.29213-0. [DOI] [PubMed] [Google Scholar]
- 28.Lee CP, Chen JY, Wang JT, Kimura K, Takemoto A, Lu CC, Chen MR. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2007;81:5166–5180. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulimo WD, Miskin JE, Dixon LK. An ARID family protein binds to the African swine fever virus encoded ubiquitin conjugating enzyme, UBCv1. FEBS Lett. 2000;471:17–22. doi: 10.1016/s0014-5793(00)01352-1. [DOI] [PubMed] [Google Scholar]
- 30.Wilsker D, Patsialou A, Dallas PB, Moran E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002;13:95–106. [PubMed] [Google Scholar]
- 31.Hingamp PM, Arnold JE, Mayer RJ, Dixon LK. A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO J. 1992;11:361–366. doi: 10.1002/j.1460-2075.1992.tb05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser P, Mandl S, Schweiger M, Schneider R. Characterization of functionally independent domains in the human ubiquitin conjugating enzyme UbcH2. FEBS Lett. 1995;377:193–196. doi: 10.1016/0014-5793(95)01323-7. [DOI] [PubMed] [Google Scholar]
- 33.Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J Biol Chem. 2002;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- 34.Daikoku T, Shibata S, Goshima F, Oshima S, Tsurumi T, Yamada H, Yamashita Y, Nishiyama Y. Purification and characterization of the protein kinase encoded by the UL13 gene of herpes simplex virus type 2. Virology. 1997;235:82–93. doi: 10.1006/viro.1997.8653. [DOI] [PubMed] [Google Scholar]
- 35.Ansari A, Emery VC. The U69 gene of human herpesvirus 6 encodes a protein kinase which can confer ganciclovir sensitivity to baculoviruses. J Virol. 1999;73:3284–3291. doi: 10.1128/jvi.73.4.3284-3291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon A, Gu H, Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc Natl Acad Sci U S A. 2006;103:9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen MR, Chang SJ, Huang H, Chen JY. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J Virol. 2000;74:3093–3104. doi: 10.1128/jvi.74.7.3093-3104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asai R, Kato A, Kato K, Kanamori-Koyama M, Sugimoto K, Sairenji T, Nishiyama Y, Kawaguchi Y. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J Virol. 2006;80:5125–5134. doi: 10.1128/JVI.02674-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawaguchi Y, Kato K, Tanaka M, Kanamori M, Nishiyama Y, Yamanashi Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J Virol. 2003;77:2359–2368. doi: 10.1128/JVI.77.4.2359-2368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gassmann R, Vagnarelli P, Hudson D, Earnshaw WC. Mitotic chromosome formation and the condensin paradox. Exp Cell Res. 2004;296:35–42. doi: 10.1016/j.yexcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Cross RA. Molecular motors: Walking talking heads. Curr Biol. 1999;9:R854–856. doi: 10.1016/s0960-9822(00)80045-7. [DOI] [PubMed] [Google Scholar]


