Synopsis
An exquisite interplay of developmental cues, transcription factors, coregulatory and signaling proteins support formation of skeletal elements of the jaw during embryogenesis and the dynamic remodeling of alveolar bone in the post-natal life. These molecules promote initial condensation of the mesenchyme, commitment of the mesenchymal progenitor to osteogenic lineage cells, and differentiation of committed osteoblast to mature osteocyte within mineralized bone. Parallel regulatory network promote formation of the functional ostoclast from mononuclear cells to support continuous bone remodeling within the alveolar bone. With an ever expanding list of new regulatory factors, the complexities of the molecular mechanisms that control gene expression in skeletal cells are being further appreciated. This review examines the multifunctional roles of prominent nuclear proteins, cytokines, hormones and paracrine factors that control osteogenesis.
Keywords: Bone Development, Skeletal Remodeling, Osteoblast, Molecular Signaling
Skeletogenesis in mammals requires coordinated activities of multiple cell types and is formed by two distinct developmental processes. 1) Endochondral ossification; majority of the skeletal elements in the body including all long bones are derived by this process. Sequential maturation and degradation of chondrocyte produced cartilaginous template is a pre-requisite for osteoblast recruitment. The second step in endochondral ossification is the eventual replacement of cartilage matrix with the mineralized matrix synthesized by osteoblast. 2) Intramembranous ossification; craniofacial skeletal elements are primarily derived through this process, whereby cells in condensed mesenchyme directly differentiate into mineralizing osteoblast.
1. Developmental origin of Alveolar Bone
During embryonic development, blocks of condensed mesenchyme are modeled into precisely shaped cartilaginous elements (1). In humans, this process of skeletal patterning is completed within first trimester of pregnancy (9th week after conception). Subsequently, the skeletal tissue template undergoes a dramatic increase in size and ossification but with relatively small change in basic shape of bones. In mammals, both the mandibular and maxillary bones develop from the first branchial arch during embryonic skeletal patterning (2).The alveolar bone and processes in the maxilla and mandible are formed by intramembranous ossification. However, cellular components of the craniofacial skeleton are unique and include cranial neural crest-derived ectomesenchyme (3, 4). The mandibular and maxillary alveolar process houses and supports the dentition. Tooth development initiates as a local thickening of oral epithelium that subsequently this thickened epithelium grows into the underlying neural crest-derived mesenchyme of the first branchial arch (5, 6). Tooth development proceeds through multiple stages of differentiation which are morphologically distinct such as, dental lamina, bud, cap, bell, crown and root. In human deciduous teeth, formation of dental lamina is noted by 7 weeks of gestation, with the ultimate cytodifferentiation of odontoblast that produce the dentin extracellular matrix by 18 weeks. Although, the alveolar bone is formed in relation to the teeth but structurally it is similar to the basal bone and the alveolar bone cells closely resemble skeletal osteoblasts (3).
2. Osteoblast Biology
During embryonic development osteoblasts originate from local mesenchyme, and postnataly from bone marrow stromal stem cell or connective tissue mesenchymal stem cell (MSC). In response to specific stimuli, these precursor cells commit to osteogenic lineage and differentiate into mature osteoblasts. Extensive research in the past 20-years by many laboratories have defined the sequence of events that results in the maturation of osteoblasts (7–13). Distinct stages of osteoblast that are characterized by the expression of specific genes and functional properties have been established using in vitro cell culture and in vivo models, and by determining modifications in gene expression in normal and affected bone tissues. Profiles of gene expression in vivo further define the sub-stages of osteoblast maturation, and these sub-stages are altered as a result of genetic mutations.
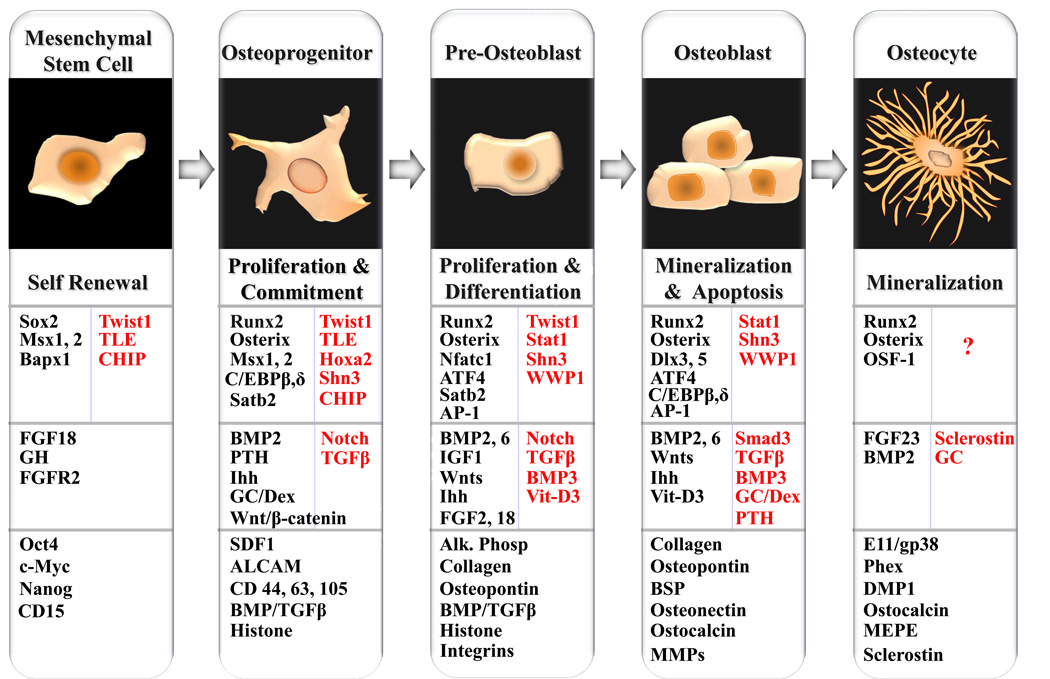
In general, osteoblastogenesis is defined by four major phases: lineage commitment, proliferative expansion, synthesis of extracellular matrix (ECM) and mineralization. All these stages are characterized by sequentially expressed genes that support the progression of osteoblast differentiation through developmental transition points (Fig. 1). The first transition requires MSC commitment to osteogenic lineage, second transition is associated with mitotic duplication and expansion of osteoprogenitor, the third transition requires exit from the cell cycle and robust production of extracellular matrix by osteoblast, the final stage is marked by mineralization of the extracellular matrix and establishment of osteocyte.
Figure 1. Ontogeny of osteoblast and regulatory control of osteoblast lineage progression and phenotypic features.
Sequence and stages of the osteoblast lineage from a self-renewing, pluripotent mesenchymal stem cell to terminally differentiated osteocyte is diagrammatically illustrated. The characteristic feature of each developmental stage is indicated below the cell morphology. Next row summarizes the key transcription factor and co-regulatory protein involved in genetic control of osteoblast differentiation. Factors that negatively regulate Runx2 activity and osteoblast differentiation are indicated in red. Several physiologic mediators influencing osteoblast development, including transforming growth factor β (TGFβ), the bone morphogenetic proteins (BMPs), and fibroblast growth factors (FGFs), Wnt/β-catenin signaling and hormones are also indicated. Secretory molecules, receptor and signal transducer that inhibit osteoblast maturation are highlighted in red. Last row summarize phenotypic marker genes expressed at different developmental stages of osteoblast differentiation.
Self renewing mesenchymal stem cells are pluripotent and can give rise to multiple tissue type lineages such as osteoblast, odontoblast, chondrocytes, myoblasts, adipocytes and tendon cells. However, due to lack of distinct morphologic feature and histochemical markers, the mesenchymal stem cells and osteoprogenitor are not readily identifiable. Initially, the Stro-1 antibody was used to isolate adherent population from adult bone marrow cells but the surface antigen is ubiquitously expressed (14). Recent reports shows that a combination of multiple cell surface markers and absence of hematopoietic stem cells lineage markers significantly enrich MSC population from bone marrow. However, to date no single specific marker has been identified that can select and enrich osteoprogenitors from all tissues.
The first step in development of the osteoblast phenotype is the lineage commitment and formation of osteoprogenitors. This commitment is regulated by master transcription factors and their co-regulators. This step involves expression of lineage inducing factors as well as inhibition of factors that maintain lineage plasticity. The key factors for commitment of osteogenic cell type include Runx2, Osterix, Sox9, and morphogens TGFβ/BMPs, FGFs. The initial progenitor are still bipotential and can commit to either osteo or chondro lineage depending upon the threshold of Sox and Runx2/Osterix. Vitamin D3, glucocorticoids, parathyroid hormone, and estrogen are shown to regulate parameters of the proliferation/growth in a dosage and time dependent fashion (15–20). These hormones and cytokines influence the progression of MSCs into the osteoblast lineage, either directly or indirectly by suppressing signaling that promote commitment to other lineages such as adipocyte and myoblast. (21–22).
In the second stages of osteoblast development there is extensive proliferation of progenitor cells with expression of growth related genes (histones, c-myc and c-fos). What does this mean The proliferation stage encompasses a wide range of osteogenic cell types, from the pluripotent MSC through the more committed chondro-osteoprogenitor and preosteoblast. During this proliferative phase, a number of matrix genes begin to be expressed (e.g., type-I collagen, fibronectin and some growth factors such as BMP2/TGFβ). Transcription factors that support proliferation of multipotential MSCs also suppress phenotypic genes that are usually induced at post-proliferative stage of osteoblast differentiation such as helix-loop-helix factors (HLH), Twist, Groucho/TLE, and Id. These factors maintain proliferating progenitor cells in an undifferentiated state (9, 23).
The third stage of osteoblast differentiation corresponds to a period of osteoblasts clustering and multilayering in cultures as well as synthesis and maturation of extracellular matrix. This stage is characterized by the expression of alkaline phosphatase, an early marker of the postproliferative osteoblast phenotype and production of a collagen matrix. The accumulation of type I collagen ECM, initiates marked reduction in histone expression, an exit from cell cycle and the cessation of cell proliferation. Moreover, collagen ECM promotes signaling cascade by both cell-matrix and cell-cell interactions to support the expression of osteoblast-related genes (24–27). During this stage, the maximal levels of collagen synthesis and assembly is coupled with induction of non-collagenous ECM proteins such as osteopontin, osteonectin, bone sialoprotein, and osteocalcin (9).
The final stage of osteoblastogenesis is characterized by deposition of minerals in the ECM, concomitant with peak expression of genes considered markers of mature osteoblast (28– 30). These include, but are not limited to, bone sialoprotein, osteocalcin, and osteopontin. This phase begins with down regulation of matrix maturing proteins, and robust expression of gene products associated with the mineralization that leads to formation and accumulation of hydroxyapatite crystals. During this period, marked morphological changes are evident in vivo, with transition of some cuboidal osteoblast to either fibroblast looking lining cells or the stellate osteocyte (Figure 1). Majority of osteoblasts (~85%) do not achieve the end stage phenotype of an osteocyte and undergo programmed cell death. Apoptotic osteoblast show increased expression of Bad, Bax genes, matrix degrading enzymes and suppression of Bcl2, a pro-cell survival gene (31–33). Osteocytes are spatially and morphologically distinct from osteoblasts, as they are completely encased in bone and possess dendritic processes. The long branched cellular processes allow them to contact each other, as well as osteoblasts and lining cells present on the bone surface and marrow stromal cells on the endosteum. Some of the genes considered marker of mature osteoblasts are also expressed in osteocytes (Osteocalcin, MEPE, DMP1). However, in the recent years, few signature genes of osteocytes have been identified such as E111/gp38, Phex, and Sclerostin (34). It is estimated that there are ten thousands osteocyte per cubic millimeter of bone and about 50 dendritic processes are present on each cell (35). Therefore, osteocytes make up >90–95% of the cellular component of the adult skeleton compared with 4–6% of osteoblast and only 1–2% osteoclast. The long-lived osteocytes function as the mechanosensors of skeletal tissues (34).
In summary, profiles of differentially expressed genes and their responses to physiological cues have provided important conceptual framework for defining distinct stages of osteoblast maturation and cellular understanding of bone development and bone remodeling.
2.1 Stem Cells, Mesenchymal Stem Cells and Induced Pluripotent Stem (iPS) Cells
Unlike pluripotent embryonic stem cells (ES), the stem cells derived from post-natal tissues are mutlipotent. They self-renew through potent proliferative capacity, and can give rise to at least more than one type of highly differentiated lineage (36). Various adult tissues contain reservoirs of specific stem cells that support both maintenance and regeneration of the parental tissue. The ever expanding list of tissues containing specific stem cells include epidermis and intestinal crypts for epithelial stem cells, brain neural tissues for neural stem cells, skeletal muscle with satellite stem cells and number of connective tissues for mesenchymal stem cells (36–39).
Due to convenient isolation, historically, the adult bone marrow has been used as a source of different types of stem cells such as hematopoietic and mesenchymal stem cells (40).The marrow derived mesenchymal stem cells are capable of differentiating into mesenchymal (bone, cartilage, muscle, tendon, adipose and other connective tissues), and non-mesenchymal tissues in vitro and in vivo (36–39). Progeny of mesenchymal stem cells are responsible for post-natal bone formation and adult fracture repair and remodeling.
Commitment and differentiation of these cells to the bone, cartilage, tendon, ligament, and muscle lineage, is a complex process involving multiple discrete cellular transitions. Progression from one stage to the next requires presence of specific bioactive factors, and dynamic environmental cues whose exquisitely controlled contributions orchestrate the entire differentiation program (9, 11). Mesenchymal stem cells have been successfully purified and expanded in invitro culture from both animals and humans. The relative ease of isolation, in vitro mitotic expansion, multi-lineage differentiation and autologous nature are the key features for the emergence of human MSCs as potential source for cell based therapy to treat genetic disorders and for reconstruction and bone tissue engineering. However, the widespread use of MSC for the treatment of many clinically challenging conditions is hampered by their limited supply, significant loss of differentiation capacity in long-term cultures.
Cellular differentiation and lineage commitment are considered to be robust and irreversible processes during development. The diverse cell types present in the adult organism are produced by embryonic stem cells during development by lineage-specific transcription factors. This has fueled attempts to study transcriptional circuitry that control pluripotency and self-renewal of embryonic stem cells. The master regulators and signaling pathways of ES population are beginning to be identified (41). Cocktail of four transcription factors that can induce pluripotency (iPS), promote and maintain the undifferentiated state of mouse and human adult somatic cells have recently been characterized (41). This approach of iPS will allow maintinace of phenotypic plasticity, exvivo expansion and directed differentiation of mesenchymal stem cells or osteoprogenitor population for use in tissue engineering and reconstruction of bone tissues. Human iPS cells, produced either by expression of Oct4, Sox2, c-Myc, and Klf4 or by Oct4, Sox2, Nanog, and Lin28 are remarkably similar to human ES cells. These cells are also morphologically similar to human ES cells, express typical human ES cell-specific cell surface antigens and genes, differentiate into multiple lineages in vitro, and form teratomas containing differentiated derivatives of all three primary germ layers when injected into immunocompromised mice (42).
More importantly, iPS cells can be derived from skin cells from patients with a variety of diseases and without c-Myc oncogene and viral integration (42–44). These discoveries eliminates a major roadblock for developing customized iPS cell tailored to patient specific needs and translating therapeutic usage these of cells. Although these specific factors are sufficient for induced pluripotency, however, it is still unclear how these genes orchestrate the erasure of the differentiated state and cause lineage reprogramming of adult somatic cells.
The use of iPS technology is still quite some distance away but the ability to produce stem cells by induced pluripotency has provided therapeutic options that have never before been available. This has also rekindled hopes for treating generalized conditions such as osteoporosis by systemic administration of culture-expanded autologous iPS/MSCs or to repair local bone defects through site directed delivery of reprogrammed cells.
2.2 Transcriptional regulation of osteoblast differentiation
Aided by combined developmental, molecular, genetic, biochemical and cellular approaches, the transcriptional regulation is the most actively investigated area of bone research. Osteoblast commitment, differentiation and functional activity are all governed by several regulatory proteins and specific transcription factors that promote expression of phenotypic genes and establishment of the osteoblast phenotype. The recent development of molecular genetic studies in human skeletal pathobiology coupled with the genetic mouse models have contributed to better understanding of regulatory machinery involved in the control of osteoblastogenesis (7–10). The role of key signaling molecules, transcription factors and chromatin remodeling enzymes that control the formation and maturation of osteoblasts are summarized below.
Regulatory factors that influence various aspects of osteoblast biology can be divided into four major categories. These include; 1) cis-acting DNA binding transcription factors 2) signal transducers and non-DNA binding coregulatory proteins 3) post-translational regulatory proteins 4) chromatin modifying enzymes. Regulatory factor in the last two categories are functional through all stages of osteoblast differentiation, however their target genes and proteins are restricted to specific stage of osteoblast differentiation.
Two transcription factors that are predominantly expressed in osteoblasts are obligatory for commitment of mesenchymal progenitors to osteoblast lineage and for development of the functional osteoblast. The first protein, Runx2 is a member of the runt domain family of transcription factors and regulates various aspects of osteoblast biology (8,9). Developmentally, Runx2 expression is first noted in osteochondroprogenitor cells of the condensing mesenchyme at the onset of skeletal development. Levels of Runx2 gradually increases in subsequent stages of osteoblast differentiation, with maximum expression observed in the mature osteoblast. Runx2 gene deletion in mice results in complete absence of bone formation and failure of osteoblast and chondrocyte maturation (45). Consistently, in humans Runx2 haploinsufficiency results in cleidocranial dysplasia, a disease characterized by defective bone formation and supernumerary teeth (46). Runx2 is both necessary and sufficient for mesenchymal cell differentiation towards the osteoblast lineage as Runx2 over-expression can convert mesenchymal cells of other lineages to osteogenic phenotype and inhibition of Runx2 blocks the differentiation of mesenchymal cells to osteoblasts (47). Runx2 control bone lineage cells by binding to the Runx regulatory element in promoters of genes considered to be markers for different stages of osteoblast differentiation (9,48). Runx2 target genes include genes expressed by immature and differentiated osteoblasts, such as TGF-β receptor, alkaline phosphatase, collagen type I alpha 1 and alpha 2 chain, osteopontin, osteonectin, Vitamin D receptor, galectin-3, bone sialoprotein, osteocalcin and collagenase (9).
The second DNA binding transcription factor that is absolutely required for osteoblast differentiation is Osterix explain this term.. Osterix/SP7 is a member of the zinc-finger containing SP family and is abundantly expressed throughout osteoblast differentiation (49). Genetic inactivation of Osterix in mice results in absence of mineralized bone matrix, defective osteoblasts and perinatal lethality (49). Similar to Runx2, forced expression of Osterix in non-bone cells, promote expression of both early and late stage marker genes of osteoblast. However, molecular and genetic studies revealed Runx2 is expressed in mesenchymal tissues of Osterix null mice (49). Thus, Osterix acts downstream of Runx2 in the transcriptional cascade of osteoblast differentiation. Consistently, Osterix expression is positively regulated by direct binding of Runx2 to a responsive element in the promoter of the Osterix gene.
Multiple transcription factors that are expressed ubiquitously also plays critical role in bone homeostasis and development of functional osteoblasts. This group consists of proteins that bind DNA in a sequence specific manner, as well as proteins that do not directly interact with the DNA such as C/EBPs, ATF4, NFATc, AP-1, Dlx5, Smads, CBFβ, Satb2 (50–54). They regulate osteoblast differentiation either by directly inducing the expression of target genes and/or by modulating the activities of Runx2 and Osterix transcription factors. For example, ATF4 and CCAAT/enhancer-binding proteins (C/EBPs) induce expression of osteoblast genes through direct binding to their respective cis-element in the promoters (51,52). Both proteins also form physical complex with Runx2 to synergistically regulate the transcription of osteocalcin. Therefore it is not surprising that disruption of ATF4 or C/EBP genes in mice results in delayed skeletal development, decreased bone formation and osteopenia phenotype (52). Similalry, Osterix is recently shown to interact physically and funcationally with NFATc, to induce osteoblast gene expression and NFATc deficiency cause a severe low-bone-mass phenotype due to decreased bone formation (55) . List of transcription factors that functions as co-activator of Runx2 is rapidly growing and includes AP-1, Dlx3, Dlx5, Menin, Smads and TAZ (9).
The examples of non-DNA binding proteins that cooperatively regulate Runx2-dependent osteoblast differentiation include Satb2 and Cbfβ. Satb2 is the nuclear matrix protein with affinity for matrix attachment regions of chromatin loops. Satb2 physically interact with Runx2 and ATF4 to enhance their promoter occupancy and transcriptional function of Runx2 to regulate osteocalcin expression and promote osteoblast differentiation. Satb2 null mice have defective craniofacial bones due to inhibited osteoblast differentiation (54). All members of the Runx family proteins usually interact with CBFβ cofactor through runt domain. CBFβ donot directly interact with DNA but enhances DNA binding affinity of Runx2 by several folds. This leads to increased functional competency of Runx2 (53). Consistent with this notion, CBFβ deletion affects skeletal development and osteoblast maturation (53).
Runx2 mediated commitment of mesenchymal cell to osteoblast lineage is also supported by homeodomain containing transcription factor Bapx1 and Msx2. Both proteins act upstream of Runx2 in a transcriptional cascade that regulates osteoblast differentiation. Accordingly, Msx2 and Bapx1-deficient mice display severe dysplasia of the ossified skeleton and a strong reduction in expression of Runx2 in early mesenchymal progenitor cells (56).
As a master transcriptional regulator, Runx2 is capable of both activating and repressing expression of multiple genes required for the progression of osteoblast differentiation. Runx2 functional activity is actively suppressed during early stages of mesenchymal cell commitment toward osteoblast lineage by Groucho/TLE, Stat1, and Twist. The Groucho/TLE and Twist proteins are expressed early during skeletogenesis and in mesenchymal progenitors. Twist proteins prevent Runx2 protein to bind target gene DNA by physically associating with DNA binding domain of Runx2 protein (58). TLE protein on the other hand inhibits Runx2 transcriptional activity by interacting with C-terminal transcriptional activation domain (57). Dramatic decreases in expression of the Twist and TLE genes result in relieving Runx2 inhibition and progression of osteoblast differentiation. Stat1transcription factor inhibit Runx2 function during osteoblast differentiation by preventing nuclear import of Runx2. Stat1 interaction with Runx2 results in cytoplasmic sequestration of Runx2 (59). Enhanced bone formation seen in Stat1 deficient mice is linked to robust nuclear translocation of Runx2 and increased osteoblast differentiation.
Post-translational modification of Runx2 that results in ubiquitination mediated degradation is yet another mechanism by which osteoblast differentiation is regulated. Runx2 interacts with enzymes responsible for protein stability such as Shn3, WWP1, Smurf1 and CHIP (60,61). Smurf1 was the first factor identified as an E3 ligase for Runx2 ubiquitination and degradation. Similarly, Schnurri 3, a zinc-finger adaptor protein in osteoblast links Runx2 to the E3 ubiquitin ligase WW domain-containing protein 1. These interaction initiates proteasomal degradation of Runx2 (60). Runx2 protein is stabilized in Shn3-deficient osteoblasts and Shn3-null mice shows severe increase in bone mass (60). CHIP, a cochaperone protein identified recently promotes the ubiquitination and degradation of chaperone-bound proteins. CHIP interacts with Runx2 in vitro and in vivo and regulates Runx2 protein stability (61). Decrease in CHIP expression results in a concomitant increase in Runx2 protein and osteoblast differentiation (61).
Finally, ability of Runx2 to activate or repress transcription of multiple genes required for the progression of osteoblast differentiation is regulated through chromatin modification of target gene promoter (9). Transcriptional activity is dependent upon the state of chromatin condensation and histone modifications that decondens chromatin facilitate recruitment of transcription factors and expression. Runx2 directly interacts and recruits p300 acetyltransferase and histone deacetylase to regulate osteoblast genes (9,62).
Thus Runx2, Osterix and associated regulatory protein complexes are important for directing mesenchymal precursor cells toward the osteoblast lineage. This is a very good clinical point!!!!
2.3 Regulation of osteoblast differentiation by secreted molecules
Spatio-temporal organization of the osteoprogenitors in developing bones and the multiple stages of osteoblast differentiation are regulated by a coordinated expression of complex series of signaling molecules and associated transcription factors. Skeletal cells produce numerous growth factors and cytokines that signal in both auto- and paracrine fashion to control cell proliferation, differentiation and survival. These secreted factors and signaling pathways either promote or suppress the expression and/or function of transcription factors essential for osteoblast differentiation. The major pathways involved in skeletal morphogenesis and development of the osteoblast phenotype include members of the fibroblast growth factor (FGFs), transforming growth factor beta (TGFβ), and hedgehog families, Wnt signaling and notch pathway.
The FGF pathway consists of 23 ligands that transduce their signal through one of the four FGF receptors (FGFR). FGFs initiate condensation of the mesenchyme and proliferation of progenitor cells. Temporal expression and activity of FGFR is critical for membranous bone formation and regulate the proliferation, differentiation, and apoptosis of osteoblasts (63). FGFR contribute to normal skeletal development, as mutation or gene deletions are associated with severe dwarfism (64). FGF ligands such as 2, 3, 4, 9 and 18 are associated with normal skeletal development, mutation or gene deletion in these is associated with skeletal deformities and delayed suture closure (64). FGF18 plays a critical role in maturation of osteoblast and FGF-2 increases Runx2 phosphorylation and functional activity (65). Similarly, Activation of FGFR2 signaling results in increased Runx2 expression and enhanced osteoblast differentiation (65). The widely known secreted molecules with potent capacity to induce osteogenesis are the bone morphogenetic proteins (BMP). BMPs are members of the TGFβ superfamily of growth factors which are produced by almost all skeletal cell types and induce mesenchymal condensations for the formation of many organs and skeleton patterning (66). BMPs signal through homomeric or heteromeric type I and type II receptors, which are expressed in all cell types including osoteoblasts. Specific BMP receptors influence lineage direction. Stimulation of mesenchymal progenitor cells by BMP2 dramatically induces expression of both Runx2 and Osterix leading to osteoblast differentiation (9, 50,67). Induction of Runx2 and Osterix by BMP2 and subsequent upregulation of osteoblast specific genes involves Dlx5, Smad transducers and MAPK pathway. Though, BMPs when applied locally, induce de novo bone formation by recapitulating osteoblast differentation but not all fall under this category. For example, BMP3 strongly inhibits osteoblast differentiation and proliferation (68). TGFβ plays a complex role during bone remodeling, with inhibition of Runx2 and osteoblast differentiation in vitro but promotes bone formation in vivo (9, 66,69).
Wnt proteins have recently emerged as central regulators of bone synthesis and bone mass. The canonical pathway that works through intra-cellular transducer β-catenin, control differentiation of osteoblast progenitor cells into mature osteoblasts. β-catenin is expressed in mesenchymal precursor cells and its inactivation favors their differentiation into chondrocytes instead of osteoblast (70). In human receptor mutations that renders a constitutively active Wnt signal, results in a generalized increase in bone mass throughout the skeleton (71). Wnt signaling controls osteoblast differentiation by modulating several transcription factors including Runx2. Wnt signaling promotes Runx2 expression and activity. LEF/TCF transcription factors, the end point of Wnt signal in the nucleus promotes Runx2 and Osterix expression and interacts with Runx2 to regulate its function during osteoblast differentiation (72).
Indian hedgehog (Ihh), a secreted molecule of the hedgehog family is widely known for its role in chondrocytes during endochondral bone formation (73). Ihh and its receptor are also expressed in osteoblast. Mice where Ihh gene is deleted, lacks osteoblast progenitor cells (73). Ihh is also needed for osteoblast proliferation and survival. Ihh controls osteoblast differentiation, first by inducing expression of Runx2 in mesenchymal cells. Secondly, Ihh enhance osteogenic action of Runx2 through an interaction between signal transdcucer Gli2 and Runx2 in osteoblast (74).
Notch receptors are the latest addition to signaling molecules vital for osteoblast differentiation and bone remodeling. Notch signling requires cell-cell interaction between the four Notch receptors and their ligands present on the cell surface. In human, mutations in the Notch signaling cause skeletal patterning defects. Notch deficient mice develop sever osteoporosis (75). Notch inhibits osteoblast differentiation by physically associating with Runx2 and interfering with functional activity of Runx2. Through its expression in osteoblasts, Notch exerts dimorphic effect during bone remodeling. Notch inhibits osteoclast differentiation by controlling production of “decoy" receptor osteoprotegerin by osteoblast (75).
Although, a number of hormones regulates osteoblast differentiation and influence bone formation and bone remodeling, a brief review of two hormones that are responsible for regulating calcium metabolism. Parathyroid hormone (PTH) produced by the chief cells of parathyroid gland, plays a central role in calcium homeostasis through its action on bone, kidney and through enhanced synthesis of another hypercalcemic hormone, 1,25(OH)2 vitamin D3 (76). The capacity of exogenous PTH to function as an anabolic agent is dependent on direct and indirect stimulation of cells of the osteoblastic lineage. PTH promotes bone formation in part through phosphorylation and activation of Runx2, resulting in expression of osteoblast genes (77). PTH also inhibits proteasome mediated degradation of Runx2 and increases expression of osterix to enhance osteoprogenitor lineage determination (78). Another regulatory action of PTH is to promote β-Catenin pathway which controls osteoblast survival (79). Thus, PTH regulates bone development, bone maturation, and bone maintenance through the regulation of Runx2 function.
The other major hypercalcemic hormone is 1,25(OH)2 vitamin D3, a steroid hormone that favors intestinal absorption of calcium (80). Deletion or inactivation of the vitamin D receptor (VDR) in mice and in humans leads to rickets, a phenotype completely reversible in both organisms by treatment with calcium. Vitamin D3 positively regulates the expression of osteoblastic phenotype markers.
3. Remodeling of Alveolar Bone
During replacement of the primary dentition with permanent teeth, the alveolar bone undergoes a complete remodeling. The alveolar bone associated with the primary tooth is completely resorbed together with the roots of the tooth while new alveolar bone is formed to support the newly erupted tooth (81). Significant remodeling of the alveolar process also occurs as part of this process. The ability of the alveolar bone to remodel rapidly also facilitates positional adaptation of teeth in response to functional forces and in the physiological drift of teeth that occurs with the development of jaw bones (81).
Bone remodeling involves the co-ordination of activities of cells from two distinct lineages, the bone synthesizing osteoblasts and the bone resorbing osteoclasts. Each step in the remodeling process is regulated by specific hormones, local factors and mechanical forces. Osteoclasts, the exclusive bone resorptive cells, are derived from the bone marrow macrophage lineage but osteoblast are required for their differentiation. Macrophage colony stimulating factor (M-CSF) and the RANKL, the two cytokines produced by osteoblasts are essential and sufficient for osteoclastogenesis (82). These soluble proteins are produced by both mesenchymal progenitors in marrow and osteoblasts. M-CSF contributes to the proliferation, survival and differentiation of osteoclast precursors as well as the survival and cytoskeletal rearrangement required for efficient bone resorption. Formation of macrophage requires an interaction of M-CSF ligand with receptor present on the surface of mononuclear precursor. The M-CSF-receptor interaction also induces the expression of receptor activator of nuclear factor-kB (RANK) on the surface of macrophage (82). The second cytokine produced by osteoblasts is the RANK-ligand (RANKL). Once linked to its receptor, RANK on the surface of macrophage, it induces their fusion and differentiation. Thus the bone microenvironment is essential for two components of osteoclastogenesis. 1) maturation and fusion of the mononuclear precursor to the multinucleated osteoclast and 2) regulation of the activity of the functional osteoclast.
Osteoblast also produces osteoprotegrin (OPG), a high affinity ligand and physiologic inhibitor of RANKL (83). OPG is secreted by cells of mesenchymal origin both basically and in response to other regulatory signals including cytokines and bone targeting steroids. Pro-inflammatory cytokines suppress OPG expression while simultaneously enhancing that of RANKL, with the net effect being a marked increase in osteoclast formation and function. Thus circulating OPG modulates the bone resorptive acitivity of RANKL. Gene deletion of OPG in both human and mice leads to profound osteopetrosis, whereas overexpression of OPG results in sever osteoporosis.
Another factor that influences bone mass is prostaglandins. Excess prostaglandins stimulate bone loss by increasing expression of RANKL and suppressing that of OPG in stromal and osteoblastic cell (84). This increase in OPG/RANKL ratio is sufficient for increased osteoclast activity. Therefore, the balance between RANKL and OPG determines the formation and activity of osteoclasts.
Finally, sex steroids are central to maintenance of bone mass and bone remodeling. This action indirectly relates to the regulation of the production of multiple cytokines by different cell types in the bone marrow microenvironment. Estrogen mediates its effect by suppression of cytokines that are involved in the regulation of osteoclast formation. For example estrogen inhibits production of IL1 and TNFα by monocyte and IL6, GM-CSF and MCSF by osteoblasts (85). Estrogen can also suppress osteoclastogenesis by enhanced production of osteoprotegrin and decreased RANKL production by osteoblast (86). Similalry, testosterone is shown to decrease osteoclast formation and resorption through increased production of OPG by osteoblast (87). Thus osteoprogenitors and osteoblast are essential regulator of the cellular and molecular events involved in bone remodeling.
Summary
Bone is of crucial importance for the human body, providing skeletal support, and serving as a home for the formation of hematopoietic cells and as a reservoir for calcium and phosphate. Over the years we have come to understand how large number of morphogens, signaling molecules, and transcriptional regulators forms the complex tissue of bone. Key factors that regulate the gene expression program that underlies the induction, proliferation, differentiation, and maturation of osteoblasts are presented here. Secreted growth factors and hormones determine the competency of osteoblast for establishing and maintaining the structural and functional properties of bone. It has become increasingly clear that these diverse transcription factors cannot be viewed as discrete, separate signaling pathways; rather they form a highly interconnected, cooperative network to allow progression of osteoblast differentiation. Bone is also continuously remodeled by hematopoietic lineage osteoclasts. Osteoblasts produced factors are essential regulators of osteoclasts differentation and their bone resorbing function. A better understanding of molecular mechanisms behind osteogenic differentiation and bone remodeling would not only help us to identify pathogenic causes of bone and skeletal diseases but also lead to the development of targeted therapies for these diseases.
Acknowledgments
This work was supported by Grant No. RO1 AG030228 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shubin NH, Alberch PA. Morphogenic approach to the origin and basic organization of the tetrapod limb. Evol. Biol. 1986;20:319–387. [Google Scholar]
- 2.Osumi-Yamashita N, Ninomiya Y, Eto K, et al. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev. Biol. 1994;164(2):409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- 3.Sodek J, McKee MD. Molecular and cellular biology of alveolar bone. Periodontology. 2000;2000(24):99–126. doi: 10.1034/j.1600-0757.2000.2240106.x. [DOI] [PubMed] [Google Scholar]
- 4.Zernik JH, Nowroozi N, Liu YH, et al. Development, maturation, and aging of the alveolar bone. New insights. Dent Clin North Am. 1997;41(1):1–15. [PubMed] [Google Scholar]
- 5.Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 6.Thesleff I, Aberg T. Molecular regulation of tooth development. Bone. 1999;25(1):123–125. doi: 10.1016/s8756-3282(99)00119-2. [DOI] [PubMed] [Google Scholar]
- 7.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289(5484):1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 8.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 9.Lian JB, Javed A, Zaidi SK, et al. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14(1–2):1–41. [PubMed] [Google Scholar]
- 10.Stein GS, Lian JB. Molecular mechanisms mediating proliferation/differentiation interrelationships during progressive development of the osteoblast phenotype. Endocr Rev. 1993;14(4):424–442. doi: 10.1210/edrv-14-4-424. [DOI] [PubMed] [Google Scholar]
- 11.Aubin JE. Regulation of osteoblast formation and function. Rev Endocr Metab Disord. 2001;2(1):81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- 12.de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13(6):721–727. doi: 10.1016/s0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 13.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 14.Gronthos S, Zannettino AC, Graves SE, et al. Differential cell surface expression of the STRO-1 and alkaline phosphatase antigens on discrete developmental stages in primary cultures of human bone cells. J Bone Miner Res. 1999;14(1):47–56. doi: 10.1359/jbmr.1999.14.1.47. [DOI] [PubMed] [Google Scholar]
- 15.Canalis E, Delany AM. Mechanisms of glucocorticoid action in bone. Ann N Y Acad Sci. 2002;966:73–81. doi: 10.1111/j.1749-6632.2002.tb04204.x. [DOI] [PubMed] [Google Scholar]
- 16.Nijweide PJ, Burger EH, Feyen JH. Cells of bone:proliferation, differentiation, and hormonal regulation. Physiol Rev. 1986;66(4):855–886. doi: 10.1152/physrev.1986.66.4.855. [DOI] [PubMed] [Google Scholar]
- 17.Suda T, Udagawa N, Nakamura I, et al. Modulation of osteoclast differentiation by local factors. Bone. 1995;17(2 Suppl):87S–91S. doi: 10.1016/8756-3282(95)00185-g. [DOI] [PubMed] [Google Scholar]
- 18.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 19.Jilka RL, O'Brien CA, Ali AA, et al. Intermittent PTH stimulates periosteal bone formation by actions on post-mitotic preosteoblasts. Bone. 2009;44(2):275–286. doi: 10.1016/j.bone.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parfitt AM. Parathyroid hormone and periosteal bone expansion. J Bone Miner Res. 2002;17(10):1741–1743. doi: 10.1359/jbmr.2002.17.10.1741. [DOI] [PubMed] [Google Scholar]
- 21.Kato S, Suzawa M, Takada I, et al. The function of nuclear receptors in bone tissues. Bone Miner Metab. 2003;21(6):323–336. doi: 10.1007/s00774-003-0453-3. [DOI] [PubMed] [Google Scholar]
- 22.Canalis E. Update in new anabolic therapies for osteoporosis. J Clin Endocrinol Metab. 2010;95(4):1496–1504. doi: 10.1210/jc.2009-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. doi: 10.1002/biof.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damsky CH. Extracellular matrix-integrin interactions in osteoblast function and tissue remodeling. Bone. 1999;25(1):95–96. doi: 10.1016/s8756-3282(99)00106-4. [DOI] [PubMed] [Google Scholar]
- 25.Franceschi RT, Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J Cell Biochem. 2003;88(3):446–454. doi: 10.1002/jcb.10369. [DOI] [PubMed] [Google Scholar]
- 26.Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115(Pt 19):3729–3738. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- 27.Cheresh DA, Stupack DG. Integrin-mediated death: an explanation of the integrin-knockout phenotype? Nat Med. 2002;8(3):193–194. doi: 10.1038/nm0302-193. [DOI] [PubMed] [Google Scholar]
- 28.Boskey AL, Wright TM, Blank RD. Collagen and bone strength. J Bone Miner Res. 1999;14(3):330–335. doi: 10.1359/jbmr.1999.14.3.330. [DOI] [PubMed] [Google Scholar]
- 29.Glimcher MJ. Mechanism of calcification: role of collagen fibrils and collagen-phosphoprotein complexes in vitro and in vivo. Anat Rec. 1989;224(2):139–153. doi: 10.1002/ar.1092240205. [DOI] [PubMed] [Google Scholar]
- 30.Gericke A, Qin C, Sun Y, et al. Different forms of DMP1 play distinct roles in mineralization. J Dent Res. 2010;89(4):355–359. doi: 10.1177/0022034510363250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robey PG. Bone proteoglycans and glycoproteins. In: Bilezikian JP, Raisz LA, Rodan GA, editors. Principal of Bone Biology. San Diego, CA, USA: Academic Press; 2002. pp. 225–238. [Google Scholar]
- 31.Xing L, Boyce BF. Regulation of apoptosis in osteoclasts and osteoblastic cells. Biochem Biophys Res Commun. 2005;328(3):709–720. doi: 10.1016/j.bbrc.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 32.Jilka RL, Weinstein RS, Bellido T, et al. Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J Bone Miner Res. 1998;13(5):793–802. doi: 10.1359/jbmr.1998.13.5.793. [DOI] [PubMed] [Google Scholar]
- 33.Lynch MP, Capparelli C, Stein JL, et al. Apoptosis during bone-like tissue development in vitro. J Cell Biochem. 1998;68(1):31–49. [PubMed] [Google Scholar]
- 34.Dallas SL, Bonewald LF. Dynamics of the transition from osteoblast to osteocyte. Ann N Y Acad Sci. 2010;1192(1):437–443. doi: 10.1111/j.1749-6632.2009.05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. 2008;42(4):606–615. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287(5457):1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 37.Slack JM. Stem cells in epithelial tissues. Science. 2000;287:1431–1433. doi: 10.1126/science.287.5457.1431. [DOI] [PubMed] [Google Scholar]
- 38.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 39.da Silva Meirelles L, Nardi NB. Murine marrow-derived mesenchymal stem cell: isolation, in vitro expansion, and characterization. Br. J. Haematol. 2003;123:702–711. doi: 10.1046/j.1365-2141.2003.04669.x. [DOI] [PubMed] [Google Scholar]
- 40.Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- 41.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 42.Belmonte JC, Ellis J, Hochedlinger K, et al. Induced pluripotent stem cells and reprogramming: seeing the science through the hype. Nat Rev Genet. 2009;10(12):878–883. doi: 10.1038/nrg2700. [DOI] [PubMed] [Google Scholar]
- 43.Ebert AD, Yu J, Rose FF, Jr, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee G, Papapetrou EP, Kim H, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 46.Mundlos S, Otto F, Mundlos C, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 47.Ducy P, Zhang R, Geoffroy V, et al. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 48.Javed A, Gutierrez S, Montecino M, et al. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19(11):7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 50.Javed A, Bae JS, Afzal F, et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283(13):8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez S, Javed A, Tennant DK, et al. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277(2):1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Matsuda K, Bialek P, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 53.Kundu M, Javed A, Jeon JP, et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32(4):639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 54.Dobreva G, Chahrour M, Dautzenberg M, et al. SATB2 is amultifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Koga T, Matsui Y, Asagiri M, et al. NFAT and Osterix cooperatively regulate bone formation. Nat. Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- 56.Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 57.Javed A, Guo B, Hiebert S, et al. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/ PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113(Pt 12):2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 58.Bialek P, Kern B, Yang X, et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 59.Kim S, Koga T, Isobe M, et al. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes. Dev. 2003;17:1979–1791. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DC, Wein MN, Oukka M, et al. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 61.Li X, Huang M, Zheng H, et al. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol. 2008;181(6):959–972. doi: 10.1083/jcb.200711044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 63.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16(12):1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 64.Liu Z, Xu J, Colvin JS, et al. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HJ, Kim JH, Bae SC, et al. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J Biol Chem. 2003;278(1):319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- 66.Wu X, Shi W, Cao X. Multiplicity of BMP signaling in skeletal development. Ann. N. Y. Acad. Sci. 2007;1116:29–49. doi: 10.1196/annals.1402.053. [DOI] [PubMed] [Google Scholar]
- 67.Celil AB, Campbell PG. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem. 2005;280(36):31353–31359. doi: 10.1074/jbc.M503845200. [DOI] [PubMed] [Google Scholar]
- 68.Daluiski A, Engstrand T, Bahamonde ME, et al. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat. Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 69.Kang JS, Alliston T, Delston R, et al. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24(14):2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Day TF, Guo X, Garrett-Beal L, et al. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 71.Boyden LM, Mao J, Belsky J, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 72.Bennett CN, Longo KA, Wright WS, et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102(9):3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maeda Y, Nakamura E, Nguyen MT, et al. Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc. Natl. Acad. Sci. USA. 2007;104:6382–6387. doi: 10.1073/pnas.0608449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimoyama A, Wada M, Ikeda F, et al. Ihh/Gli2 Signaling Promotes Osteoblast Differentiation by Regulating Runx2 Expression and Function. Mol Biol Cell. 2007;18(7):2411–2418. doi: 10.1091/mbc.E06-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hilton MJ, Tu X, Wu X, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chorev M, Rosenblatt M. Parathyroid hormone:Structure-function relations and analog design. In: Bilezikian JP, Raisz LG, Rodan GA, editors. Principles of bone biology. san Diego, CA: Academic Press; 1996. pp. 305–323. [Google Scholar]
- 77.Krishnan V, Moore TL, Ma YL, et al. Parathyroid hormone bone anabolic action requires Cbfa1/Runx2-dependent signaling. Mol. Endocrinol. 2003;17(3):423–435. doi: 10.1210/me.2002-0225. [DOI] [PubMed] [Google Scholar]
- 78.Bellido T, Ali AA, Plotkin LI, et al. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts. A putative explanation for why intermittent administration is needed for bone anabolism. J. Biol. Chem. 2003;278(50):50259–50272. doi: 10.1074/jbc.M307444200. [DOI] [PubMed] [Google Scholar]
- 79.Tobimatsu T, Kaji H, Sowa H, et al. Parathyroid hormone increases beta-Catenin levels through Smad3 in mouse osteoblastic cells. Endocrinology. 2006;147(5):2583–2590. doi: 10.1210/en.2005-1627. [DOI] [PubMed] [Google Scholar]
- 80.Dusso AS, Brown AJ. Mechanism of vitamin D action and its regulation. Am. J. Kidney Dis. 1998;32:S13–S24. doi: 10.1053/ajkd.1998.v32.pm9808140. [DOI] [PubMed] [Google Scholar]
- 81.Sodek J. A comparison of the rates of synthesis and turnover of collagen and non-collagen proteins in adult rat periodontal tissues and skin using a microassay. Arch Oral Biol. 1977;22:655–665. doi: 10.1016/0003-9969(77)90095-4. [DOI] [PubMed] [Google Scholar]
- 82.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 83.Simonet WS, Lacey DL, Dunstan CR, et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 84.Kobayashi T, Narumiya S. Function of prostanoid receptors: studies on knockout mice. Prostaglandins Other Lipid Mediat. 2002;68–69:557–573. doi: 10.1016/s0090-6980(02)00055-2. [DOI] [PubMed] [Google Scholar]
- 85.Riggs BL. The mechanisms of estrogen regulation of bone resorption. J Clin Invest. 2000;106(10):1203–1204. doi: 10.1172/JCI11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michael H, Härkönen PL, Väänänen HK, et al. Estrogen and testosterone use different cellular pathways to inhibit osteoclastogenesis and bone resorption. J Bone Miner Res. 2005;20(12):2224–2232. doi: 10.1359/JBMR.050803. [DOI] [PubMed] [Google Scholar]
- 87.Chen Q, Kaji H, Kanatani M, et al. Testosterone increases osteoprotegerin mRNA expression in mouse osteoblast cells. Horm Metab Res. 2004;36(10):674–678. doi: 10.1055/s-2004-826013. [DOI] [PubMed] [Google Scholar]