Summary
An emerging concept in development is that transcriptional poising pre-sets patterns of gene expression in a manner that reflects a cell’s developmental potential. However, it is not known how certain loci are specified in the embryo to establish poised chromatin architecture as the developmental program unfolds. We find that, in the context of transcriptional quiescence prior to the midblastula transition in Xenopus, dorsal specification by the Wnt/β-catenin pathway is temporally uncoupled from the onset of dorsal target gene expression, and that β-catenin establishes poised chromatin architecture at target promoters. β-catenin recruits the arginine methyltransferase Prmt2 to target promoters, thereby establishing asymmetrically dimethylated H3 arginine 8 (R8). Recruitment of Prmt2 to β-catenin target genes is necessary and sufficient to establish the dorsal developmental program, indicating that Prmt2-mediated histone H3R8 methylation plays a critical role downstream of β-catenin in establishing poised chromatin architecture and marking key organizer genes for later expression.
Introduction
Transcriptional poising represents a widespread mechanism of post-initiation control of gene expression that is observed in metazoan biological model systems [see (Margaritis and Holstege, 2008; Saunders et al., 2006) for reviews]. Establishment of poised chromatin architecture at genetic loci allows for a rapid and synchronous transcriptional response to environmental and biological stimuli (Baugh et al., 2009; Boettiger and Levine, 2009; Hargreaves et al., 2009; Muse et al., 2007; Radonjic et al., 2005; Rougvie and Lis, 1988). Poised loci have undergone successful pre-initiation complex formation, yet are stalled at the transition from transcriptional initiation to elongation (Saunders et al., 2006). Thus, they are marked by covalent histone modifications (acetylation of lysine 9 and 14, and trimethylation of lysine 4 on Histone H3, H3K9/14ac and H3K4me3, respectively) and a phosphorylated form of the large subunit of the RNA Polymerase holoenzyme (Pol II CTDpSer5) that correlate with transcriptional initiation prior to the onset of mRNA expression (Guenther et al., 2007; Margaritis and Holstege, 2008). Remarkably, in the context of embryonic development, poised chromatin architecture is established within multipotent precursor cells in a manner that reflects the developmental potential of the lineage (Bernstein et al., 2006; Guenther et al., 2007; Hammoud et al., 2009; Vastenhouw et al., 2010; Zeitlinger et al., 2007). However, it is not well understood how certain loci are specified to establish poised chromatin architecture as the developmental program unfolds.
The earliest events in embryogenesis are controlled by maternal factors until the activation of the zygotic genome. In Xenopus, Drosophila, and Zebrafish, zygotic genome activation occurs several hours and cell divisions after fertilization, at the midblastula transition (MBT) (Edgar and Schubiger, 1986; Kane and Kimmel, 1993; Newport and Kirschner, 1982). However, while zygotic transcription is constrained before the MBT, essential steps in embryonic patterning are accomplished before the MBT and embryos emerge from this period having begun the process of regional specification. In particular, the Wnt/β-catenin pathway mediates the earliest cell fate decision in amphibian (and teleost) embryogenesis, the establishment of the dorso-ventral axis. Dorsal specification by the Wnt/β-catenin pathway takes place under conditions of global transcriptional repression, prior to the MBT (Heasman et al., 2000; Kao et al., 1986; Yamaguchi and Shinagawa, 1989; Yang et al., 2002b). While β-catenin is required for the transcription of a small set of genes that are expressed before the MBT (Takahashi et al., 2000; Yang et al., 2002b), the critical Wnt target genes that direct dorsal development are silent until the MBT. Notably, β-catenin can interact with numerous factors that direct both chromatin modification and RNA Pol II recruitment to promoters [reviewed in (Mosimann et al., 2009)], including factors that establish both H3K9/14ac and H3K4me3. These observations raise the possibility that β-catenin functions during the preMBT period to establish a heritable, transcriptionally poised state that results in the later expression of dorsal determinants such as siamois and xnr3.
We have investigated the chromatin architecture of β-catenin target genes before the MBT, and report that β-catenin contributes to the establishment of poised chromatin architecture, thus priming target promoters for activation at the onset of zygotic gene expression. Before the MBT, β-catenin target promoters associate with RNA Pol II (CTDpSer5) and are marked by H3K9/14ac and H3K4me3, independently of their level of mRNA expression. Deposition of H3K4me3, in particular, requires both preMBT β-catenin and RNA Pol II function. Importantly, during dorsal specification, β-catenin recruits the arginine methyltransferase Prmt2 to target gene promoters, which results in the asymmetric dimethylation of Histone H3 arginine 8. Recruitment of Prmt2 to β-catenin target gene promoters is both necessary and sufficient to establish the dorsal gene expression program. We therefore provide direct evidence for a complex pre-transcriptional mechanism at work in early embryos to pre-set patterns of gene expression, and provide an initial analysis of chromatin architecture during this critical period of development.
Results
Dorsal specification by β-catenin is temporally uncoupled from the onset of target gene expression
The maternal Wnt/β-catenin pathway in Xenopus (and zebrafish) specifies dorsal cell fates before the MBT under conditions of global transcriptional repression. Two classes of dorsal genes are expressed in response to maternal β-catenin (Yang et al., 2002b): genes such as siamois and xnr3 are expressed at the MBT (Figure 1A), whereas genes exemplified by xnr5 and xnr6 are transcribed as early as the 256-cell stage, bypassing preMBT global transcriptional repression (Figure 1A).
Figure 1. β-catenin target genes are poised for expression before the MBT.
(A) Onset of expression of maternal β-catenin target genes (Siamois, Xnr3, Xnr5, and Xnr6). Maternally expressed Ornithine decarboxylase (Odc) is shown as a control for loading. Odc (-RT) indicates no reverse transcriptase, as a control for genomic DNA contamination. (B) Embryos were injected at the 2-cell stage with the β-catenin morpholino (ii-iv) and subsequently at the 4-cell stage (2 dorsal blastomeres) with 2pg siamois (iii) or 300pg β-catenin (iv) mRNAs. The frequency of each representative phenotype is indicated. (C) Embryos were injected into two dorsal blastomeres at the 4-cell stage with 500pg ΔNTcf3-GR mRNA. Wnt/β-catenin activity was inhibited by addition of dexamethasone (Dex) to the culture medium at the indicated stages. Siamois, Xnr3, and ODC mRNA expression was measured by RT-PCR at stage 10. (D) Left panel: occupancy of initiating (CTD pSer5) RNA Pol II in the promoter-proximal regions of the Siamois and Xnr3 loci before (1000-cell stage) or after (Stage 9) the onset of expression as measured by ChIP-QPCR. Right panel: elongating (CTD pSer2) RNA Pol II associated with the 3’ CDS of the same panel of genes at the same timepoints. Binding of RNA Pol II at Xnr6, which is expressed at both stages, is a positive control. Pooled data from three independent experiments are presented as a percentage of input chromatin to facilitate comparison between the 1000-cell stage and Stage 9. The average signal (0.02% input) from a negative control (IgG) ChIP is marked as a dotted line. Error bars are S.E.M. (E) PreMBT occupancy of maternal β-catenin target promoters (Siamois, Xnr3, and Xnr5) or a zygotic β-catenin target promoter (Myf5) by either H3K9/14ac or β-catenin was observed by ChIP on 1000-cell stage embryos. Myosin light chain 2 (Mlc2): negative control locus. “Input” indicates chromatin prior to ChIP (1%). (F) Promoter occupancy by β-catenin or H3K4me3 measured by ChIP on 1000-cell embryos. See also Figure S1.
Maternal β-catenin is required to activate the dorsal gene expression program (Heasman et al., 2000), but it is not yet clear which β-catenin targets are required for dorsal development. Siamois induces complete secondary axes and rescues dorsal development in ventralized embryos (Lemaire et al., 1995), whereas combined loss of siamois and the closely related gene twin blocks dorsal development (Ishibashi et al., 2008; Laurent et al., 1997), indicating that siamois and twin play essential, instructive roles in dorsal induction. However, these experiments leave open the possibility that dorsal development depends on additional targets of maternal Wnt/β-catenin signaling. To test whether siamois expression is sufficient for dorsal development in β-catenin-deficient embryos, β-catenin was depleted by injection of a morpholino oligonucleotide (βMO), which blocked dorsal development (Figure 1B, panel ii), as described (Heasman et al., 2000); expression of siamois in these embryos rescued dorsal development to the same extent as β-catenin itself (Figure 1B, panels iii and iv). Therefore, establishment of zygotic siamois expression represents an essential patterning event driven by the maternal Wnt/β-catenin pathway.
It is paradoxical, then, to consider how a transcription factor such as β-catenin functions during a period of global transcriptional repression. One possibility is that, while β-catenin is present throughout cleavage stages, it only functions at the MBT to activate target genes such as siamois and xnr3. This seems unlikely, as β-catenin is required for preMBT transcription of xnr5 and xnr6 and dorsal specification by β-catenin is complete by the 32-cell stage (Yang et al., 2002b). However, those experiments did not directly address the requirement for early β-catenin function in the induction of siamois and xnr3 at the MBT. We therefore inhibited Wnt pathway activity at discrete times before the MBT using a dexamethasone-inducible form of Tcf3 that is unable to bind β-catenin (ΔNTcf3-GR) and thereby inhibits endogenous β-catenin/Tcf3 transactivation. While inhibition of β-catenin/Tcf3 transactivation at the 4-cell stage significantly reduces siamois and xnr3 expression after the MBT, inhibition at the 32-cell stage has little or no effect (Figure 1C). This indicates that, as early as the 32-cell stage, dorsal cell fates have been specified and “locked-in”, as the embryo is no longer sensitive to inhibition of β-catenin/Tcf3 transactivation. Notably, six cell divisions separate the 32-cell stage from the MBT, indicating that the information imparted to promoters must be inherited through multiple cell divisions following dorsal specification. We thus conclude that dorsal specification by β-catenin is temporally uncoupled from the onset of dorsal target gene expression.
β-catenin establishes poised chromatin architecture at target promoters before the MBT
We next hypothesized that β-catenin binds to target promoters and poises them for activation before the MBT so that they may be expressed following the large-scale activation of the zygotic genome. In general, transcriptionally poised loci are bound by initiating (CTD pSer5) RNA Pol II and marked by H3K9/14ac and H3K4me3 prior to the onset of transcription (Guenther et al., 2007). To test whether these promoters bear marks of poised chromatin architecture before the onset of mRNA expression, we performed chromatin immunoprecipitation (ChIP) assays (Blythe et al., 2009) for these marks of poised loci and for β-catenin in 1000-cell stage embryos, the earliest preMBT time-point (prior to the onset of siamois and xnr3 expression) where it is feasible, in our hands, to perform ChIP (Figure 1D-F).
Although transcription is constrained before the MBT, phosphorylated forms of RNA Pol II corresponding to both initiating (CTD pSer5) and elongating (CTD pSer2) are detected before the MBT in whole embryo lysates (Figure S1), albeit at lower levels than after the MBT. To assess whether initiating or elongating RNA Pol II associates with β-catenin target genes, we performed ChIP against CTD pSer5- and CTD pSer2-RNA Pol II (Figure 1D), comparing samples collected before the onset of siamois and xnr3 expression (1000-cell) with those collected after the onset of expression (Stage 9, blastula, see Figure 1A). As a positive control, we compared RNA Pol II occupancy at the xnr6 locus, which is transcribed during both the 1000-cell stage and at Stage 9 (see Figure 1A). As expected, similar levels of both forms of RNA Pol II are found associated with the xnr6 locus before and after the MBT (Figure 1D). Consistent with our hypothesis, the siamois and xnr3 loci are bound by initiating RNA Pol II before the onset of their expression, at levels similar to those found after the MBT (Figure 1D, left panel); however the amount of elongating RNA Pol II associated with siamois and xnr3 increases after the MBT (Figure 1D, right panel). These observations indicate that the siamois and xnr3 loci are bound by initiating RNA Pol II prior to the onset of their expression, as expected for poised loci.
In addition to binding the promoters of preMBT expressed genes, xnr5 and xnr6, β-catenin also binds the siamois and xnr3 promoters before the MBT (Figure 1E and F). We also tested whether β-catenin could bind later (zygotic Wnt) targets during preMBT stages. Before the MBT, β-catenin does not bind a cluster of Tcf/Lef binding sites flanking the zygotic Wnt target gene myf5 (Yang et al., 2002a), although it binds these sites after the MBT (Figure 1E and data not shown), indicating that β-catenin binding is restricted to maternal Wnt target genes before the MBT. The siamois and xnr3 promoters are also marked by H3K9/14ac before the onset of their expression (Figure 1E). However, while β-catenin binding is restricted to maternal Wnt target genes, H3K9/14ac binding is more widepread: the promoters of all genes tested—maternal and zygotic β-catenin target genes and a negative control locus, myosin light chain 2 (mlc2)—contain H3K9/14ac before the MBT. Finally, maternal Wnt target promoters are marked before the MBT with another marker of poised loci, H3K4me3 (Figure 1F). Taken together, these data demonstrate that the β-catenin target genes siamois and xnr3 arrive at the MBT poised for expression.
These observations raise the possibility that β-catenin plays an instructive role in establishing poised chromatin architecture at the siamois and xnr3 loci. In support of this hypothesis, depletion of β-catenin reduces H3K4me3 at the siamois and xnr3 promoters (Figure 2A). In addition, inhibition of β-catenin/Tcf3 transactivation by ΔNTcf3 reduces β-catenin binding to the siamois and xnr3 promoters and blocks H3K4me3 (Figure 2B). In contrast, ΔNTcf3 has less of an effect on H3K9/14ac at the poised siamois and xnr3 promoters, suggesting that preMBT acetylation at siamois and xnr3 is established, at least in part, independently of β-catenin.
Figure 2. β-catenin and RNA Pol II establish H3K4me3 at poised promoters.
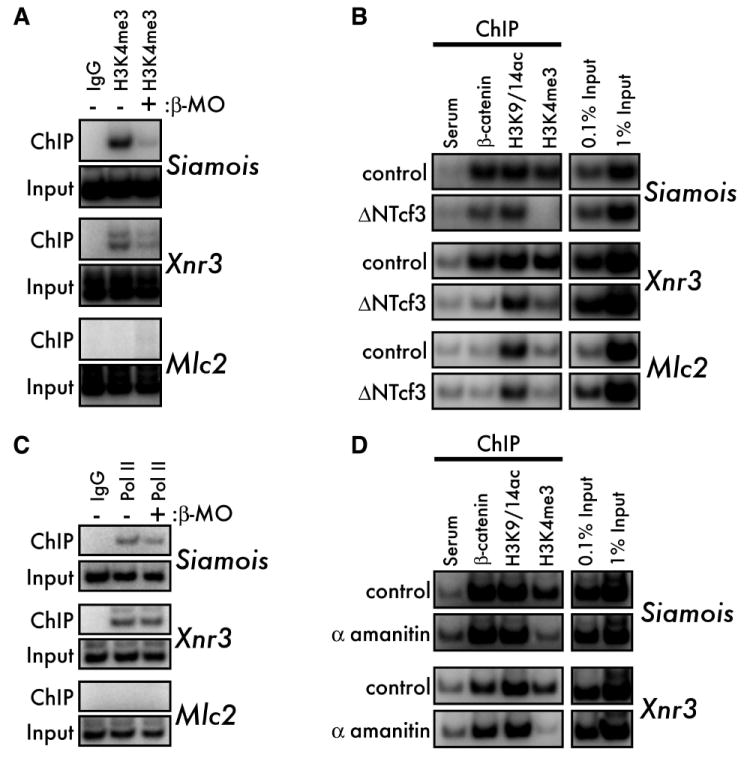
(A) ChIP for H3K4me3 in 1000-cell control and β-catenin depleted (β-MO) embryos. All sibling β-MO embryos were completely ventralized at later stages (not shown). (B) H3K4me3, but not H3K9/14ac, requires β-catenin function. MBT-stage control (upper panel for each gene) and ΔNTcf3 mRNA-injected (500pg, lower panels) embryos were subjected to ChIP for β-catenin, H3K9/14ac, and H3K4me3. (C) 1000-cell stage control and β-catenin depleted (β-MO) embryos were subjected to ChIP for RNA Pol II (CTD pSer5). All sibling β-MO embryos were completely ventralized at later stages (not shown). (D) MBT-stage control (upper panels) and α-amanitin injected (10ng, lower panels) embryos were subjected to ChIP for β-catenin, H3K9/14ac, and H3K4me3.
Dorsal specification by β-catenin prior to the 32-cell stage is sensitive to inhibition of RNA Pol II (Yang et al., 2002b). Deposition of H3K4me3 at promoters also correlates with occupancy of initiating polymerase (Ng et al., 2003), and RNA Pol II is also required for β-catenin-mediated H3K4me3 in Drosophila (Parker et al., 2008). We therefore tested whether preMBT RNA Pol II function is necessary for the establishment of other marks of poised loci. Knocking down β-catenin has no effect on the occupancy of initiating RNA Pol II at the poised siamois and xnr3 promoters (Figure 2C), indicating that initiating RNA Pol II is established at poised loci independently of β-catenin. In addition, RNA Pol II function is not required for binding of β-catenin; however, inhibition of Pol II greatly reduces H3K4me3 at the siamois and xnr3 promoters, but does not affect H3K9/14ac (Figure 2D).
In summary, we find that the maternal Wnt target genes siamois and xnr3 arrive at the MBT poised for activation. β-catenin functions, in collaboration with RNA Pol II, to establish the H3K4me3 mark in particular, whereas these promoters are bound by initiating RNA Pol II and H3K9/14ac even when Wnt/β-catenin signaling is inhibited. We conclude that β-catenin is required for the establishment of poised chromatin architecture at these loci, thereby priming them for activation at the MBT.
β-catenin associates with a Histone H3 (R8) methyltransferase activity in early Xenopus embryos
We next hypothesized that, during dorsal specification (between the 4- and 32-cell stage), β-catenin interacts with a chromatin-modifying activity that functions to establish poised chromatin architecture at target promoters. To identify such a factor, we immunoprecipitated β-catenin from 8- to 32-cell stage embryos and performed in vitro histone acetyl- or methyl-transferase (HAT or HMT) assays. During dorsal specification, β-catenin interacts with a HMT activity that specifically methylates Histone H3 but not H4 (Figure 3A). Under these assay conditions, we were unable to detect an associated HAT activity (Figure S2A). β-catenin is predicted to interact with several functionally different macromolecular complexes (Gottardi and Gumbiner, 2001). We determined that β-catenin interacts with this HMT activity in a high molecular weight complex that is unique to a subset of cellular β-catenin (Figure S2B-D).
Figure 3. β-catenin associates with a Histone H3(R8) methyltransferase before the MBT.

(A) β-catenin was immunoprecipitated from 16-cell embryos and HMT activity was visualized by fluorography (1 day exposure) for incorporation of [3H] methyl groups into calf thymus histones (top panel). Equal loading of histones is shown by coomassie staining (lower panel). “Input” represents HMT activity in embryo lysates, with activity toward both H3 and H4. (B) β-catenin IP/HMT assays were performed on either wild type (WT) recombinant H3.3 or H3.3 with the indicated point mutations. (C) β-catenin IP/HMT assays were performed on peptides corresponding to unmodified H3 (aa 1-15, lanes 1&2), asymmetrically dimethylated R8 (aa 1-15, lane 3), unmodified H3 (aa 1-21, lanes 4&5), acetylated K9 (aa 1-20, lane 6) and trimethylated K9 (aa 1-24, lane 7). (D) MBT-stage control and lithium chloride treated (LiCl, 300mM for 10 minutes, 1 hour prior to harvest) embryos were subjected to ChIP for either H3K9me1 or H3K9me3. (E) ChIP was performed as described in panel D, using instead antibodies to either H3R8me2a or H3R8me2s (Pal et al., 2004). (F) ChIP was performed on 1000-cell stage control and β-catenin knockdown (β-MO) embryos with the Active Motif H3R8me2a antiserum. See also Figure S2.
To identify the residue on Histone H3 targeted by the β-cat/HMT complex, we performed β-catenin IP/HMT assays using as the substrate recombinant H3.3 (rH3.3) with alanine point mutations at candidate target residues (Figure 3B) on the H3 N-terminal tail previously shown to be methylated: arginines (R) 2, 8, 17, 26 and lysines (K) 4 and 9 (Bedford and Clarke, 2009; Kouzarides, 2007). As with H3, the β-cat/HMT significantly methylates rH3.3(WT) over background. Importantly, mutation of K4 has no effect on H3 methylation, suggesting that the H3K4me3 observed at the poised siamois and xnr3 promoters is indirectly established by β-catenin (Figure 1F, 2A and 2B, see Discussion). These observations also rule out R2, 17, and 26 as the major methyl acceptor sites for the β-cat/HMT. In contrast, mutation of either R8 or K9 prevents H3 methylation by the β-cat/HMT. Similarly, while the β-cat/HMT methylates an unmodified H3 (1-15) peptide (Figure 3C, lane 2) to a similar level as full-length H3 (data not shown), modification of H3 peptides pre-modified at either R8 (asymmetric dimethyl) or K9 (acetyl and trimethyl) prevents methylation by the β-cat/HMT (Figure 3C, lanes 3, 6, and 7). Thus, in addition to targeting either position R8 or K9, the β-cat/HMT is also sensitive to the modification status of these residues.
H3K9 methylation is generally associated with heterochromatin and transcriptional repression (Kouzarides, 2007), and is therefore an unlikely target residue for the β-cat/HMT. Several ChIP experiments failed to detect H3K9me2 and –me3 at the siamois and xnr3 promoters between the 1000-cell stage and MBT, and H3K9me1 levels were not sensitive to stabilization of β-catenin by LiCl treatment (Figure 3D and data not shown). On the other hand, the effect of H3R8 methylation on transcriptional control is poorly understood. To test whether β-catenin activity regulates H3R8 methylation at target promoters, we generated an antibody that specifically detects asymmetrically dimethylated H3R8 (H3R8me2a) (Figure S2E and F), and confirmed that the H3R8me2a modification occurs in vivo. Subsequently, we measured H3R8 methylation at the siamois promoter by ChIP. H3R8me2a associates with the siamois promoter at the MBT and symmetric H3R8 dimethylation (H3R8me2s) is not detected (Figure 3E). This result was confirmed with an independent source of anti-H3R8me2a antibody (Figure 3F and Figure S2G). To test whether association of H3R8me2a correlates with β-catenin activity, we exposed preMBT embryos to a pulse of LiCl, which stabilizes β-catenin throughout the embryo. One hour after the LiCl pulse, H3R8me2a increased dramatically at the siamois and xnr3 promoters (Figure 3E and data not shown), indicating that asymmetric H3R8 methylation correlates with preMBT β-catenin activity. Furthermore, knockdown of β-catenin reduced H3R8me2a at the siamois and xnr3 promoters before the MBT (Figure 3F). Thus, the β-cat/HMT asymmetrically dimethylates H3R8 and is sensitive to the modification state of H3K9. Our results also indicate that β-catenin interacts with a type I (asymmetric) arginine HMT in early Xenopus embryos.
β-catenin recruits the H3R8 methyltransferase Prmt2 to target loci
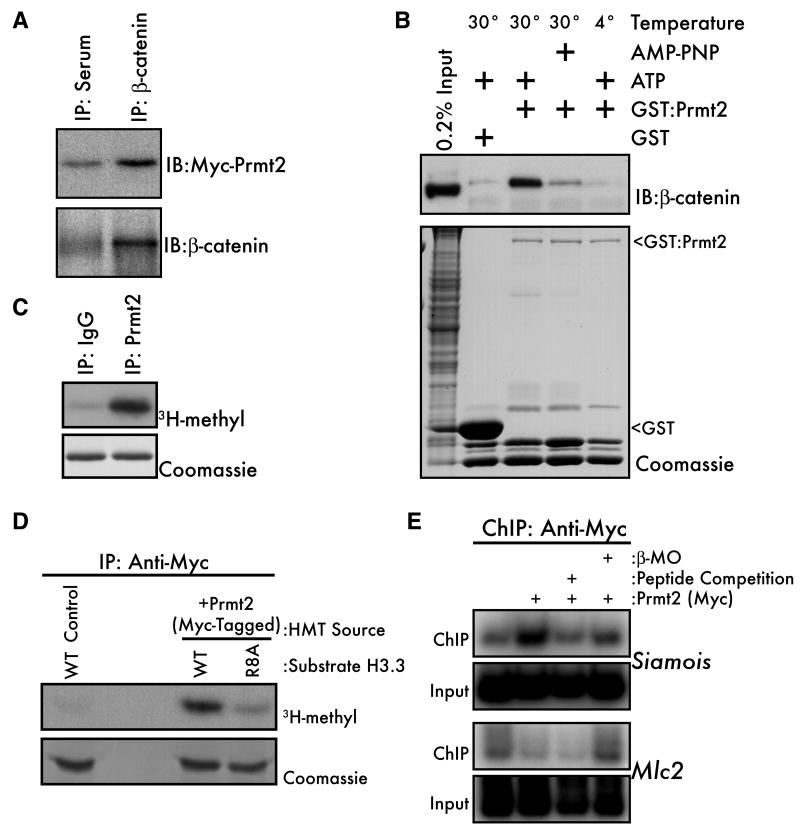
We undertook a candidate-based approach to identify the β-catenin-associated arginine HMT. Of the ten members of the protein arginine methyltransferase (Prmt) family, three have been shown to methylate Histone H3 in vitro: Carm1, Prmt5, and Prmt6 (Guccione et al., 2007; Hyllus et al., 2007; Pal et al., 2004; Schurter et al., 2001). Of these, only the type II, symmetric HMT Prmt5 specifically targets R8 (Pal et al., 2004). By expressing myc-tagged Prmts in Xenopus embryos and immunoprecipitating β-catenin, we found that none of the Prmts known to methylate H3 (Carm1, Prmt5, and Prmt6) co-purified with β-catenin (data not shown). However, Prmt2, which is most closely related to Carm1 and Prmt6, co-immunoprecipitates with β-catenin (Figure 4A). Although recombinant β-catenin and Prmt2 do not interact directly in vitro (data not shown), GST-tagged Prmt2 interacts with β-catenin in Xenopus embryo lysates in a temperature and ATP-dependent manner (Figure 4B). Coupled with our observation that the β-cat/HMT complex is large (Figure S2B-D), we propose that an unknown catalytic activity is required for Prmt2 and β-catenin to interact within a large macromolecular complex.
Figure 4. β-catenin interacts with the Histone H3(R8) methyltransferase Prmt2.
(A) Myc-tagged mouse Prmt2 (500pg) was expressed in Xenopus embryos and embryo lysates (early blastula) were immunoprecipitated with anti-β-catenin or pre-immune serum and subjected to western blot with either anti-myc (upper panel) or anti-β-catenin antibodies. (B) PreMBT embryo lysates were incubated for 1 hour with GST-Prmt2 beads at 4° or 30°C and with ATP or the nonhydrolyzable ATP analog AMP-PNP. Bound proteins were eluted and β-catenin was visualized by western blot (upper panel). GST beads alone were used as a negative control and the relative amounts of bait proteins in each lane were visualized by coomassie staining (lower panel, arrowheads). “Input” indicates non-fractionated input embryo lysate. (C) Endogenous Prmt2 from mouse embryonic stem cells was subjected to an IP/HMT assay using recombinant H3.3 as a substrate. Rabbit IgG was the negative control. (D) Activity of Myc-Prmt2 from 32- to 128-cell Xenopus embryos was measured by IP/HMT assay using wild-type (WT) or R8A histone H3.3 as substrates. Non-injected embryos served as negative controls. (E) Wild type or β-MO injected 1000-cell embryos expressing Myc-Prmt2 were subjected to ChIP using the anti-Myc-tag antibody. Non-injected embryos served as a negative control. The specificity of ChIP was confirmed by including a 200-fold excess of the Myc-peptide (lane 3) in the IP.
By sequence, Prmt2 is most closely related to the type I HMTs Carm1 and Prmt6, but its substrate preference has not been determined because recombinant Prmt2 has little to no activity in vitro (Lakowski and Frankel, 2009; Scott et al., 1998). Also, to our knowledge, HMT activity associated with endogenous Prmt2 has not been reported. Interestingly, endogenous Prmt2 immunoprecipitated from mouse embryonic stem cells methylates Histone H3 (Figure 4C). Likewise, myc-Prmt2 expressed in Xenopus embryos methylates Histone H3, and this activity requires H3R8 (Figure 4D). In these experiments, purified Prmt2 has HMT activity towards Histone H3, reflecting either the activity of Prmt2 itself or the activity of another HMT that co-immunoprecipitates with Prmt2. Further investigation will be required to determine the factors that regulate endogenous Prmt2 catalysis.
Importantly, Prmt2 binds the siamois promoter in preMBT embryos (Figure 4E) as determined by ChIP. Furthermore, β-catenin knockdown reduced Prmt2 occupancy at the siamois promoter, suggesting that β-catenin recruits Prmt2 to target promoters. We therefore conclude that Prmt2 represents the HMT activity that associates with β-catenin in early Xenopus embryos based on the observations that β-catenin interacts with Prmt2, Prmt2 has a HMT activity directed at H3 that is sensitive to the R8A mutation, and β-catenin recruits Prmt2 to target genes during the preMBT period.
Maternal Prmt2 is necessary for dorsal specification
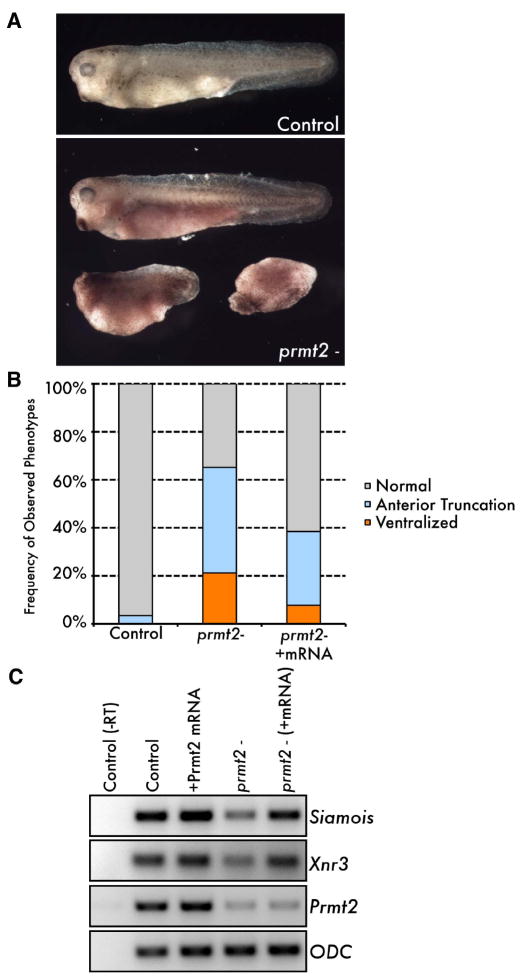
Based on the above observations, we predicted that recruitment of Prmt2 by β-catenin to dorsal target genes would be essential for specifying the dorsal developmental program. Therefore, we tested whether loss of Prmt2 function in early embryos would affect dorsal specification. Xenopus Prmt2 is expressed maternally, and knockdown of Prmt2 in fertilized embryos only weakly affects expression of siamois and xnr3 (data not shown). Therefore, we knocked down Prmt2 in oocytes and generated maternally depleted Prmt2 (prmt2-) embryos by the host transfer method (Mir and Heasman, 2008). Prmt2- embryos generated by antisense DNA are impaired in dorsal development (Figure 5A). These embryos develop with a range of dorso-ventral morphologies, displaying completely ventralized, anterior-truncated (partially ventralized), and normal phenotypes within single clutches (Figure 5A and B, N=66 embryos from six independent experiments). Importantly, depletion of Prmt2 reduces the expression of both siamois and xnr3 following the MBT (Figure 5C and Figure S3A and B), indicating that Prmt2 function is necessary for the activation of maternal Wnt/β-catenin dependent transcription. These ventralized phenotypes are specific to loss of Prmt2 function, as expression of mouse Prmt2 mRNA in prmt2- embryos rescues siamois and xnr3 expression and partially rescues the morphological phenotype (Figure 5C and B, N=26 from three independent experiments). We also replicated the knockdown of siamois and xnr3 with a translational blocking morpholino oligonucleotide against Prmt2 (Figure S3A). However, while the morpholino generated a more severe knockdown of siamois and xnr3, it was more difficult to generate viable MBT-stage embryos, possibly due to a general requirement for Prmt2 in the regulation of chromatin structure beyond its proposed role in the Wnt pathway. Alternatively, the antisense DNA yielded a partial knockdown of maternal prmt2 message that allowed for recovery of viable embryos at the expense of the severity of the phenotype (Figure 5 and Figure S3). Nonetheless, these observations strongly support the conclusion that recruitment of Prmt2 to dorsal gene promoters is a necessary step in establishing the dorsal gene expression program.
Figure 5. Maternal Prmt2 is necessary for dorsal specification.
(A and B) Transplantation and fertilization of maternal Prmt2-depleted (prmt2-) oocytes results in a range of ventralized tadpole-stage phenotypes (A, lower panel) compared to controls, which develop normally (A, top panel). The mean frequency of phenotypes arising from maternal Prmt2 depletion is plotted in B (see text for details). The frequency of ventralized embryos (both partial and complete) is reduced by co-injection of Prmt2 mRNA (“prmt2-/+mRNA”). (C) Blastula stage (stage 9) siamois and xnr3 expression was measured in prmt2- and rescued (“prmt2- (+mRNA”)) host transfer embryos as compared to control (non-depleted) and Prmt2 mRNA injected embryos. As a control for the efficiency of knockdown, prmt2 was measured. Note that the rescuing mRNA (1ng of mouse Prmt2, injected into oocytes) is not amplified by the Xenopus prmt2 primers used here. Expression of rescuing mRNA was confirmed by western blot (not shown). Embryos expressing Prmt2 mRNA alone developed identically to controls, with no dorsoventral defects (not shown). See also Figure S3.
Directing Prmt2 to β-catenin target promoters is sufficient to drive dorsal specification in the absence of β-catenin
β-catenin interacts with chromatin via the Tcf/Lef family of DNA-binding factors (Behrens et al., 1996; Molenaar et al., 1996), and previous investigations have exploited this interaction to target factors of interest to Tcf/Lef binding sites and test their effects on target gene expression (Vleminckx et al., 1999). Therefore, we generated chimeric proteins (Figure 6A) between Prmt2 and the DNA binding domain of Lef-1 (ΔNLef1) to direct Prmt2 to target genes and evaluate its effect on dorsal specification. Chimeras between ΔNLef1 and a SAM-binding mutant of Prmt2 (Prmt2GG)(Qi et al., 2002), or wild type Carm1, Prmt5, and Prmt6 were also generated as controls. All chimeric proteins were expressed to similar levels in blastula stage embryos (Figure 6B and data not shown).
Figure 6. Directing Prmt2 to β-catenin target gene promoters is sufficient to drive dorsal specification.
(A) Schematic of the Prmt2:ΔNLef1 chimeric construct. To direct Prmt2 to Tcf/Lef DNA binding sites, the DNA-binding HMG domain of mouse Lef1 was fused to the C-terminus of mouse Myc-Prmt2. (B) 4-cell embryos were injected with either wild-type (WT) or catalytically inactive SAM-binding mutant G159,161R (GG) Prmt2:ΔNLef1 into two ventral blastomeres. Phenotypes (left panel) were scored at late-neurula / early tailbud stages. Mean frequency of secondary axis formation (both fully and partially extended) resulting from expression of wild-type or GG mutant Prmt2:ΔNLef1 is plotted on the right. N=197, 172, and 81 embryos for control, WT, and GG mutant, respectively. Error bars are SEM. P=0.017 (two-tailed Student’s T-Test) for the 4 independent trials where WT and mutant were compared directly. Equal expression of wild type and GG mutant Prmt2:ΔNLef1 was verified by western blot for the myc tag (inset). (C) Embryos were depleted for β-catenin (β-MO, panels ii-iv) and subsequently injected with 500pg of either Prmt2:ΔNLef1 (iii) or Prmt5:ΔNLef1 (iv) mRNA. Rescue of β-MO-induced ventralization (ii) was measured at tadpole stages. Note the rescue of the anterior-most, dorsally derived cement gland and eye in panel iii, compared to control, non-injected embryos (i). The percentages in the upper right corner of each panel indicate the frequency at which the phenotypes shown were observed. (D) Embryos were depleted for β-catenin (β-MO) and subsequently injected with Prmt2:ΔNLef1 or ΔNLef1 mRNA as in (C). Expression of Siamois and Xnr3 was measured by RT-PCR at stage 10. EF1α expression is shown as a loading control. (E) Prmt2:ΔNLef1 and Carm1:ΔNLef1 mRNAs (500pg) were injected and RT-PCR was performed as described for C and D.
Recruitment of Prmt2 to β-catenin target genes is sufficient to specify the dorsal developmental program, as expression of the Prmt2:ΔNLef1 chimera in ventral blastomeres converts them to dorsal progenitors (N=172, Figure 6B), similar to activation of Wnt/β-catenin signaling. Ventral expression of the ΔNLef1 DNA binding domain alone did not induce secondary axes (N=31, data not shown). To determine whether Prmt2-induced dorsal cell fates are dependent on its catalytic activity, we tested the ability of the Prmt2GG mutant to induce secondary axes. At similar levels of expression, Prmt2GG:ΔNLef1 induced significantly fewer secondary axes (N=81, Figure 6B, histogram), and these axes were typically truncated (Figure 6B, photo) compared to those observed with Prmt2:ΔNLef1, indicating that dorsal specification by Prmt2 is dependent on its HMT activity. However, Prmt2GG nonetheless conferred some dorsal axis inducing activity; therefore we cannot rule out that additional factors that interact with Prmt2 contribute to Prmt2-dependent dorsal specification.
In a complementary approach, we tested whether Prmt2:ΔNLef1 could rescue β-catenin loss of function. Embryos depleted for β-catenin develop with a ventralized phenotype (Figure 6C ii; also figure 1B ii), whereas expression of Prmt2:ΔNLef1 in β-catenin-depleted embryos restores the full range of dorsal and anterior structures (Figure 6C iii), remarkably similar to control embryos (Figure 6C i), albeit typically with a single eye (86% rescue, N=172). The anterior defects in Prmt2:ΔNLef1-rescued β-MO embryos could result from activation of the posteriorizing zygotic Wnt target genes, as expression of Prmt2:ΔNLef1 alone in dorsal blastomeres did not affect dorsal specification, but did induce a weak posteriorized phenotype (data not shown, N=29). Importantly, expression of Prmt2:ΔNLef1 in β-MO embryos rescues organizer gene expression, whereas the ΔNLef1 DNA binding domain alone has minimal activity (Figure 6D).
To assess the specificity of Prmt2:ΔNLef1, we also tested whether other Prmts could rescue dorsal specification in β-catenin depleted embryos. Prmt5 shares target residue specificity with Prmt2, but symmetrically dimethylates H3R8 (Pal et al., 2004), and Prmt5:ΔNLef1 is unable to rescue dorsal specification in β-catenin depleted embryos (Figure 6B iv—0% Rescue, N=31)). On the other hand, the robust transcriptional activator Carm1 asymmetrically methylates H3 R2, R17, and R26, with only a weak activity towards R8 (Chen et al., 2000; Schurter et al., 2001). Expression of Carm1:ΔNLef1 is toxic to embryos shortly after gastrulation, so phenotypic rescue could not be scored. However, at gastrula stages, Carm1:ΔNLef1 does not rescue siamois and xnr3 expression (Figure 6D). Finally, Prmt6, which targets H3R2 (Guccione et al., 2007; Hyllus et al., 2007), also did not rescue dorsal specification (not shown, 0% rescue, N=30). Thus, of the Prmts tested, only the recruitment of Prmt2 to β-catenin target promoters is sufficient to rescue dorsal specification, demonstrating the unique role of Prmt2 in the regulatory events that establish the transcriptional network driving dorsal development in Xenopus embryogenesis.
Discussion
In this work, we demonstrate that dorsal specification by the Wnt/β-catenin pathway drives transcriptional poising at Wnt regulated dorsal target genes. This accounts for the uncoupling of dorsal specification (prior to the 32-cell stage) from the onset of target gene expression (MBT for siamois and xnr3). We have investigated this from two perspectives. First, ChIP analysis demonstrates that the β-catenin target genes siamois and xnr3 arrive at the MBT poised for activation, marked by initiating RNA Pol II, H3K4me3, and H3K9/14ac. β-catenin activity, in particular, is required for the ultimate establishment of H3K4me3. Second, biochemical and functional analyses show that β-catenin interacts with the H3R8 methyltransferase Prmt2 during dorsal specification and that Prmt2 activity is both necessary and sufficient to drive dorsal development downstream of β-catenin. Transcriptional poising occurs in eukaryotes from yeast to mammals, including in several embryonic contexts (Akkers et al., 2009; Baugh et al., 2009; Bernstein et al., 2006; Guenther et al., 2007; Hargreaves et al., 2009; Muse et al., 2007; Radonjic et al., 2005; Rougvie and Lis, 1988; Vastenhouw et al., 2010; Zeitlinger et al., 2007); here we illustrate how a critical developmental signaling pathway, acting through a gene-specific transcription factor, poises genetic loci for expression at a later time in development.
Drosophila heat shock genes are the most well understood examples of poised genetic loci. Initially, “pioneer factors” (such as trithorax-like/GAGA factor) function to open chromatin and nucleate the assembly of the initiated (but stalled) RNA Pol II holoenzyme [(Lee et al., 2008; Lee et al., 1992; Shopland et al., 1995) and references therein]. Following binding of “release factors” (such as Heat Shock Factor) that recruit elongation-promoting factors such as CyclinT/Cdk9, RNA Pol II enters into productive elongation [(Lis et al., 2000) and references therein]. Our work does not suggest that β-catenin functions as a pioneer factor: in the absence of β-catenin function, target gene promoters are nonetheless maintained in an “incompletely poised” state containing stalled RNA Pol II and H3K9/14ac but not H3K4me3. Rather, our work suggests that β-catenin functions as part of the mechanism that establishes a “fully poised” state, recruiting additional chromatin modifying activities (such as Prmt2), ultimately resulting in the downstream establishment of H3K4me3 in collaboration with RNA Pol II. Upon establishment of a fully poised promoter, β-catenin target genes thereby become receptive to transcriptional activation at the MBT. We further speculate that the incompletely poised state of dorsal target genes seen in the absence of β-catenin reflects the embryonic competency to activate the dorsal gene expression program throughout the embryo.
Pre-setting patterns of gene expression in the embryo
The targets of the maternal Wnt/β-catenin pathway demonstrate different latencies between dorsal specification and the onset of gene expression. The preMBT genes xnr5 and xnr6 require additional input from the maternal transcription factor VegT (Takahashi et al., 2000). Thus, VegT could function as a “release factor” for these genes, recruiting elongation-promoting factors to the xnr5 and xnr6 loci downstream of β-catenin. On the other hand, factors that regulate the global activation of the zygotic genome at the MBT could be responsible for the release of the later responding siamois and xnr3 genes. Indeed, transcriptional poising is an attractive mechanism to account for the synchronous activation of large-scale zygotic gene expression at the MBT. While such global activating factors have yet to be identified in Xenopus, in Drosophila, both Smaug and Zelda have been shown to be essential factors for zygotic genome activation (Benoit et al., 2009; Liang et al., 2008). Smaug activity is essential for the establishment of elongating (CTD pSer2) RNA Pol II at the MBT, and could thereby promote “release” of such preMBT poised loci, albeit indirectly (Benoit et al., 2009). The mechanism of action for Zelda is unknown, but it binds DNA sequences present in the majority of immediate-early zygotic transcripts (Liang et al., 2008). Further investigation is needed to determine the extent of transcriptional poising at immediate-early zygotic loci and the mechanism of action for such global zygotic gene activators at the MBT.
While preMBT Xenopus embryos are transcriptionally competent (Prioleau et al., 1994; Toyoda and Wolffe, 1992), several overlapping mechanisms dominantly suppress zygotic gene expression (Veenstra, 2002). Interfering with these repressive activities can reveal a suppressed pro-transcriptional activity. Depleting embryos of the DNA methyltransferase Dnmt1 causes precocious expression of many genes, suggesting that these genes are poised for activation prior to the MBT but are repressed by Dnmt1-mediated DNA methylation (Stancheva and Meehan, 2000). Also, embryos generated from transplantation of transcriptionally active nuclei will display preMBT expression of genes that were active in the original donor cells (Ng and Gurdon, 2005). This transcriptional memory is linked to chromatin modifications that correlate with active transcription, particularly the incorporation of the histone variant H3.3 (Ng and Gurdon, 2008). These observations demonstrate the competency of preMBT embryos to establish and maintain active-but-repressed chromatin. We further speculate that transcriptional poising is a major mechanism underlying the activation of the zygotic genome at the MBT.
Context-dependent chromatin modifying activities for β-catenin?
β-catenin can interact with a number of chromatin modifying enzymes and may recruit additional factors to poised loci, including H3K4 methyltransferases or, at the MBT, elongation promoting factors (Mosimann et al., 2006; Sierra et al., 2006). As hypothesized by Mosimann and Basler (2009), “cofactor switching” of β-catenin is an attractive model to account for the numerous possible interactions between β-catenin and its coactivators, yet has proven difficult to dissect at the molecular level. A recent genome-wide screen for β-catenin interactors in colorectal carcinoma cell lines (Major et al., 2008) confirmed several of these interactions and identified novel complex members, including the H4 arginine HMT Prmt1. Carm1 also interacts with β-catenin in the context of androgen receptor signaling (Koh et al., 2002). In preMBT embryos, β-catenin interacts with Prmt2, but not with Carm1 or Prmt1, suggesting that the β-catenin/chromatin remodeling complex associates with different Prmt family members in a context-dependent manner. Additionally, in colon cancer cell lines, the β-catenin/chromatin remodeling complex contains the MLL family of H3K4 HMTs (Major et al., 2008; Sierra et al., 2006). Our analysis indicates that, in early Xenopus embryos, the primary HMT activity associated with β-catenin is directed at H3R8, but the possibility remains that β-catenin interacts with MLL family members in later developmental stages. A systematic comparison of these complexes from diverse developmental stages will be necessary to investigate this possibility further.
Notably, after the 32-cell stage, siamois and xnr3 expression becomes “locked in” and resistant to inhibition of β-catenin/Tcf transactivation. Establishment of poised chromatin architecture is an attractive model to account for this observation. ΔNTcf3 functions by inhibiting Tcf3 binding to chromatin (Tutter et al., 2001), thereby excluding β-catenin from its genomic loci. However, another co-factor of β-catenin, Pygopus, binds methylated H3K4 via its PHD domain (Fiedler et al., 2008) and is also essential for dorsal specification (Belenkaya et al., 2002). This suggests an attractive model where, following establishment of poised chromatin architecture prior to the 32-cell stage, Pygopus could function as a Tcf3-independent tether for β-catenin at poised target genes (de la Roche and Bienz, 2007; Fiedler et al., 2008). Additionally, since poised loci can also behave as chromatin insulators (Chopra et al., 2009), the poised chromatin architecture of the siamois and xnr3 loci could serve the dual purpose of marking these genes for activation at the MBT and insulating them from down-regulation by transcriptional repressors, such as non-liganded Tcf3.
H3R8 methylation and the Histone Code
A remaining question is what role asymmetric H3R8 methylation plays in either the establishment or maintenance of poised chromatin architecture, in particular H3K4 methylation. The Histone Code Hypothesis postulates that histone modifications function combinatorially to regulate events such as transcriptional activation (Strahl and Allis, 2000). Thus, H3R8 methylation could recruit downstream complexes containing an H3K4 HMT, resulting in the subsequent methylation of H3K4. Although factors that “read” H3R8me2a have yet to be identified, histone arginine methylation influences the patterns of histone tail modifications (Bedford and Clarke, 2009). Alternatively, H3R8 methylation could inhibit the activity of a repressive factor, such as an H3K4 demethylase, which would otherwise maintain transcriptional repression. In support of this model, H3R8 methylation inhibits demethylation of K4 by LSD-1 (Forneris et al., 2006). In addition, we observe that β-catenin mediated H3R8 methylation is sensitive to the modification state of K9, as also observed for the symmetric H3R8 HMT Prmt5 (Pal et al., 2004). Similarly, Prmt1, whose methylation of Histone H4R3 potentiates downstream H4 acetylation, will not methylate a pre-acetylated H4 tail (Wang et al., 2001). Conversely, H3K9 methylation by the HMT G9a is inhibited by methylation of H3R8 (Rathert et al., 2008). These observations suggest that H3 R8 and K9 antagonism represents a critical regulatory node underlying the interpretation of the Histone Code, where asymmetric methylation of R8 activates and K9 represses gene expression. Antagonistic H3K9 methylation could thus represent a mechanism to restrict the expression domains of dorsal determinants in early embryos.
In conclusion, we have demonstrated how a developmental signaling pathway plays an instructive role in the establishment of poised chromatin architecture, linking with chromatin regulatory mechanisms to pre-set gene expression programs in the embryo. Future investigations will focus on how the constellation of chromatin marks established at promoters before the MBT function to regulate the initial patterns of embryonic gene expression.
Experimental Procedures
Extensive details are provided as a supplement
Embryo Manipulations
Embryos were obtained, cultured, and microinjected as described (Sive et al., 2000). Capped mRNAs were produced by in vitro transcription using the SP6 mMessage mMachine Kit (Ambion). β-catenin morpholino (Heasman et al., 2000) was injected separately from mRNA to prevent precipitation.
Plasmid Construction
cDNAs for mouse Prmt2, 5, 6, and Carm1 were PCR-amplified from I.M.A.G.E. Consortium [LLNL] clones (Open Biosystems) (Lennon et al., 1996) and subcloned into pCS2+MT.
RT-PCR
Radiolabeled RT-PCR was performed as described (Yang et al., 2002b) using GoTaq Flexi polymerase (Promega). Purified RNA (5 embryos/sample) was DNase treated and re-purified by RNeasy (Qiagen). 2.5μg total RNA was used per cDNA synthesis reaction.
Chromatin Immunoprecipitation
ChIP was performed as described (Blythe et al., 2009) with 50 embryos per IP and either radiolabeled PCR or SYBR green QPCR. Anti N-β-catenin was described in (Blythe et al., 2009). Purchased antibodies included anti-H3acK9/14 and -H3K4me3 (Millipore #06-599 & #07-473), anti-Pol II p-Ser5 (Active Motif #39233), anti-Pol II p-Ser2 (Abcam #Ab5095), H3K9me1 and H3K9me3 (Millipore 07-450 & 07-442), H3R8me2a (Active Motif #39651), and anti-Myc epitope (Sigma #C3956). The anti-H3R8me2s antibody was kindly provided by Dr. Said Sif (Ohio State University) (Pal et al., 2004). For primer information, see Supplemental Methods.
Peptide Synthesis
Unmodified and R8me2a H3 (1-15+C) peptides were synthesized by the Proteomics Resource Center of the Rockefeller University. Unmodified H3 1-21, H3K9ac 1-20, and H3K9me3 1-24 peptides were from AnaSpec.
Anti-H3R8me2a
H3R8me2a antiserum was generated by immunizing rabbits with H3R8me2a (1-15+C) peptide coupled to KLH (Cocalico Biologicals). This antiserum was depleted of antibodies to unmodified H3 and affinity purified with the R8me2a peptide. The limit of detection was ~8pmol H3R8me2a (1-15) by dot blot. Results were duplicated with H3R8me2a antiserum from Active Motif (39651) (see Figure S2G).
Immunoprecipitations
Embryos were homogenized in HM 0.1M buffer (Sierra et al., 2006), supplemented with Protease Inhibitor Cocktail (Sigma P8340) and Phosphatase Inhibitor Cocktail I and II (Sigma P2850 & P5726). Insoluble material was pelleted by centrifugation (14k × g) for 10 minutes at 4°C. Supernatants were adjusted to 0.15mM KCl and 0.2% NP-40 and filtered through a 0.45mm cellulose acetate syringe filter (Millipore) prior to addition of antibodies. Immune complexes precipitated with recombinant protein G agarose were washed 5x in wash buffer (HMZ 0.15M) (Sierra et al., 2006) prior to IP/HMT analysis or western blot.
Histone Methyltransferase Assays
HM 0.1M lysates of 8- to 32-cell embyos were immunoprecipitated and processed for IP/HMT assays as described (Sierra et al., 2006)(see supplement) using 50mM Tris-HCl (pH8.8) in the reaction buffer. 5μg Calf Thymus Histones, 2.5μg recombinant HA-H3.3, or 5μg synthetic peptide was used as substrates. IP/HMT reactions typically generated <1pmol of in vitro labeled substrate.
Maternal Depletion of Prmt2
Maternal depletions were performed as described (Mir and Heasman, 2008). Antisense DNA (1ng/nl) against Prmt2 mRNA (Genbank HM205111) was injected vegetally into defolliculated oocytes at the indicated doses. Control and antisense injected oocytes were incubated for 24 hours prior to in vitro maturation (2μM Progesterone, 10 hours) and transfer to an egg-laying host. Transferred eggs were collected in high-salt buffer for 4 hours following transplantation prior to in vitro fertilization. For mRNA rescue experiments, 10nl of mouse Prmt2 mRNA (100pg/nl) was injected vegetally shortly before oocyte maturation. The sequence of the Prmt2 antisense oligo is: 5’- TCC GTT CTG TAT CTC TCC —3’.
Supplementary Material
Acknowledgments
We thank Daniel Kessler, Steve DiNardo, Marisa Bartolomei, Gerd Blobel, Tom Kadesch, Chris Wylie, Matt Kofron and the members of the Klein Lab for helpful discussions and encouragement. We also thank Dr. Said Sif (Ohio State University) for his gift of the H3R8me2s antiserum, and Scott Paschke (Active Motif) for providing the H3R8me2a antiserum. This work was supported by National Institutes of Health grants (T32-GM007229) and (T32-HD007516) to SAB and (R01-GM76621) to PSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II Accumulates at Promoters of Growth Genes During Developmental Arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein Arginine Methylation in Mammals: Who, What, and Why. Molecular Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J, Lin X. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development. 2002;129:4089–4101. doi: 10.1242/dev.129.17.4089. [DOI] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Developmental Dynamics. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- Chopra VS, Cande J, Hong J, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes & Development. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M, Bienz M. Wingless-independent association of Pygopus with dTCF target genes. Curr Biol. 2007;17:556–561. doi: 10.1016/j.cub.2007.01.063. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Fiedler M, Sánchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Müller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Molecular Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Dall’Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J Biol Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Gumbiner BM. Adhesion signaling: how beta-catenin interacts with its partners. Curr Biol. 2001;11:R792–794. doi: 10.1016/s0960-9822(01)00473-0. [DOI] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young R. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes & Development. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi H, Matsumura N, Hanafusa H, Matsumoto K, De Robertis EM, Kuroda H. Expression of Siamois and Twin in the blastula Chordin/Noggin signaling center is required for brain formation in Xenopus laevis embryos. Mech Dev. 2008;125:58–66. doi: 10.1016/j.mod.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kao KR, Masui Y, Elinson RP. Lithium-induced respecification of pattern in Xenopus laevis embryos. Nature. 1986;322:371–373. doi: 10.1038/322371a0. [DOI] [PubMed] [Google Scholar]
- Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lakowski TM, Frankel A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J. 2009;421:253–261. doi: 10.1042/BJ20090268. [DOI] [PubMed] [Google Scholar]
- Laurent MN, Blitz IL, Hashimoto C, Rothbächer U, Cho KW. The Xenopus homeobox gene twin mediates Wnt induction of goosecoid in establishment of Spemann’s organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Development. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Development. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, Ferrer M, Yi X, Stoick-Cooper CL, von Haller PD, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Science signaling. 2008;1:ra12. doi: 10.1126/scisignal.2000037. [DOI] [PubMed] [Google Scholar]
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Mir A, Heasman J. How the mother can help: studying maternal Wnt signaling by anti-sense-mediated depletion of maternal mRNAs and the host transfer technique. Methods Mol Biol. 2008;469:417–429. doi: 10.1007/978-1-60327-469-2_26. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Muse G, Gilchrist D, Nechaev S, Shah R, Parker J, Grissom S, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Molecular Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Ng R, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci USA. 2005;102:1957–1962. doi: 10.1073/pnas.0409813102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DS, Ni YY, Chang JL, Li J, Cadigan KM. Wingless signaling induces widespread chromatin remodeling of target loci. Mol Cell Biol. 2008;28:1815–1828. doi: 10.1128/MCB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Molecular Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5’ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Development. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes & Development. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & Development. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, Asashima M. Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development. 2000;127:5319–5329. doi: 10.1242/dev.127.24.5319. [DOI] [PubMed] [Google Scholar]
- Toyoda T, Wolffe AP. Characterization of RNA polymerase II-dependent transcription in Xenopus extracts. Dev Biol. 1992;153:150–157. doi: 10.1016/0012-1606(92)90099-3. [DOI] [PubMed] [Google Scholar]
- Tutter AV, Fryer CJ, Jones KA. Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro. Genes & Development. 2001;15:3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ. Early Embryonic Gene Transcription in Xenopus. Adv Dev Biol Biochem. 2002;12:85–105. [Google Scholar]
- Vleminckx K, Kemler R, Hecht A. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech Dev. 1999;81:65–74. doi: 10.1016/s0925-4773(98)00225-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Shinagawa A. Marked alteration at midblastula transition in the effect of lithium on formation of the larval body pattern of Xenopus laevis. Dev Growth Differ. 1989;31:531–541. doi: 10.1111/j.1440-169X.1989.00531.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Mei W, Otto A, Xiao L, Tao Q, Geng X, Rupp RA, Ding X. Repression through a distal TCF-3 binding site restricts Xenopus myf-5 expression in gastrula mesoderm. Mech Dev. 2002a;115:79–89. doi: 10.1016/s0925-4773(02)00121-1. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002b;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J, Nechaev S, Adelman K, Levine M, Young R. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.