Summary
Chlamydiae are gram-negative obligate intracellular bacteria that cause diseases with significant medical and economic impact. Chlamydia trachomatis replicates within a vacuole termed an inclusion, which is extensively modified by the insertion of a number of bacterial effector proteins known as inclusion membrane proteins (Incs). Once modified, the inclusion is trafficked in a dynein-dependent manner to the microtubule organizing center (MTOC), where it associates with host centrosomes. Here we describe a novel structure on the inclusion membrane comprised of both host and bacterial proteins. Members of the Src family of kinases are recruited to the chlamydial inclusion in an active form. These kinases display a distinct, localized punctate microdomain-like staining pattern on the inclusion membrane that colocalizes with four chlamydial inclusion membrane proteins (Incs) and is enriched in cholesterol. Biochemical studies show that at least two of these Incs stably interact with one another. Furthermore, host centrosomes associate with these microdomain proteins in C. trachomatis-infected cells and in uninfected cells exogenously expressing one of the chlamydial effectors. Together, the data suggest that a specific structure on the C. trachomatis inclusion membrane may be responsible for the known interactions of chlamydiae with the microtubule network and resultant effects on centrosome stability.
Keywords: Chlamydia, kinase, centrosome, microtubule, dynein
Introduction
Chlamydia trachomatis is a Gram-negative obligate intracellular bacterium responsible for a number of significant human diseases. C. trachomatis is comprised of over fifteen unique serovars, among which are the etiological agents of trachoma, the leading cause of infectious blindness worldwide, while other serovars are the most common cause of sexually transmitted diseases (Schachter, 1999).
C. trachomatis is characterized by a biphasic lifecycle, alternating between infectious elementary bodies (EBs) and replicative reticulate bodies (RBs). Following endocytosis by a host cell, C. trachomatis resides within a parasitophorous vacuole called an inclusion, which is modified by bacterial type III secreted effector proteins, termed Incs. The chlamydial inclusion is non-fusogenic with the endosomal/lysosomal pathway but acquires sphingomyelin and cholesterol from the Golgi apparatus via a subset of host cell transport vesicles (Hackstadt et al., 1995; Hackstadt et al., 1996; Heinzen et al., 1996; Carabeo et al., 2003; Scidmore et al., 2003; Beatty, 2006; Moore et al., 2008). Once modified by de novo synthesized chlamydial proteins, the nascent inclusion is trafficked to the microtubule organizing center (MTOC) where it is typically observed in close apposition with the host cell nucleus and centrosomes (Higashi, 1965; Campbell et al., 1989a; Campbell et al., 1989b; McBride and Wilde, 1990; Majeed and Kihlstrom, 1991; Hackstadt et al., 1996; Scidmore et al., 1996). This initial trafficking is dependent upon the host microtubule network and the minus-end microtubule motor, dynein (Clausen et al., 1997; Grieshaber et al., 2003). The association of the chlamydial inclusion with centrosomes is maintained throughout mitosis and can lead to an increase in chromosomal abnormalities including abnormal spindle pole formation and supernumerary centrosomes (Grieshaber et al., 2006; Johnson et al., 2009). These effects may explain the positive correlation between C. trachomatis infection and cervical cancer (Hakama et al., 1993; Koskela et al., 2000; Anttila et al., 2001; Wallin et al., 2002; Smith et al., 2004). Although the interactions of the chlamydial inclusion with microtubules require chlamydial protein synthesis (Scidmore et al., 1996; Grieshaber et al., 2003), the bacterial effectors involved in mediating microtubule interactions are currently unknown.
Incs are characterized by bilobed hydrophobic domains and are presumably secreted by chlamydia’s type III secretion apparatus (Bannantine et al., 2000; Subtil et al., 2001; Fields et al., 2003; Muschiol et al., 2006). Incs have attracted a good deal of attention, however, for most of the 50 Incs predicted to be encoded in the C. trachomatis genome a definitive function has yet to be described (Scidmore-Carlson et al., 1999; Bannantine et al., 2000; Shaw et al., 2000; Fields et al., 2003; Li et al., 2008). IncA is thought to govern homotypic fusion of chlamydial inclusions (Hackstadt et al., 1999; Suchland et al., 2000), presumably via SNARE mimicry (Delevoye et al., 2004; Delevoye et al., 2008; Paumet et al., 2009). IncG and CT229 have been shown to interact with host proteins 14-3-3β and Rab4, respectively (Scidmore and Hackstadt, 2001; Rzomp et al., 2006).
Given their localization within the inclusion membrane and access to the host cell cytoplasm, Incs are a logical choice for putative effectors responsible for mediating microtubule/centrosomal interaction. Here we describe microdomains present in the inclusion membrane that are comprised of host cell kinases and at least four Incs. Furthermore, we present data implicating these microdomains in interactions of the chlamydial inclusion with the microtubule network and centrosomes.
Results
Src Family kinases are recruited to the chlamydial inclusion
The Src family kinases are non-receptor membrane associated tyrosine kinases that are involved in a number of cellular signaling pathways regulating such diverse functions as differentiation, motility, adhesion, apoptosis and membrane trafficking. Src, Fyn and Yes are ubiquitously expressed in mammalian cells and some functional redundancy is present, while the other family members are expressed in more specialized cells (Stein et al., 1994; Roche et al., 1995; Thomas and Brugge, 1997; Sandilands and Frame, 2008). Src family kinases are regulated by the phosphorylation state of multiple tyrosines within each kinase. Phosphorylation at Tyr216 and Tyr419 of human Src appears to be necessary for upregulated kinase activity, with autophosphorylation at 419 thought to be the predominant activation site. Conversely, phosphorylation at Tyr530 results in down-regulated kinase activity (Roskoski, 2005). We examined the recruitment of the ubiquitously expressed Src family kinases (Src, Yes, Fyn) to the mature chlamydial inclusion. Fyn and Src, but not Yes, were detected in association with the C. trachomatis serovar L2 inclusion membrane (Fig. 1 and data not shown). When antibodies that recognize all forms of individual kinases regardless of activation state are used, the staining typically appears as a diffuse pattern around the periphery of the inclusion. However, when an antibody specific for phosphorylated Tyr419, which only recognizes enzymatically active forms of Src family kinases was used, a defined punctate staining pattern was observed that when viewed tangentially appeared as discrete bar-like microdomains on the inclusion membrane (Fig. 1). Immunostaining followed by electron-tomography was used to confirm the microdomain-like localization of active kinase at the C. trachomatis serovar L2 inclusion membrane. A single slice from electron tomographs of active kinase labeled samples (Fig. 1) show electron dense diaminobenzidine (DAB) reaction product localized in microdomains at the inclusion membrane. Negative controls lacking active kinase antibody show no such labeling at the inclusion membrane. Typically, one to three microdomains were observed on each inclusion.
Figure 1. Host cell kinases colocalize with the inclusion membrane.
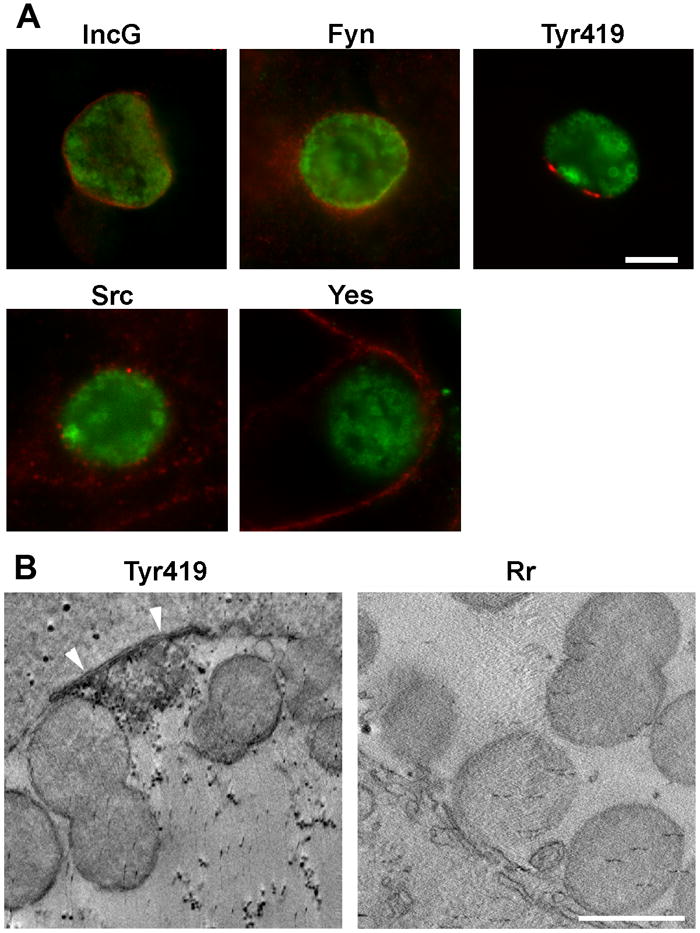
(A) Hela cells were infected with C. trachomatis serovar L2. At 24 hrs post infection cells were fixed, permeabilized and labeled with anti-EB (green) and either anti-IncG, anti-Fyn, anti-Src, anti-Yes, or anti-active kinase (Tyr419) (red). Images were acquired on a Nikon FXA epifluorescent microscope. IncG demonstrates typical inclusion membrane staining. Fyn appears as a diffuse staining pattern around the inclusion, Active kinase colocalizes with the inclusion membrane in a discrete punctate pattern. Scale bar = 10 μm. (B) Electron tomographs show active kinase at the inclusion membrane in samples labeled with anti-active kinase (Tyr419) (white arrowheads), but not with the negative control anti-R. rickettsii (Rr) antibody. Reticulate bodies can be seen attached to the inclusion membrane. Scale bar = 1 μm.
To further confirm this microdomain-like localization at the inclusion membrane an antibody targeting the secondary phospho-activation site (Tyr216) was used to label C. trachomatis L2 infected cells and showed an identical microdomain-like staining pattern as anti-Tyr419 (data not shown). This morphology suggests that the active Src family kinases are localized to specific microdomains within the inclusion membrane.
Because the Tyr419 activation site is conserved among Src, Yes and Fyn, the anti-active kinase antibody could potentially recognize the active form of any of these Src family kinases. Therefore, we used siRNA depletion of the three ubiquitously expressed kinases to unambiguously determine which were recruited to the chlamydial inclusion microdomains (Fig. 2). When the kinases were knocked-down individually and compared to a non-targeting siRNA, depletion of Fyn had the most significant effect on active kinase recruitment to the inclusion membrane, although faint staining of some inclusions was still observed. Src depletion had a lesser effect and Yes had no observable effect. Additionally, simultaneous depletion of both Src and Fyn eliminated active kinase staining at the chlamydial inclusion indicating that either kinase may be recruited and thus potentially provide functional redundancy. No significant effect on inclusion development was observed in the double kinase knockdown (data not shown).
Figure 2. Fyn and Src are the primary Src family kinases recruited to the inclusion.
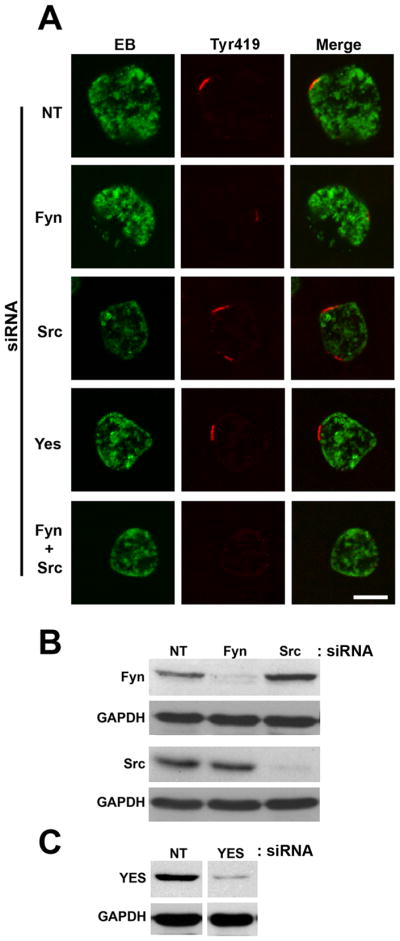
(A) HeLa cells were transfected with non-targeting (NT), Fyn, Src, Yes, or Fyn + Src siRNA. 48 hrs post transfection cells were infected with C. trachomatis L2. 24 hrs post-infection cells were fixed and labeled with anti-EB (green) and anti-active kinase (Tyr419) (red) antibodies. Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Discrete active kinase labeled microdomains are observed in the non-targeting siRNA cells. Fyn depletion significantly decreases the active kinase staining at the inclusion membrane. Src knockdown decreases the active kinase staining only slightly, while Yes depletion has no visible effect. Depletion of both Fyn and Src results in a lack of active kinase staining at the inclusion membrane. Scale bar = 10 μm. (B). Immunoblots of siRNA knockdowns of Fyn and Src probed with anti-Fyn and anti-Src with anti-GAPDH as a loading control. (C) Immunoblot of siRNA knockdown of Yes probed with anti-Yes and anti-GAPDH as a loading control. Lanes were excised from the same gel and exposure and arranged for presentation.
Active kinase recruitment is not conserved in all species of chlamydiae
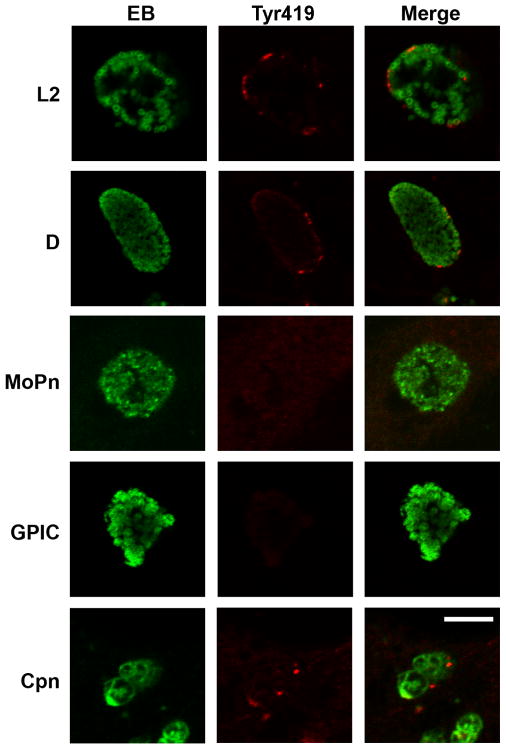
We examined the inclusions of other chlamydial species and serovars to investigate whether active kinase recruitment represents a conserved process. Active kinase colocalized with the C. trachomatis serovar D inclusion membranes in a similar staining pattern as that observed for serovar L2 (Fig. 3), suggesting that active kinase recruitment may be conserved in the C. trachomatis LGV and oculo-urogenital biovars. HeLa monolayers were also infected with C. muridarum, C. caviae, or C. pneumoniae and stained with active kinase antibody. Punctate staining was observed in association with the C. pneumoniae inclusion membrane, but not with C. muridarum or C. caviae inclusion membranes. Therefore, Src family kinase recruitment is not conserved in all species of chlamydiae.
Figure 3. Active kinase recruitment is not conserved in all species of chlamydiae.
Hela cells were infected with C. trachomatis serovars L2 or D, C. muridarum Mouse pneumonitis (MoPn), C. caviae (GPIC), or C. pneumoniae AR-39 (Cpn). At various times post infection (C. trachomatis L2, C. caviae = 24 hrs; C. trachomatis serovar D, C. muridarum = 36hrs; C. pneumoniae = 72hrs) cells were fixed, permeabilized and labeled with an species-appropriate anti-EB (green) and anti-active kinase (Tyr419) (red) antibodies. Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Active kinase is recruited to the C. trachomatis and C. pneumoniae inclusions, but not to C. muridarum or C. caviae. Scale bar = 10 μm.
Specific Incs colocalize with active kinase
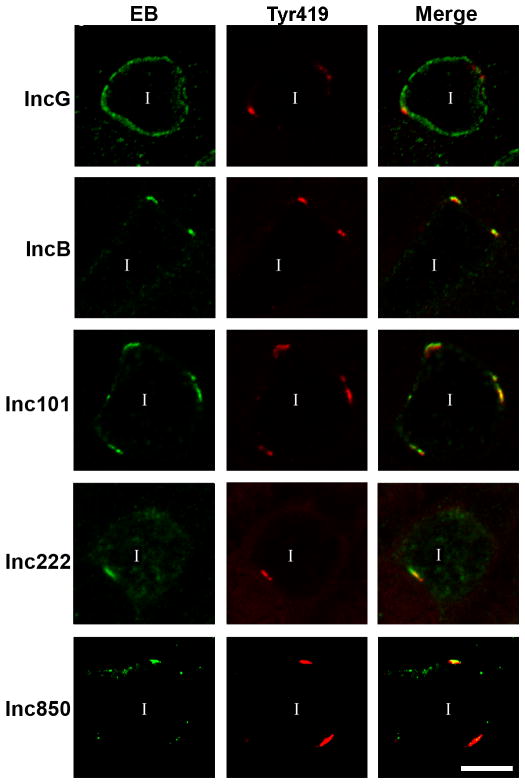
Incs are expressed at the interface of the inclusion with the host cell cytosol (Rockey et al., 1995; Bannantine et al., 1998; Hackstadt et al., 1999; Bannantine et al., 2000) and are thus positioned to specifically regulate the interactions of the inclusion with the host cell. We therefore explored the possibility of Inc proteins being involved in the structure or function of the observed inclusion membrane microdomains. Antibodies against a panel Incs (Shaw et al., 2000) were screened for colocalization with active kinase in C. trachomatis L2 infected cells (Fig. 4). As previously described (Scidmore-Carlson et al., 1999), IncG displayed a uniform staining pattern around the periphery of the inclusion membrane. There is some overlap with active kinase but no apparent enrichment in association with the active kinase. However, four Incs (IncB, Inc101, Inc222 and Inc850) showed a discrete punctate microdomain-like staining pattern that colocalized with the active kinase. These Inc/active kinase microdomains varied in size and number but this variability did not correlate with MOI or time post infection (data not shown). With few exceptions, the Inc protein overlap with active kinase is complete, in that microdomains contain both Incs and active kinase and both classes of protein are present along the entirety of the microdomain.
Figure 4. A subset of Incs colocalizes with active kinase.
HeLa cells were infected with C. trachomatis L2. 24 hrs post infection cells were fixed, permeabilized, and immunolabeled with anti-active kinase (Tyr419) (red) and anti-IncG, anti-IncB, anti-Inc101, anti-Inc222, or anti-Inc850 (green). Specimens were examined by confocal microscopy. IncG has a uniform staining pattern around the periphery of the inclusion membrane, while Incs B, 101, 222 and 850 show discrete microdomain-like staining patterns that colocalizes with active kinase. Inclusions are indicated by “I”. Scale bar = 10 μm.
IncB, 101, 222 and 850 colocalize with each another
Each member of this subset of four Incs colocalized individually with active kinase. The observation that the individual Inc’s colocalization with active kinase was virtually complete suggests that they exist in common microdomains. To confirm this hypothesis, Inc antibodies were directly conjugated to complementary Alexa Fluor dyes and pairs of Incs were observed for colocalization on the inclusion membrane. When IncB was used as the common Inc, Incs 101, 222 and 850 colocalized within IncB microdomains (Fig. 5). Identical results were obtained when Inc222 was used as the common antibody (data not shown).
Figure 5. The four microdomain Incs colocalize with each other.
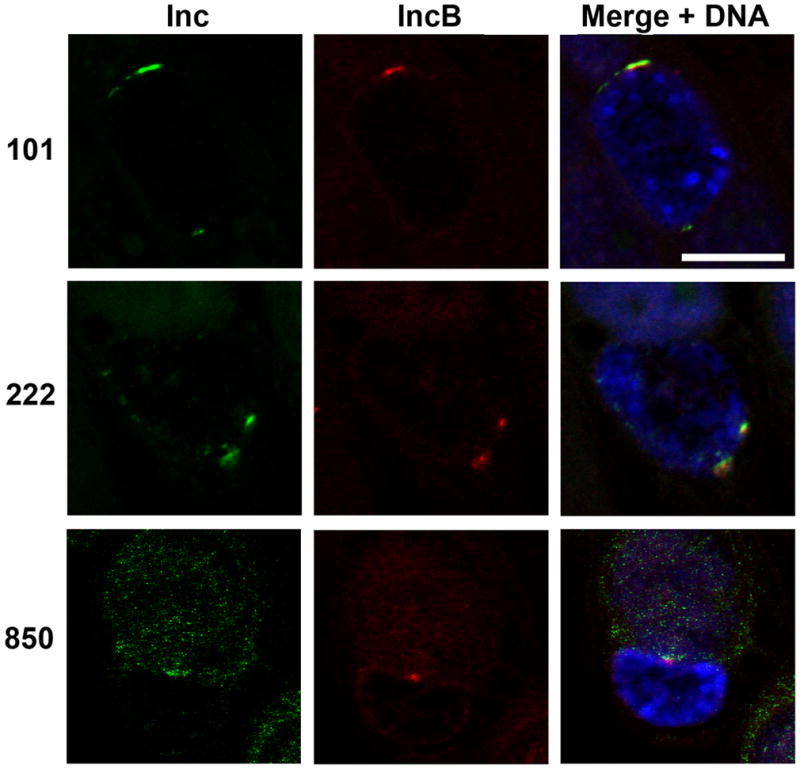
Antibodies against IncB, 101, 222 and 850 were directly conjugated to Alexa Fluor-488 or Alexa Fluor-568. HeLa cells were infected with C. trachomatis L2. 24 hrs post infection cells were fixed, permeabilized and immunolabeled with anti-IncB-Alexa568 (red) paired with either anti-Inc101, 222 or 850-AlexaFluor 488 (green). Monolayers were counterstained with Draq5 to label DNA and visualize inclusions. Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Incs 101, 222 and 850 colocalize with IncB at the inclusion membrane. Scale bar = 10 μm.
Stable interaction of two of the microdomain Incs
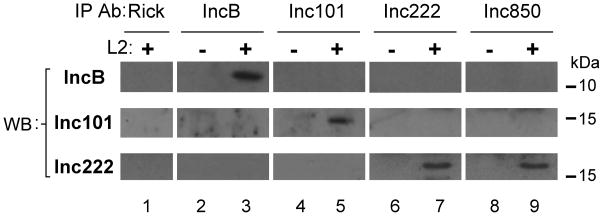
We investigated the possibility of any direct interaction of the four microdomain Incs with each other. Co-immunoprecipitation (Co-IP) experiments were performed using an antibody to each individual Inc followed by immunoblotting with antibodies to all Incs of this sub-class (Fig. 6). An anti-rickettsia antibody was used as a negative control. Zwittergent 3–14 was used to solubilize the infected cells as it has previously been used to successfully preserve the interaction between an Inc and a host protein (Scidmore and Hackstadt, 2001). Western blots probed with Inc222 confirmed that Inc222 was pulled down using an Inc222 antibody (Fig. 6, lane 7). Inc222 was also pulled down when an Inc850 antibody was used for the immunoprecipitation (Fig. 6, lane 9). Inc222 was not precipitated from uninfected lysate or when anti-rickettsia, anti-IncB, or anti-Inc101 antibodies were used for the immunoprecipitations. Conversely, Incs B and 101 were only observed in lanes where the corresponding antibody was used for the immunoprecipitation (Fig. 6, lanes 3 and 5) and was not immunoprecipitated with antibodies to any other Inc proteins or the negative control. These results suggest that Inc222 and Inc850 physically interact with each other individually or as a complex but not with IncB or Inc101.
Figure 6. Two microdomain Incs interact with each other.
T150 flasks of HeLa cells were infected with C. trachomatis L2 or mock infected. 24 hrs post infection cells were harvested and lysate prepared for co-immunoprecipitation. The supernatant from infected lysate (lanes 1, 3, 5, 7, and 9) or uninfected lysate (lanes 2, 4, 6, and 8) was added to Protein-A beads that had been pre-loaded with antibody: anti-Rickettsia negative control antibody (lane 1), anti-IncB (lanes 2 and 3), anti-Inc101 (lanes 4 and 5), anti-Inc222 (lanes 6 and 7) or anti-Inc850 (lanes 8 and 9). Bands corresponding to IncB and Inc101 are only observed in the infected IncB and Inc101 immunprecipitates respectively (lanes 3 and 5). A band corresponding to Inc222 is not observed in the anti-Rickettsia, anti-IncB, anti-Inc101 or mock infected lanes, but is observed in the anti-Inc222 and anti-Inc850 immunoprecipitated samples only (lanes 7 and 9).
Inc microdomains are enriched in cholesterol
The chlamydial inclusion membrane contains cholesterol derived from the host cell (Carabeo et al., 2003). We questioned whether these localized microdomains on the inclusion membrane might have some altered lipid composition related to the recruitment of active Src family kinases and specific chlamydial inclusion membrane proteins. We therefore examined Inc microdomains for the presence of cholesterol. Fluorescent microscopy using filipin as a cholesterol probe showed an enrichment of cholesterol at the microdomains within the inclusion membrane (Fig. 7). These results are consistent with a unique structure and composition of the observed microdomains.
Figure 7. Inc microdomains are enriched in cholesterol.
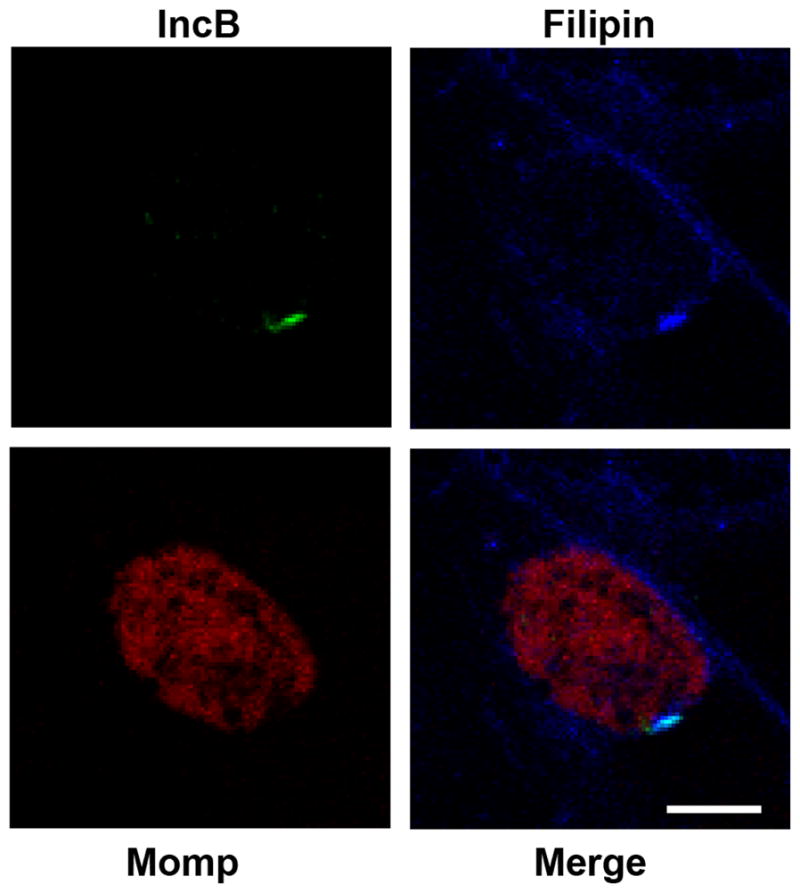
HeLa cells were infected with C. trachomatis L2. 24 hrs post infection cells were fixed, lightly permeabilized and immunolabeled with anti-Momp (red), anti-IncB (green) and filipin (blue). The entire inclusion membrane typically stains with filipin although Inc microdomains appear enriched in cholesterol. Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Scale bar = 10 μm.
Inc microdomains associate with centrosomes
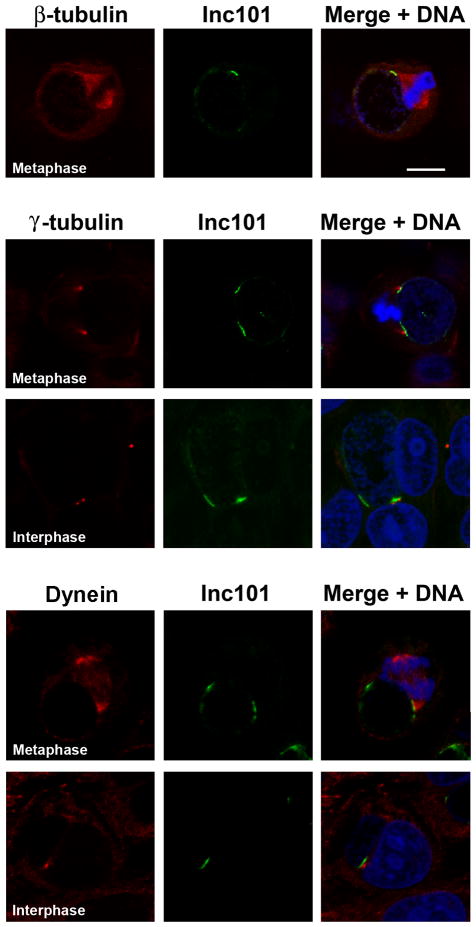
In order to elucidate possible functions of the Inc microdomains, a panel of antibodies against various cellular organelles or cytoskeletal structures was examined by fluorescence microscopy for association with the Inc microdomains. Only one antibody, to β-tubulin, appeared to indicate an interaction with Inc microdomains. It is known that chlamydial inclusions use interactions with the minus-end microtubule motor complex dynein to traffic to the MTOC where they associate with centrosomes (Grieshaber et al., 2006). Fluorescent microscopy revealed that Inc microdomains were consistently associated with the mitotic spindle pole in cells undergoing mitosis. When a section of the inclusion is viewed tangentially (Fig. 8), Inc microdomains appear as well-defined narrow bar-like structures on the inclusion membrane and the mitotic spindle appears to terminate at the Inc microdomain. During mitosis, one or both spindle poles may be observed in association with Inc microdomains.
Figure 8. Inc microdomains colocalize with the host cell centrosomes and the mitotic spindle apparatus.
HeLa cells were infected with C. trachomatis L2. 24 hrs post infection cells were fixed and permeabilized. Cells were labeled with anti-β tubulin (red), anti-γ tubulin (red), or anti-dynein intermediate chain (red) in conjunction with anti-Inc101 (green) and counterstained with Draq5 (blue) for DNA. Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Spindle poles and centrosomes of metaphase and interphase cells colocalize with Inc microdomains at the inclusion membrane. This colocalization can occur with one or both spindle poles/centrosomes. Scale bar = 10 μm.
Centrosomes are located at the convergence of the mitotic spindle apparatus, therefore we labeled infected cells with γ-tubulin, which preferentially stains the centrosome. Centrosomes are observed in close apposition to the inclusion microdomains in both interphase and metaphase cells (Fig. 8), indicating that this association is not limited to mitosis. Similar to what is observed in β-tubulin stained cells, one or both centrosomes can be associated with Inc microdomains.
Dynein, plays a role in multiple cellular functions including organelle positioning and vesicular transport. It is also implicated in mitosis with a role in spindle organization and chromosome movement (Vallee and Sheetz, 1996; Karki and Holzbaur, 1999; Dujardin and Vallee, 2002). Consistent with this role, dynein is observed localized at kinetochores and spindle fibers with a focus at the spindle poles and is enriched at centrosomes during the S and G2 phases of the cell cycle (Quintyne and Schroer, 2002). Because of its role in trafficking nascent chlamydial inclusions to the MTOC and maintaining a tight interaction with centrosomes (Grieshaber et al., 2006), we examined the association of dynein with chlamydial inclusion microdomains (Fig. 8). Again, dynein was localized to mitotic spindles and appeared to focus at the Inc microdomain. During mitosis, one or both spindle poles could be observed in association with Inc microdomains. In non-mitotic cells, dynein was observed less frequently in association with microdomains although this likely reflects cell cycle dependent association of dynein with centrosomes (Quintyne and Schroer, 2002).
Ectopically expressed Inc850 associates with host centrosomes
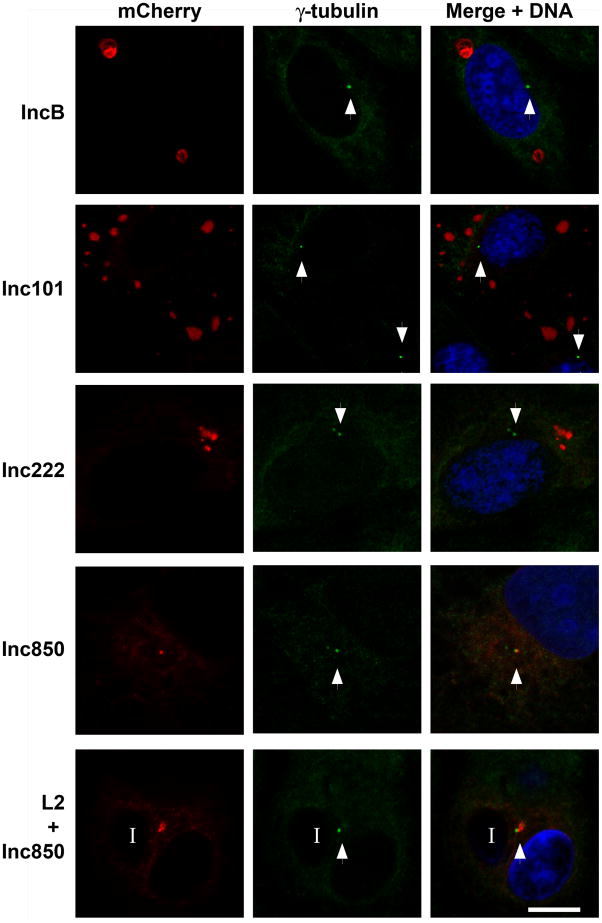
To further investigate the association between Inc microdomains and host centrosomes, N-terminal mCherry fusions of each microdomain Inc were constructed and used to transfect HeLa cells. In cells expressing mCherry-Inc850, the exogenous protein appeared as aggregates throughout the cell cytoplasm, which showed significant colocalization with the one or both host centrosomes (Fig. 9). This colocalization was observed in cells expressing high, medium or low levels of mCherry-Inc850. Low level expressing cells are shown for ease of visualization. These results indicate that Inc850, in the absence of a chlamydial inclusion membrane or other chlamydial proteins, is able to associate with host centrosomes. This colocalization persists when cells are infected with C. trachomatis L2 and then transfected with mCherry-Inc850, resulting in centrosomes that colocalized with both the inclusion membrane and exogenous Inc850. In cells expressing mCherry-IncB, -Inc101 or -Inc222, the exogenous protein could be seen in a variety of intracellular structures of different sizes throughout the cytoplasm. However, no significant association with host centrosomes was observed. These results imply that Inc850 may play a key role in the interactions of the C. trachomatis inclusion with the microtubule network.
Figure 9. Exogenously expressed Inc850 colocalizes with host centrosomes.
HeLa cells were transfected with mCherry-Inc fusions (red). 24hrs post transfection cells were fixed, permeabilized and labeled with anti-γ tubulin (green) and Draq5 for DNA (blue). Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Incs B, 101, and 222 do not colocalize with the host centrosome (white arrow heads), while Inc850 colocalizes with host centrosomes. When C. trachomatis L2 infected cells are transfected with Inc850 (L2 + Inc850), the exogenous Inc850 colocalizes with both the centrosome and the inclusion (“I”). Scale bar = 10 μm.
Quantiation of the association of ectopically expressed mCherry-Inc850 with centrosomes indicated that in approx. 98% of the cells expressing mCherry-Inc850, centrosomes were associated with Inc850 (Fig. 10). This is in comparison to approx. 2% of the cells expressing mCherry-IncB.
Figure 10.
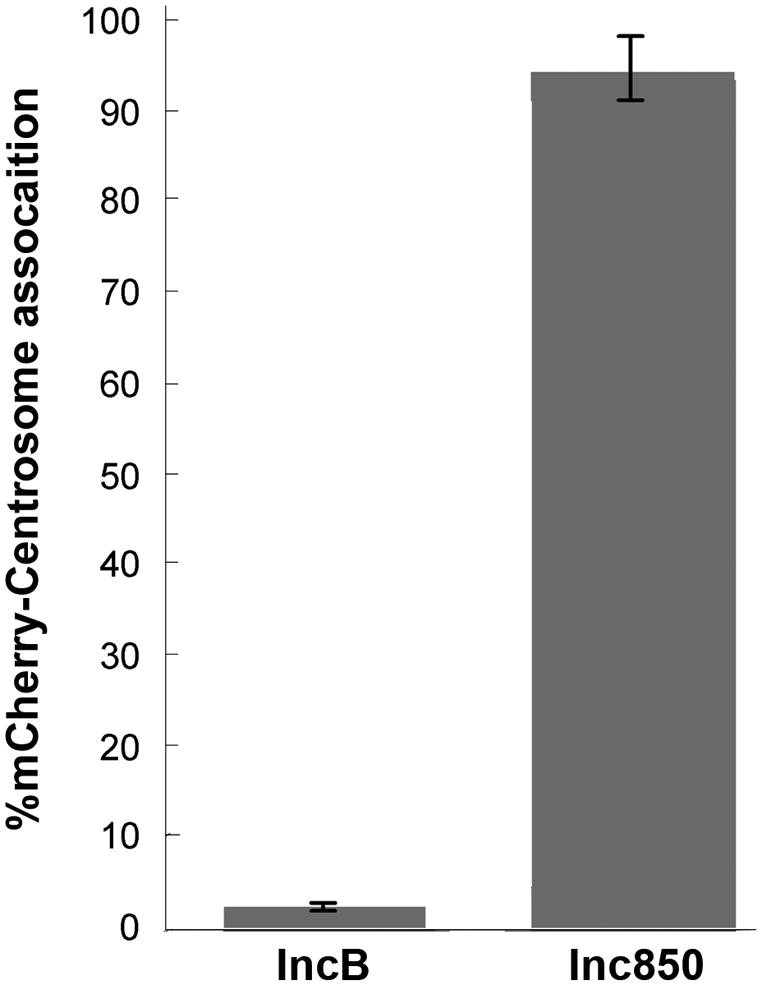
Quantitation of mCherry-IncB and mCherry-Inc850 fusions associating with host centrosomes. The percentage of transfected cells in which mCherry-IncB or mCherry-Inc850 colocalizes with host centrosomes was determined in duplicate for at least 100 cells in each experiment (n=2; error bars = standard deviation).
Discussion
Within the first few hrs following endocytosis, C. trachomatis is unidirectionally transported to a perinuclear location associated with the microtubule organizing center (MTOC) where the inclusion remains for the duration of chlamydial intracellular development (Higashi, 1965; Campbell et al., 1989a; Campbell et al., 1989b; McBride and Wilde, 1990; Majeed and Kihlstrom, 1991; Hackstadt et al., 1996). Translocation of the nascent inclusion to the MTOC follows microtubule tracks and is mediated by the minus-end directed microtubule motor complex, dynein (Clausen et al., 1997; Grieshaber et al., 2003). Microtubules are organized at the MTOC by centrosomes and the chlamydial inclusion maintains a tight association with centrosomes that is also dependent upon dynein (Grieshaber et al., 2006). Surprisingly, p50 dynamitin, an essential component of the dynactin complex that activates dynein and links vesicular cargo to the dynein motor, is not required for the transport of inclusions to the MTOC or the association of inclusions with centrosomes (Grieshaber et al., 2003; Grieshaber et al., 2006). A requirement for chlamydial de novo transcription and translation to initiate trafficking to the MTOC (Scidmore et al., 1996) has led to the suggestion that chlamydial protein(s) modifying the inclusion membrane may supplant the requirement for p50 dynamitin and an intact dynactin complex in tethering the nascent inclusion to the dynein motor (Grieshaber et al., 2003). The chlamydial proteins mediating this interaction are unknown. Here we describe a novel structure on the chlamydial inclusion membrane that is enriched with active Src-family kinases, as well as at least four inclusion membrane proteins, including one that colocalizes with centrosomes when exogenously expressed. Inc microdomains are localized at the point of contact of centrosomes with the inclusion membrane and may represent a complex of chlamydial and host proteins that mediates the interactions with dynein to direct migration along microtubule tracks to the MTOC.
C. trachomatis expresses up to fifty predicted inclusion membrane proteins characterized by a long, bilobed hydrophobic domain of approximately 40 amino acids in length (Scidmore-Carlson et al., 1999; Bannantine et al., 2000; Shaw et al., 2000; Fields et al., 2003; Li et al., 2008). Incs are exposed on the cytosolic face of the inclusion membrane and thus are likely candidates for factors controlling interactions with the host cell. Incs typically are distributed evenly around the periphery of the inclusion membrane although one has been described as localized to the inclusion membrane at the attachment site of RBs (Hackstadt et al., 1999; Scidmore-Carlson et al., 1999). One additional Inc protein has been described as localized to discrete sites on the inclusion membrane (Alzhanov et al., 2009) although this particular Inc was not one we observed in the microdomains described here. Two of the microdomain Incs, IncB and 850, are early gene products synthesized by 2 hrs post-infection. Inc 101 and 222 are considered mid-cycle genes expressed first around 8–12 hrs post-infection (Shaw et al., 2000; Belland et al., 2003). When these microdomains first form is difficult to visualize as early, nascent inclusions are quite small, however, by 12 hrs post-infection, when RBs are beginning to divide, microdomains can be clearly observed by immunofluorescent staining (data not shown). These microdomains are observed through at least 36 hrs post-infection. It is worth noting that centripetal migration of nascent inclusions has been initiated by 2 hrs post-infection thus the critical chlamydial proteins mediating interaction with dynein are present and functional by that time (Hackstadt et al., 1996; Grieshaber et al., 2003).
At least two of the Incs, Inc 222 and 850, in the microdomain stably interacted with each other in co-immunoprecipitation studies but the other two, Inc B and 101, were not similarly associated. It is unclear at this point whether the affinity of Inc 222 and 850 are involved in the formation of microdomains or whether other factors may be involved. The chlamydial inclusion is known to contain cholesterol but filipin staining indicates that the microdomains may be further enriched in cholesterol. At this point it cannot be definitively determined whether the microdomain Incs are recruited to a region of high cholesterol or whether the protein composition of these domains instigates the establishment of an altered lipid environment that is enriched in cholesterol.
These microdomains were initially identified based upon the presence of active Src-family kinases. A role for tyrosine phosphorylation in chlamydia pathogenesis had been proposed previously based upon the immunofluorescent detection of phosphotyrosine on nascent inclusions although the predicted host protein(s) were not identified (Birkelund et al., 1994; Birkelund et al., 1997; Clausen et al., 1997; Fawaz et al., 1997). C. trachomatis endocytic vacuoles are now known to be modified by a type III secreted effector protein, Tarp, that is also phosphorylated by Src family kinases (Elwell et al., 2008; Jewett et al., 2008; Lane et al., 2008; Mehlitz et al., 2008). This tyrosine phosphorylation peaks by 30 – 60 min post-infection and diminishes thereafter (Clifton et al., 2004). The function of Tarp tyrosine phosphorylation remains unclear. A clear distinction between Tarp phosphorylation and the intracellular trafficking of nascent inclusions is that the latter requires de novo chlamydial protein synthesis (Scidmore et al., 1996) whereas unphosphorylated Tarp is preloaded in EBs and is translocated and phosphorylated without a requirement for chlamydial transcription or translation (Clifton et al., 2004). It therefore appears likely that tyrosine phosphorylation of either host or chlamydial proteins by Src family kinases may play a role in multiple stages of the chlamydial developmental cycle.
Dynein appears to play an important role in the trafficking of chlamydiae to the MTOC (Clausen et al., 1997; Grieshaber et al., 2003). The absence of a need for the cargo-linking activity of p50 dynamitin to dynein coupled with a requirement for chlamydial modification of the nascent inclusion, strongly suggests that a chlamydial protein modifying the inclusion membrane is necessary for intracellular trafficking. Components of the microdomains described here appear to be good candidates based upon their colocalization with centrosomes at the MTOC. Attempts to define specific cellular interactions by immunoprecipitation of each of the four microdomain Incs and immunodetection of components of dynein or dynactin have thus far been unrevealing (data not shown). Although four chlamydial inclusion membrane proteins and host Src-family kinases are known to comprise these microdomains, there may be other host or chlamydial proteins present that are required for intracellular interactions. One of the four Incs, Inc850, showed consistent colocalization with centrosomes when ectopically expressed in HeLa cells. Inc 850 is expressed as early as 1 hour post infection (Belland et al., 2003) and thus is present at the time that trafficking to the MTOC is initiated This data suggests that Inc850 should bear increased consideration as the possible linkage of the inclusion to the dynein motor. Fyn has been linked with membrane-associated γ-tubulin complexes that can nucleate microtubule formation (Macurek et al., 2008) and has been proposed to play a role in microtubule dynamics and spindle organization (Levi and Shalig, 2010), thus Fyn is also deserving of further attention in consideration of chlamydial interactions with the microtubule network.
The association of the C. trachomatis inclusion with centrosomes is maintained throughout mitosis and can lead to centrosomal abnormalities including an increase in supernumerary chromosomes and chromosomal segregation defects (Grieshaber et al., 2006; Johnson et al., 2009). The chromosomal instability induced by C. trachomatis infection may be maintained even after curing of the infected cells (Grieshaber et al., 2006) and may be a factor in the epidemiological links between chlamydial infection and certain cancers (Hakama et al., 1993; Koskela et al., 2000; Anttila et al., 2001; Wallin et al., 2002; Smith et al., 2004). Identification of the chlamydial factors involved and the mechanisms that mediate the stable interaction of microdomains with centrosomes may shed light on the association of chlamydial infections with certain cancers. C. trachomatis infection also inhibits cytokinesis (Horoschak and Moulder, 1978; Campbell et al., 1989a; Greene and Zhong, 2003). Indeed, three Incs, CT223, 224, and 225, which share little sequence identity among themselves or with any of the microdomain Incs described here, were inhibitory to host cytokinesis when ectopically expressed in McCoy cells (Alzhanov et al., 2009). However, it has also been shown recently that defective cytokinesis cannot entirely account for the increase in centrosome numbers thus other mechanisms must be involved (Johnson et al., 2009).
The unique domain on the C. trachomatis inclusion membrane described here implies a functional interaction with the host microtubule network and the microtubule motor, dynein. These microdomains thus appear to act as a platform for interactions controlling trafficking and positioning of the chlamydial inclusion. A better understanding of this complex should promote a more complete characterization of host and chlamydial proteins involved in this domain and aid in discerning interactions with the host cell promoting infection or subsequent cellular transformation and malignancy.
Experimental procedures
Organism and Cell Culture
Chlamydia trachomatis serovar L2 (LGV 434), serovar D (UW-3-Cx), Chlamydia muridarum, Chlamydophila caviae, and C. pneumoniae (CWL029) were propagated in HeLa 229 cells and purified by Renografin density gradient centrifugation as previously described (Caldwell et al, 1981).
Immunofluorescent microscopy
HeLa cells were plated on glass coverslips in 24-well plates (Corning, Lowell, MA). C. trachomatis L2 and C. caviae infections were performed in Hanks Balanced Salt Solution (HBSS, Invitrogen/Gibco, Carlsbad, CA). After 1 hour incubation, the media was removed and replaced with pre-warmed RPMI-1640/10% FBS. C. trachomatis serovar D and C. muridarum infections were performed as above, except the HeLa cells were pretreated with 1% DEAE-Dextran in HBSS for 15 minutes prior to infection. For C. pneumoniae infections, EBs were diluted in SPG buffer (219mM sucrose, 10mM Na2HPO4, 3.8mM KH2PO4, 5mM glutamic acid, pH 7.4) and centrifuged at 900 × g for 1 hour. The media was then replaced with pre-warmed RPMI-1640/10% FBS.
For Fyn, Src and Yes labeling, cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS, permeabilized with 0.01% saponin (Sigma, St. Louis, MO) in PBS and blocked with 1% BSA (Sigma) in PBS. Cells were then stained with goat anti-Fyn (Novus, Littleton, CO), mouse anti-Src (Millipore/Upstate, Billerica, MA) or mouse anti-Yes (BD Biosciences, San Jose, CA) in conjunction with a polyclonal rabbit anti-EB serum followed by anti-mouse or anti-goat DyLight 594 and anti-rabbit DyLight 488 secondaries (Jackson ImmunoResearch Labs, West Grove, PA). For active kinase staining, cells were fixed with cold methanol and labeled with mouse anti-phospho Src family Tyr416 clone 9A6 (Millipore). Note that this antibody was produced against avian Src which is phosphorylated on Tyr416 but reacts with human Src family kinases phosphorylated on Src419. Tubulin staining was performed using mouse anti-β tubulin (BD Biosciences) and mouse anti-γ tubulin, clone GTU-88 (Sigma). Dynein intermediate chain was stained with mAb 74.1 (Covance, Emeryville, CA). The chlamydia antibodies used were polyclonal rabbit anti-C. trachomatis L2 EB, anti-C. caviae EB, and anti-C. pneumoniae AR39 EB. The anti-IncG antibody has been previously described (Scidmore-Carlson et al., 1999). Monoclonal antibody L2-I-45 against C. trachomatis L2 MOMP was kindly provided by H.D. Caldwell. Rabbit anti-peptide antibodies against IncB (CT232, LARPQVFTLSTQFSPTKPQ, Inc101 (CT101, SKSMLKQHELDAQL), Inc222 (CT222, VRTNYEEVRSSSTGDQV), Inc850 (CT850, TVKDSFLKKARRERFLA) were prepared commercially by Quality Controlled Biochemicals, Inc (Hopkinton, MA). The DNA of host cells and chlamydia was counterstained using Draq5 (Biostatus Limited). Cholesterol was visualized using 50ug/ml filipin (Sigma).
Images were acquired on a Nikon Microphot-FXA microscope using a 60× 1.4 NA oil immersion objective (Nikon, USA) or a Zeiss LSM 510 Meta laser confocal scanning microscope using a 63×, 1.4 NA oil objective (Carl Zeiss MicroImaging, Inc., Maple Grove, MN). Images are representive of typical confocal sections (approx. 0.37 μm).
Transmission Electron Microscopy
HeLa cells were grown on Thermanox coverslips (Nunc, Rochester, NY) and infected with C. trachomatis L2 for 24 hrs. Cells were rinsed twice with HBSS and fixed with periodate-lysine-paraformaldehyde (PLP fixative: 75mM lysine, 37mM sodium phosphate, 10mM sodium periodate, 2% paraformaldehyde) plus 0.25% glutaraldehyde for 2 hrs at room temperature. Samples were rinsed twice with PBS, permeabilized with 0.01% saponin in PBS for 5 minutes at room temperature, and incubated with mouse anti-active Src kinase (Tyr419) in 0.01% saponin/PBS overnight at room temperature. The cells were rinsed twice with PBS, incubated with peroxidase conjugated F(ab′)2 donkey anti-mouse IgG (Jackson Immunoresearch) in 0.01% saponin/PBS for 1 hour at room temperature and rinsed the three times with PBS. The samples were then fixed for 1 hour with 1.5% glutaraldehyde in 0.1M sodium-cacodylate pH 7.4 plus 5% sucrose and rinsed three times in 50mM Tris-HCl pH 7.4 plus 7.5% sucrose. The reactions were developed using Immunopure Metal Enhanced DAB reagent (Pierce Chemical, Rockford, IL). Samples were rinsed three times with 50mM Tris-HCl with 7.5% sucrose and fixed overnight at 4 °C in 2.5% glutaraldehyde in 100mM sodium cacodylate buffer, pH 7. Cells were post-fixed two times in 1.0% osmium tetroxide reduced with 1% potassium ferricyanide using a Pelco Biowave laboratory microwave system (Ted Pella, Inc., Redding, CA) at 250 W (2 min on, 2 min off, 2 min on) under 20- in. Hg Vacuum. The cells were later washed with water and dehydrated in graded ethanol series for 45 seconds each in a microwave at 250 W. The cultures were embedded in Spurr’s resin and sectioned with a UC6 ultramicrotome (Leica Microsystems). Tomography was performed on the 180nm thin sections using a Tecnai Spirit TEM (FEI, Hillsboro, OR) at 120 kV. Tilt series with 1 degree increments totalling 137 images (−68° to 68°) were recorded on 2048×2048 pixel Gatan CCD camera.
siRNA knockdown of Src family kinases
HeLa cells were plated to approximately 50% confluency on coverslips in 24-well plates (Corning). Cells were transfected with ON-TARGETplus SMARTpool siRNA corresponding to Fyn, Src, Yes, Fyn and Src or non-targeting sequence #1 (Thermo Scientific/Dharmacon, Lafayette, CO) according to the manufacturer’s instructions. At 48 hrs after transfection, cells were infected with C. trachomatis serovar L2 and 24 hrs post-infection cells were fixed with methanol for immunostaining. Samples were observed using confocal microscopy and images were acquired using identical settings for each set of samples.
Co-Immunoprecipitation and Western Blotting
HeLa cells were infected with C. trachomatis serovar L2 or mock-infected. At 24 hrs post infection, cells were washed 3 times in cold PBS, scraped into 1.5 mls of 1% Zwittergent 3–14 in PBS, agitated for 20 minutes on ice, and incubated with protein-A beads followed by centrifugation to pre-clear the lysate. Protein-A agarose beads were incubated with rabbit antibodies against R. rickettsii, IncB, Inc101, Inc222 or Inc850 for 6 hrs at room temperature. The beads were rinsed 3 times with PBS and mixed with the pre-cleared cell lysate. After gentle agitation overnight at 4°C, the beads were washed 3 times with 1% Zwittergent 3–14 in PBS and proteins were eluted into 1× SDS-PAGE sample buffer. The eluted proteins were separated by SDS-PAGE on a 14% acrylamide gel and electrophoretically transferred to nitrocellulose. Membranes were blocked in 2.5% non-fat dry milk in 50 mM Tris-HCl, pH 7.4–150 mM NaCl (TBS-T) for 1 hr and then incubated with anti-IncB, anti-Inc101 or anti-Inc222 in TBS-T overnight at 4°C. Unbound antibody was removed by three rinses with TBS-T and bound antibody detected with an HRP-conjugated donkey anti-rabbit IgG secondary for 45min. Blots were then rinsed 3 times with TBS-T and developed with Femto substrate (Pierce) and exposed to CL-Xposure film (Thermo Scientific).
Plasmids and Transfections
mCherry was fused to the N-terminus of full length Inc proteins using the XhoI and EcoRI sites in pmCherry-C1 (Clontech, Mountain View, CA). Each Inc gene was amplified from C. trachomatis serovar L2 DNA using forward primers (IDT, Coralville, IA) that incorporated an XhoI site (IncB: CCC CTC GAG GGA TGG TTC ATT CTG TAT ACA ATT CAT TG; Inc101: CCC CTC GAG GGA TGA TCT CCA TGA TTC CAA GG; Inc222: CCC CTC GAG GGA TGC GTT GCT GTT GTG TTC GTA C; Inc850: CCC CTC GAG GGA TGG GAT TCG GAA CTG TGA GAG G) and reverse primers (IDT) that incorporated an EcoRI site (IncB: CCC GAA TTC CTA TTC TTG AGG TTT TGT TGG GCT G; Inc101: CCC GAA TTC TCA GTA ATA ATA AAC AGA ATA TTT TGA TTT TAA C; Inc222: CCC GAA TTC TCA GTT GGA ATA CAC TAA TTG CTT TTA ATT C; Inc850: CCC GAA TTC TTA CCG ATT CTG GTT GTG AAG TAC TAA C). These plasmids were used to transfect HeLa cells in 24 well plates using Lipofectamine reagents (Invitrogen) according to the manufacturer’s instructions.
Acknowledgments
This work was supported by the intramural research program of the NIAID/NIH. We thank Janet Sager for technical assistance and Drs. R. Heinzen and J. Celli for critical review of the manuscript.
References
- Alzhanov DT, Weeks SK, Burnett JR, Rockey DD. Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223. BMC Microbiol. 2009;5:2. doi: 10.1186/1471-2180-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila T, Saikku P, Koskela P, Bloigu A, Dillner J, Ikaheimo I, et al. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA. 2001;285:47–51. doi: 10.1001/jama.285.1.47. [DOI] [PubMed] [Google Scholar]
- Bannantine JP, Stamm WE, Suchland RJ, Rockey DD. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect Immun. 1998;66:6017–6021. doi: 10.1128/iai.66.12.6017-6021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci. 2006;119:350–359. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkelund S, Johnsen H, Christiansen G. Chlamydia trachomatis serovar L2 induces protein tyrosine phosphorylation during uptake by HeLa cells. Infect Immun. 1994;62:4900–4908. doi: 10.1128/iai.62.11.4900-4908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkelund S, Bini L, Pallini V, Sanchez-Campillo M, Liberatori S, Clausen JD, et al. Characterization of Chlamydia trachomatis L2-induced tyrosine-phosphorylated HeLa cell proteins by two-dimensional gel electrophoresis. Electrophoresis. 1997;18:563–567. doi: 10.1002/elps.1150180338. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Kromhaut J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Richmond SJ, Yates P. The development of Chlamydia trachomatis inclusions within the host eukaryotic cell during interphase and mitosis. J Gen Microbiol. 1989a;135:1153–1165. doi: 10.1099/00221287-135-5-1153. [DOI] [PubMed] [Google Scholar]
- Campbell S, Richmond SJ, Yates PS. The effect of Chlamydia trachomatis infection on the host cell cytoskeleton and membrane compartments. J Gen Microbiol. 1989b;135:2379–2386. doi: 10.1099/00221287-135-9-2379. [DOI] [PubMed] [Google Scholar]
- Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci U S A. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen JD, Christiansen G, Holst HU, Birkelund S. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol. 1997;25:441–449. doi: 10.1046/j.1365-2958.1997.4591832.x. [DOI] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber S, Dooley CA, Fischer E, Mead D, et al. A chlamydial type III translocated protein is trosine phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, Nilges M, Dautry-Varsat A, Subtil A. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J Biol Chem. 2004;279:46896–46906. doi: 10.1074/jbc.M407227200. [DOI] [PubMed] [Google Scholar]
- Delevoye C, Nilges M, Dehoux P, Paumet F, Perrinet S, Dautry-Varsat A, Subtil A. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 2008;4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawaz FS, van Ooij C, Homola E, Mutka SC, Engel JN. Infection with Chlamydia trachomatis alters the tyrosine phosphorylation and/or localization of several host cell proteins including cortactin. Infect Immun. 1997;65:5301–5308. doi: 10.1128/iai.65.12.5301-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Mead D, Dooley CA, Hackstadt T. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol. 2003;48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- Greene RD, Zhong G. Inhibition of host cell cytokinesis by Chlamydia trachomatis infection. J Infect. 2003;47:45–51. doi: 10.1016/s0163-4453(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Grieshaber S, Grieshaber N, Hackstadt T. Chlamydia trachomatis uses host cell dynein to traffic to the microtube organizing center in a p50 dynamitin independent process. J Cell Biol. 2003;116:3793–3802. doi: 10.1242/jcs.00695. [DOI] [PubMed] [Google Scholar]
- Grieshaber SS, Grieshaber NA, Miller N, Hackstadt T. Chlamydia trachomatis causes centrosomal defects resulting in chromosomal segregation abnormalities. Traffic. 2006;7:940–949. doi: 10.1111/j.1600-0854.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Hakama M, Lehtinen M, Knekt P, Aromaa A, Leinikki P, Miettinen A, et al. Serum antibodies and subsequent cervical neoplasms: a prospective study with 12 years of follow-up. Am J Epidemiol. 1993;137:166–170. doi: 10.1093/oxfordjournals.aje.a116656. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi N. Electron microscopic studies on the mode of reproduction of trachoma virus and psittacosis virus in cell cultures. Exp Mol Pathol. 1965;76:24–39. doi: 10.1016/0014-4800(65)90021-3. [DOI] [PubMed] [Google Scholar]
- Horoschak KD, Moulder JW. Division of single host cells after infection with chlamydiae. Infect Immun. 1978;19:281–286. doi: 10.1128/iai.19.1.281-286.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. Chlamydia trachomatis Tarp is phosphorylated by src family tyrosine kinases. Biochem Biophys Res Commun. 2008;371:339–344. doi: 10.1016/j.bbrc.2008.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Tan M, Sütterlin C. Centrosome abnormalities during a Chlamydia trachomatis infection are caused by dysregulation of the normal duplication pathway. Cell Microbiol. 2009;11:1064–1073. doi: 10.1111/j.1462-5822.2009.01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur ELF. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- Koskela P, Anttila T, Bjørge T, Brunsvig A, Dillner J, Hakama MHT, et al. Chlamydia trachomatis infection as a risk factor for invasive cervical cancer. Int J Cancer. 2000;85:35–39. doi: 10.1002/(sici)1097-0215(20000101)85:1<35::aid-ijc6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Lane BJ, Mutchier C, Al Khodor S, Grieshaber SS, Carabeo RA. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M, Shalig R. The role of Fyn kinase in the release from metaphase in mammalian oocytes. Mol Cell Endocrinol. 2010;2:228–233. doi: 10.1016/j.mce.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Dráberová E, Richterová V, Sulimenko V, Sulimenko T, Dráberová L, et al. Regulation of microtubule nucleation from membranes by complexes of membrane-bound gamma-tubulin with Fyn kinase and phosphoinositide 3-kinase. Biochem J. 2008;416:421–430. doi: 10.1042/BJ20080909. [DOI] [PubMed] [Google Scholar]
- Majeed M, Kihlstrom E. Mobilization of F-actin and clathrin during redistribution of Chlamydia trachomatis to an intracellular site in eucaryotic cells. Infect Immun. 1991;59:4465–4472. doi: 10.1128/iai.59.12.4465-4472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride T, Wilde E., III . Intracellular translocation of Chlamydia trachomatis. In: Bowie WR, Caldwell HD, Jones RP, Mardh P-A, Ridgway GL, Schachter J, et al., editors. Chlamydial Infections. Cambridge: Cambridge University Press; 1990. pp. 36–39. [Google Scholar]
- Mehlitz A, Banhart S, Hess S, Selbach M, Meyer TF. Complex kinase requirements for Clamydia trachomatis Tarp phosphorylation. FEMS Microbiol Lett. 2008;289:233–240. doi: 10.1111/j.1574-6968.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- Moore ER, Fischer ER, Mead DJ, Hackstadt T. The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic. 2008;9:2130–2140. doi: 10.1111/j.1600-0854.2008.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, et al. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2006;103:14566–14571. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet F, Wesolowski J, Garcia-Diaz A, Delevoye C, Aulner N, Shuman HA, et al. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS ONE. 2009;4:e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Shroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Fumagalli S, Courtneidge SA. Requirement for Src family protein tyrosine kinases in G2 for fibroblast cell division. Science. 1995;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Heinzen RA, Hackstadt T. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- Roskoski RJ. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrance protein CT229. Infect Immun. 2006;74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Frame MC. Endosomal trafficking of Src tyrosine kinase. Trends Cell Biol. 2008;18:322–329. doi: 10.1016/j.tcb.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia; Intracellular biology, pathogenesis, and immunity. Washington, D.C.: ASM Press; 1999. pp. 139–169. [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Hackstadt T. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol Microbiol. 2001;39:1638–1650. doi: 10.1046/j.1365-2958.2001.02355.x. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Fischer ER, Hackstadt T. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun. 2003;71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun. 1996;64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- Smith JS, Bosetti C, Muñoz N, Herrero R, Bosch FX, Eluf-Neto J, et al. Chlamydia trachomatis and invasive cervical cancer: a pooled analysis of the IARC multicentric case-control study. Int J Cancer. 2004;111:431–439. doi: 10.1002/ijc.20257. [DOI] [PubMed] [Google Scholar]
- Stein PL, Vogel H, Soriano P. Combined deficiencies of Src, Fyn, and Yes tyrosine kinases in mutant mice. Genes Dev. 1994;8:1999–2007. doi: 10.1101/gad.8.17.1999. [DOI] [PubMed] [Google Scholar]
- Subtil A, Parsot C, Dautry-Varsat A. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol Microbiol. 2001;39:792–800. doi: 10.1046/j.1365-2958.2001.02272.x. [DOI] [PubMed] [Google Scholar]
- Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun. 2000;68:360–367. doi: 10.1128/iai.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Sheetz MP. Targeting of motor proteins. Science. 1996;271:1539–1544. doi: 10.1126/science.271.5255.1539. [DOI] [PubMed] [Google Scholar]
- Wallin KL, Wiklund F, Luostarinen T, Angström T, Anttila T, Bergman F, et al. A population-based prospective study of Chlamydia trachomatis infection and cervical carcinoma. Int J Cancer. 2002;101:371–374. doi: 10.1002/ijc.10639. [DOI] [PubMed] [Google Scholar]