Abstract
A major obstacle for the effective treatment of cancer is the invasive capacity of the tumor cells. Previous studies have demonstrated the capability of mesenchymal stem cells (MSCs) to target these disseminated tumor cells and to serve as therapeutic delivery vehicles. However, the molecular mechanisms that would enhance the migration of MSCs towards tumor areas are not well understood. In particular, very little is known about the role that epigenetic mechanisms play in cell migration and tropism of MSCs. In this study, we investigated whether histone deacetylation was involved in repression of urokinase plasminogen activator (uPA) expression in MSCs derived from umbilical cord blood (CB) and bone marrow (BM). Induction of uPA expression by histone deacetylase (HDAC) inhibitors trichostatin A (TSA) and sodium butyrate (SB) was observed in both CB- and BM-derived MSCs examined. In vitro migration assays showed that induction of uPA expression by HDAC inhibitors in CB- and BM-derived MSCs significantly enhanced tumor tropism of these cells. Furthermore, overexpression of uPA in CB-MSCs induced migration capacity toward human cancer cells in vitro. In addition, our results demonstrated that uPA-uPAR knockdown in PC3 prostate cancer cells significantly inhibited tumor-specific migration of uPA-overexpressing MSCs. These results have significant implications for development of MSC-mediated, tumor-selective gene therapies.
Keywords: Mesenchymal stem cells, Histone modifications, Urokinase plasminogen activator, Cell migration, Tumor tropism
INTRODUCTION
Tumor cells deregulate the expression of growth factors, proteases, and extracellular matrix and cell surface proteins to gain their devastating invasion capacity (1). Several studies suggest that MSCs can be used as vehicles for delivering therapeutic genes to treat invasive tumors (2–8). MSCs derived from CB and BM exhibit tropism for experimental tumors and migrate toward outgrowing microsatellites (2, 6, 8). A thorough understanding of the molecular events that regulate migration of MSCs to tumors is necessary to optimize the use of MSCs as therapeutic delivery vehicles.
Recently, several groups have shown that chemokines and growth factors overexpressed by tumor cells are able to attract MSCs to the tumor microenvironment (9–11). However, the observation that even microsatellites and invading tumor cells distant from the main tumor mass are targeted by MSCs suggests that additional local regulators for MSCs migration exist. Tumor cell invasion depends largely on the expression of extracellular matrix-degrading proteases, and interestingly, extracellular matrix-secreted soluble factors from tumor cell lines are able to promote stem cell migration (12). Among the large number of proteases involved in cell migration, uPA is of particular importance because it initiates the activation of metalloproteinases and the conversion of plasminogen to plasmin (1, 13). Hence, we hypothesize that uPA may have a significant influence on the cell migration and tropism of MSCs.
An increasing body of evidence indicates that changes in chromatin structure by histone modification appear to play an important role in the regulation of gene transcription (14). In the present study, we examined acetylation of histones associated with uPA promoter, uPA mRNA expression and in vitro migration of MSCs treated with inhibitors of HDACs. Our results show that histone deacetylation plays a central role in the transcriptional regulation of the uPA gene in MSCs and that use of HDAC inhibitors results in the epigenetic activation of uPA.
RESULTS
TSA induces accumulation of acetylated histones in chromatin associated with the uPA gene in mesenchymal stem cells
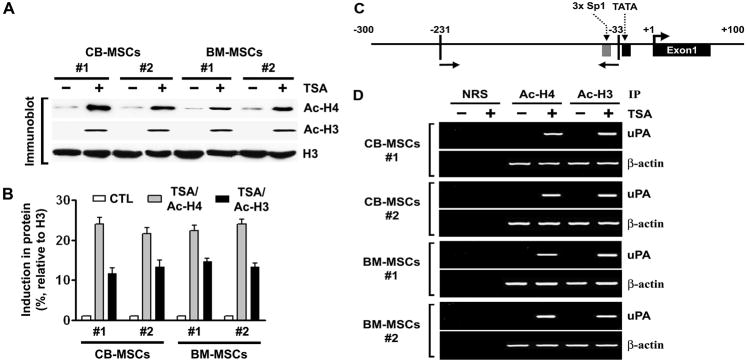
We have previously shown that the levels of uPA mRNA expression in human cancer cell lines correlate with their state of histone acetylation and that TSA, an inhibitor of HDACs, induces uPA in the human cancer cell lines (15). In the present study, we found that uPA mRNA expression was undetectable in MSCs isolated from CB and BM (Supplementary Fig. S1A). We treated uPA-silenced MSCs isolated from CB (n = 2) and BM (n = 2) with 100 nM TSA for 16 hrs and performed immunoblot analysis on the nuclear extracts using antibodies to acetylated histones H3 and H4. Accumulation of acetylated histones was observed in TSA-treated CB- and BM-MSCs (Fig. 1A–B). A similar trend was observed by immunofluorescence assay as well (Supplementary Fig. S2).
Figure 1. TSA induces accumulation of acetylated histones H3 and H4 in chromatin associated with the uPA gene.
(A) Nuclear extracts were isolated from control and TSA-treated CB- and BM-MSCs, and immunoblot analysis was performed using anti-acetyl histone H3, anti-acetyl histone H4, and histone H3 antibodies. Histone H3 was utilized as a loading control.
(B) Densitometric analysis of immunoblots. Data are normalized to H3, averaged, and expressed as percentage of control (CTL = 1).
(C) Schematic representation of the uPA promoter region and the location of primers used for PCR amplification in the ChIP assay. Bent arrow, transcriptional start site.
(D) Chromatin fragments from CB- and BM-MSCs cultured with (+) or without (−) TSA for 16 hrs were immunoprecipitated with antibody to acetylated (Ac) histones H3 and H4 or control normal rabbit serum (NRS).
Next, we performed the ChIP assay on the uPA promoter to determine whether the patterns of histone acetylation were altered upon TSA treatment. Chromatin fragments from MSCs cultured with or without TSA for 16 hrs were immunoprecipitated with antibodies to acetylated histones H3 and H4. DNA from the immunoprecipitates was isolated, and PCR was performed using uPA promoter primers (Fig. 1C). Acetylation of histones H3 and H4 associated with the uPA promoter region in MSCs isolated from CB and BM was undetectable before TSA treatment. However, we observed remarkable increases in the acetylation of histones H3 and H4 in the promoter region of both CB- and BM-MSCs after treatment with TSA (100 nM, 16 hrs) (Fig. 1D). We also carried out PCR on the same set of immunoprecipitated DNA fractions for β-actin promoter as a control. The relative levels of acetylated histones H3 and H4 at the β-actin promoter was similar in all TSA-treated and untreated cells. These results suggest the potential involvement of histone deacetylation in loss of uPA expression in MSCs isolated from CB and BM. Our observations are in line with the previously shown data that TSA, as well as other HDACIs, induce the accumulation of acetylated histones in different stem cell populations (16, 17).
Effects of TSA on uPA expression and migration capacity of mesenchymal stem cells
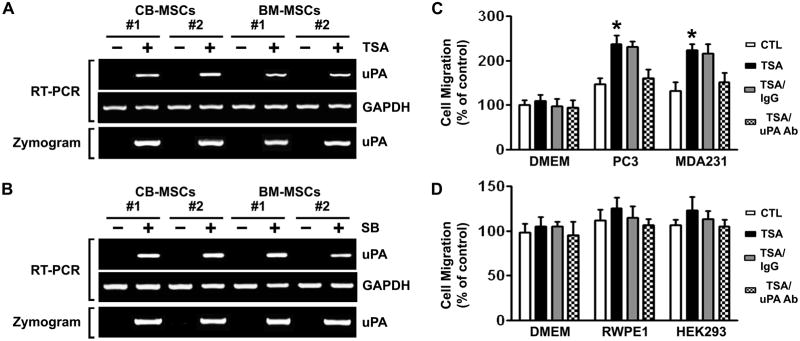
If histone deacetylation is associated with transcriptional repression, then histone acetylation following TSA treatment should lead to uPA expression. According to the RT-PCR results, treatment with TSA did induce uPA mRNA expression in MSCs isolated from CB and BM (Fig. 2A, top). We then investigated whether the resulting increases in mRNA level were sufficient to increase uPA activity levels. As expected, a corresponding increase in uPA activity levels was also observed after treatment of CB- and BM-MSCs with TSA (Fig. 2A, bottom). SB, a HDAC inhibitor differing from TSA in structure, showed similar results in all the examined MSCs (Fig. 2B). These results indicate that the effect of TSA on uPA expression can be extended to other HDAC inhibitor SB. They also suggest that the induction of uPA expression by HDAC inhibitors was not confined to a single cell source of MSC. In line with our findings, previous studies showed that HDAC inhibition was associated with gene activation by increasing H3 and H4 acetylation levels in different stem cell populations (16, 17). Because it is known that uPA can be silenced by promoter DNA methylation (15, 18), we examined the effects of the DNA methylation inhibitor 5-aza on the re-activation of uPA in CB- and BM-MSCs by RT-PCR. However, treatment with higher doses (1–10 μM) of 5-aza for five days did not restore the expression of uPA in all MSCs analyzed (data not shown). The absence of reexpression following 5-aza treatment was consistent with the observation that the uPA promoter was unmethyated in CB- and BM-MSC (Supplementary Fig. S1B).
Figure 2. Effects of trichostatin A (TSA) on uPA expression and cellular migration in MSCs.
(A) uPA mRNA expression (top) and activity levels (bottom) in control and TSA-treated CB- and BM-MSCs were analyzed by RT-PCR and fibrin zymography, respectively.
(B) uPA mRNA expression (top) and activity levels (bottom) in control and SB-treated CB- and BM-MSCs were analyzed by RT-PCR and fibrin zymography, respectively.
(C) The migration capacity of the control and TSA-treated CB-MSCs toward human cancer cells PC3 and MDA231 were assessed in vitro by transwell migration assay. Columns, percentage of the DMEM control (*p <0.05); bars, SD.
(D) The migration capacity of the control and TSA-treated CB-MSCs toward non-tumor cells RWPE1 and HEK293 were assessed as described in (C).
To determine whether HDACI-induced uPA functionally contributes to the migration of MSCs, the migration capacity of the control and TSA-treated MSCs was determined by in vitro transwell migration assay. Induction of uPA expression in BM- and CB-MSCs treated with 100 nM TSA for 16 hrs resulted in a significant increase in the migration capacity of these cells toward human cancer cell lines PC3 and MDA-231 (Fig. 2C and Supplementary Fig. S3A). In contrast, there was no significant increase in migration of uPA re-expressing MSCs toward non-tumor cell lines RWPE1 and HEK293 cells (Fig. 2D and Supplementary Fig. S3B), further confirming the tumor-specific nature of the MSC migration. Incubation of TSA-treated MSCs with the anti-uPA antibody blocked this acquired tumor-specific migration potential. In contrast, TSA-treated MSCs incubated with an isotype-matched nonspecific antibody did not exhibit decreased migration toward tumor cells PC3 and MDA-231 (Fig. 2C and Supplementary Fig. S3). This provided further convincing evidence that this increase in BM- and CB-MSCs migration after treatment with TSA is due to induction of uPA expression. These results also suggest an important role for HDACI-induced uPA in tumor-tropic behavior exhibited by these cells.
Overexpression of uPA enhances the tumor-specific migration ability of mesenchymal stem cells
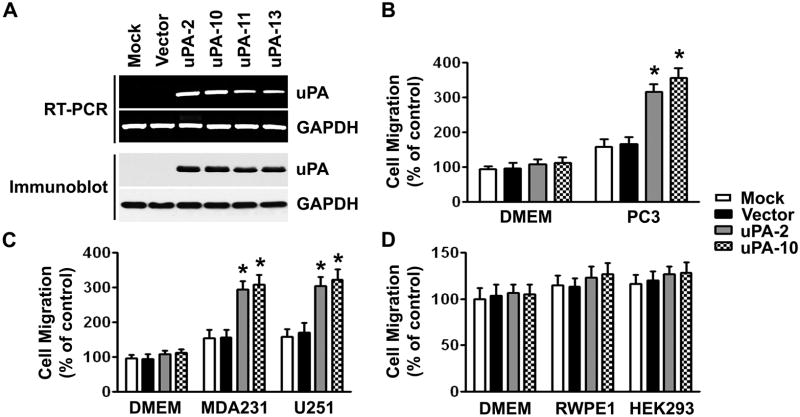
To further confirm the biological function of uPA, we investigated whether the overexpression of the uPA cDNA in MSCs would affect the migration. We used adenoviral vectors to create stable cell lines overexpressing uPA in MSCs isolated from CB. We observed a high expression level of uPA in each of the four uPA-transfected cells (uPA-2, uPA-10, uPA-11, uPA-13) but not in parental (mock) or empty vector-transfected cells as detected by RT-PCR (Fig. 3A, top) and immunoblotting (Fig. 3A, bottom). The stable clones with the highest uPA expression at the mRNA and protein levels (uPA-2 and uPA-10) were selected for further experiments. We used the in vitro transwell migration assay to examine the movement of the uPA-overexpressing cells toward PC3 cells. uPA-overexpressing MSCs (MSC/uPA) displayed significantly higher migration capacities when compared with their parental cells and empty vector controls (Fig. 3B). These cells also responded to other human cancer cell lines, MDA-231 and U251, by displaying significant migration advantage over control cells (Fig. 3C). Interestingly, the uPA-overexpressing MSCs did not change their migration capacities when non-tumor cell lines, RWPE1 and HEK293, were seeded in the bottom chamber in the assays (Fig. 3D). Moreover, the amount of cells migrating to plain DMEM cell culture medium remained similar between the uPA-expressing cells and the controls (Fig. 3). These results indicated that uPA-overexpressing MSCs were capable of stimulating tumor-specific MSCs migration and that the migratory ability of MSCs was not affected by adenoviral transduction.
Figure 3. Overexpression of uPA enhances the tumor-specific migration ability of MSCs.
(A) RT-PCR (top) and immunoblot (bottom) analyses of CB-MSCs stably transduced with mock, uPA expression vector or empty vector. GAPDH was used as loading control for RNA and protein analysis.
(B) The migration capacity of the uPA-overexpressing MSCs toward PC3 cells was assessed in vitro by transwell migration assay. Columns, percentage of the vector control (*p <0.05); bars, SD.
(C) The migration capacity of the uPA-overexpressing MSCs toward MDA-231 and U251 was assessed as described in (B).
(D) The migration capacity of the uPA-overexpressing MSCs toward RWPE1 and HEK293 was assessed as described in (B).
Activation of ERK by uPA mediates the tumor-specific migration capacity of mesenchymal stem cells
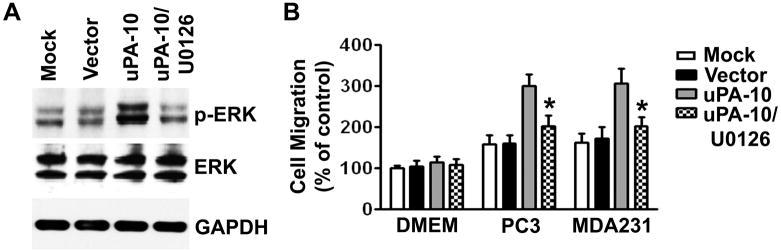
Because MAPK/ERK activation plays an important role in uPA-mediated cell migration (13, 19), we investigated whether uPA would affect ERK activation in MSCs. Immunoblot analysis showed that ERK1/2 phosphorylation significantly increased in uPA-overexpressing MSCs. Treatment of uPA-expressing MSCs with ERK inhibitor (50 μM, U0126) completely blocked the phosphorylation of ERK1/2 (Fig. 4A, Supplementary Fig. S4A). The amount of total ERK1/2 protein levels was unchanged among these cells. Treatment of MSCs with 100 nM TSA for 16 hrs also resulted significant increase in ERK1/2 phosphorylation and this effect was reversed by an ERK inhibitor (50 μM, U0126) (Supplementary Fig. S4B). We further observed that the tropism of these ERK inhibitor pretreated uPA-expressing MSCs toward PC3 cells was inhibited drastically in Transwell migration assays and the number of migrating cells went down to a level close to that of basal cell migration in response to serum-free DMEM (Fig. 4B). These results suggest that ERK activation in uPA-expressing cells might be one possible mechanism underlying the augmented tumor tropism of these cells.
Figure 4. Activation of ERK by uPA mediates the tumor-specific migration capacity of MSCs.
(A) Immunoblot analysis shows increased levels of ERK1/2 phosphorylation in uPA-expressing MSCs (uPA-10). GAPDH was used as a loading control.
(B) Pretreatment of ERK inhibitor significantly decreased uPA-expressing MSC migration toward PC3 and MDA231 cells compared with cells treated with nonspecific isotype IgG. Columns, percentage of the vector control (*p <0.05); bars, SD.
Effects of tumor cells expressing uPA and uPAR on the migration ability of MSC/uPA
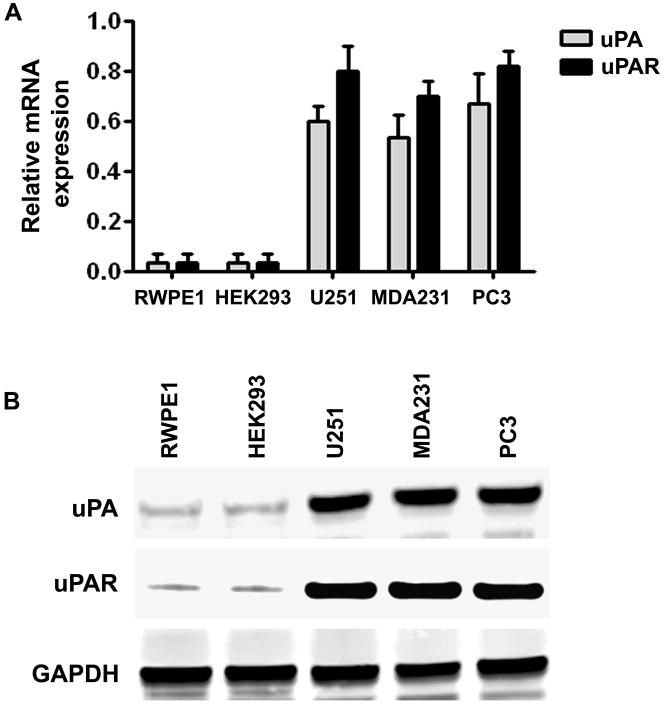
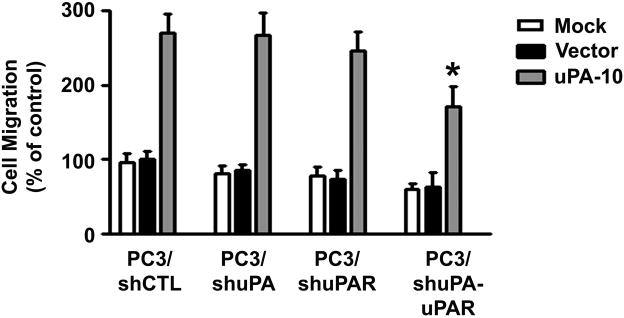
It has been reported recently that uPA and uPAR released from human tumor cells may be a potential chemoattractant involved in the tropism of MSC (20). We compared the endogenous expression levels of uPA and uPAR in the tumor and non-tumor cell lines. As shown in Fig. 5A and 5B, uPA and uPAR expression at the mRNA and protein levels were significantly higher in tumor cell lines PC3, MDA231 and U251 as compared with the non-tumor cell lines RWPE1 and HEK293, which expressed undetectable levels of these proteins. These results demonstrated a strong correlation between uPA and uPAR expression levels in tumor cell lines and the migration ability of MSCs. To determine the role of tumor cells expressing uPA and uPAR in regulating MSC migration, we used small hairpin RNAs to knockdown endogenous uPA and uPAR gene expression in the human prostate cancer cell line PC3, which has been shown to have robust expression of uPA and uPAR (Fig. 5). We developed pcDNA3-CMV vectors containing small hairpin constructs capable of generating 19- or 21-nucleotide duplex RNAi oligonucleotides corresponding to either uPA or uPAR. Also, a single bicistronic construct driven by CMV promoter to deliver dual small hairpins targeted against both uPA and uPAR was constructed to test the effectiveness of simultaneously inhibiting expression of two endogenous genes. A pcDNA3-scrambled vector with an imperfect sequence, which does not form a perfect hairpin structure, was used to develop the scrambled vector for use as a control (shCTL). The empty vector (EV) and scrambled vector (SV) controls have been tested in multiple cell lines and do not demonstrate any toxicity to cells as demonstrated by MTT assay after transfection and have no effect on the expression of housekeeping genes, GAPDH and β-actin. We previously reported that these gene-specific single and bicistronic shRNA constructs significantly inhibited uPA and uPAR expression at both the mRNA and protein levels in the prostate tumor cell line PC3 (19). These results also demonstrated that ERK1/2 phosphorylation was completely abolished in the shuPA-uPAR transfected PC3 cells but not in the cells transfected with either sh-uPA or sh-uPAR. These findings provide evidence that binding of uPA with uPAR activates ERK signaling and molecular targeting against both uPA and uPAR would be a robust way to prove its biological contribution. We then used the shRNA-transfected PC3 cells as chemoattractants for MSC/uPA in a transwell migration assay. The migration of MSC/uPA induced by shuPAuPAR-transfected PC3 cells was 55% less than that induced by shCTL-transfected PC3 cells. In contrast, the migration of MSC/uPA did not change significantly in PC3 cells transfected with either sh-uPA or sh-uPAR (Fig. 6). Taken together, these results suggest that tumor cells expressing uPA and uPAR play a major role in chemoattraction of MSC/uPA to tumors.
Figure 5. Expression levels of the uPA and uPAR in human tumor and non-tumor cell lines.
(A) uPA and uPAR mRNA expression in RWPE1, HEK293, U251, MDA231 and PC3 cells were analyzed by quantitative RT-PCR.
(B) Immunoblot analysis of uPA and uPAR proteins in HEK293, U251, MDA231 and PC3 cells. GAPDH was used as a loading control.
Figure 6. Effects of tumor cells expressing uPA and uPAR on the migration ability of MSC/uPA.
The migration capacity of the uPA-overexpressing MSCs (uPA-10) toward shRNA-transfected PC3 cells was assessed in vitro by transwell migration assay. Columns, percentage of the vector control (*p <0.05); bars, SD.
DISCUSSION
The failure of current therapies for malignant disease is mainly due to the ability of the tumor cells to extensively invade the surrounding normal tissue, hence escaping localized treatments. MSCs represent an attractive option as delivery vehicles for therapeutic genes and their products as a result of their apparent ability to both home in on the tumor site and evade the host immune response (7). Previous studies have shown the inherent tumor-tropic property of MSCs and their use as cellular vehicles for effective delivery of therapeutic agents to several types of invasive tumors (2–8). Their abilities to track infiltrating tumor cells and localize to distant tumor microfoci make MSCs attractive gene therapy vehicles with promising clinical potential. Identification of molecular mechanisms involved in MSCs tropism will be important for the development of MSC-based tumor therapies.
A limited number of stem cell attractants and cytokines emanating from solid tumors have been identified (9–11). Recent studies suggest that epigenetic mechanisms play an important role in controlling stem cell potency and cell fate decisions (21–23). However, the molecular mechanisms controlling stem cell migration to tumors remain poorly understood. In the present study, we have provided evidence for the first time that histone deacetylation is involved in the repression of uPA expression in MSCs derived from CB and BM. Histone deacetylation is a critical component of chromatin remodeling and transcriptional regulation (24). The acetylation level of core histones results from the balance between the activities of HDACs and histone acetyltransferases. Inhibition of HDACs by TSA leads to activation of only specific target genes through increased histone acetylation (25, 26). Our experiments showed that induction of uPA expression by TSA in CB- and BM-MSCs was accompanied by a remarkable increase in acetylation of histones H3 and H4 associated with the uPA promoter region (−231 to −33). The increase of core histone acetylation at the promoter region of the uPA gene after TSA treatment indicates that the chromatin structure of uPA promoter may become a loose and non-condensed structure, which is usually necessary for the start of transcription (27, 28).
Ample evidence indicated that increased levels of uPA are crucial for cell migration and invasion (29–32). Our in vitro migration assays showed that HDACI-induced uPA activation might stimulate tumor-specific migration capacity of MSCs. The importance of HDACI-induced uPA activity to stimulate MSCs migration was confirmed by using function blocking uPA antibodies. Moreover, overexpression of uPA was seen in the current study to provide MSCs an increased migration capacity toward human tumor cells in vitro. Our results also indicated that overexpression of uPA activates ERK in MSCs and inhibiting ERK activation resulted in loss of the migration activity of these cells.
Cell migration is a highly complex process and involves several factors, from sensing of environmental cues, restructuring the cytoskeleton, dynamic regulation of cell attachment and detachment to extracellular matrix, to signaling between all these processes to coordinate the movements. The results that uPA-overexpressing cells and control cells migrated at the same rate in plain cell culture medium or toward non-tumor cells suggest that uPA overexpression provided no beneficial effects on the general movement of MSCs. Most interestingly, uPA-overexpressing cells respond strongly to tumor cell-secreted cues in transwell migration assays. These data strongly suggested that uPA overexpression increases the sensitivity of MSCs to appropriate signals that stimulate cell migration.
There is considerable evidence that the ERK pathway plays a central role in uPA-uPAR system regulated cell physiology (33). Coupling of uPA with uPAR orchestrates several different signaling molecules that form a unique network of several different types of biological responses, such as proliferation, migration, invasion, angiogenesis, and metastasis. These biological responses to uPA-uPAR binding seem to be highly specific to cell type, the nature of the downstream signaling molecule, and the level of its expression. Gonias and co-workers (34) demonstrated that binding of uPA with uPAR activates ERK1/2 and that this induced ERK activity is required for uPA-induced MCF-7 breast cancer cell migration. They further showed that a signaling cascade including FAK, Src, and Shc is responsible for uPA-induced ERK activation and cell migration (35). In contrast, uPA-induced vascular smooth muscle cells migration and proliferation required activation of the Stat pathway (36). A previous study in human breast cancer cells showed that uPA-induced mitogenic activity requires activation of both Stat and ERK pathways (37).
Prior work from our laboratory has demonstrated that simultaneous silencing of uPA and uPAR in PC3 cells using a single plasmid construct expressing shRNAs for both uPA and uPAR abrogated uPA-uPAR signaling to downstream target molecules such as ERK1/2 and Stat3 and ultimately resulted in a dramatic reduction of tumor cell invasion (19). These findings provide evidence that binding of uPA with uPAR activates signaling cascades in order to regulate cell migration, invasion, proliferation, and survival. In the present study, RNAi for uPA-uPAR in PC3 cells showed significant suppression of uPA overexpression stimulated MSCs migration. Knockdown of uPA or uPAR alone in PC3 cells did not show significant decrease in MSC/uPA migration. We believe that this was because blocking either uPA or uPAR alone may not sufficiently affect the downstream signals to attract MSCs. This notion has been supported by data from our previous study that simultaneous inhibition of uPA and uPAR impair downstream signaling pathways in PC3 prostate cancer cells (19).
In summary, we found that HDACIs enhances the tropism of MSCs for tumor cells through induction of uPA expression. The role of HDACI-induced uPA in MSCs is a novel finding. Furthermore, we observed the ERK activation in uPA-expressing MSCs and treatment with the ERK inhibitor significantly reduces tumor-specific MSC migration. Further studies are warranted to determine the basic mechanisms underlying the effects of uPA to enhance MSC migration and to investigate the therapeutic utility of this observation. An adequate understanding of these epigenetic mechanisms could have important implications for effective cellular delivery of therapeutic agents for tumor therapy.
MATERIALS AND METHODS
Cell culture
Human prostate cancer cells (PC3), human breast cancer cells (MDA231), human glioma cells (U251), normal prostate epithelial cells (RWPE1) and human embryonic kidney cells (HEK293) were obtained from the American Type Culture Collection and cultured as directed. Human CB harvest and expansion of MSCs isolated from CB were conducted as previously reported (38). The separated MSCs were subcultured at a concentration of 1×106/100-mm dish in Mesencult basal medium (Stem Cell Technologies, Vancouver, Canada) and used for experiments during passages 5 to 8. The cells were incubated at 37°C in 5% CO2 in a humidified atmosphere. When the cells reached 80% confluency, they were detached with 0.25% trypsin and replated at a 1:3 ratio. Rat MSCs isolated from bone marrow were purchased from Chemicon (Temecula, CA) and maintained as per manufacturer’s instructions. Cells were treated with TSA, sodium butyrate (SB) and 5-aza as described previously (15). Total RNA and genomic DNA were isolated from the treated cells using RNA and DNA isolation kits.
Overexpression and gene silencing
Adenoviral vectors were generated and used as reported previously (39). The uPA expression vector (Ad-uPA) was prepared by inserting full-length human uPA cDNA into a pAd-CMV vector (Invitrogen, Carlsbad, CA). Empty and uPA adenoviruses were produced using ViraPower Adenoviral Expression kit (Invitrogen, Carlsbad, CA). Virus transduction in CB-MSCs to express uPA, cell selection for stable expression, and cell maintenance were carried out as recommended by the manufacturer (Invitrogen, Carlsbad, CA). For short hairpin RNA (shRNA)-mediated uPA and uPAR gene silencing, prostate cancer PC3 cells were transfected with the previously described shRNA constructs (19). Cells were plated to reach 90% confluence on the day of transfection of the shRNA expression plasmids, and plasmid DNA was transfected with Lipofectamine 2000 according to the manufacturer’s protocol.
In vitro cell migration assay
In vitro migration of MSCs toward tumor cells was examined using Transwell assays (Costar, Cambridge, MA) in 24-well plates. Each well of the plates was separated into two chambers by an insert membrane with 8-μm pores. One day before the assays were carried out, 50,000 tumor cells were seeded into each lower chamber. After a 24-h incubation period, cell culture medium in the lower chamber was removed and replaced with 500 μl of serum-free DMEM. CB-MSCs, BM-MSCs or CB-MSCs/uPA cells (50,000 in 500 μl of serum-free DMEM) were then seeded into the upper chamber. For the assay using a neutralization antibody to block cell migration, 40 μg/ml of anti-uPA monoclonal antibodies (Biomeda, Foster City, CA) were incubated with CB- and BM-MSCs for one hour at room temperature before the cell seeding. After a 12-h incubation period, the non-migrated cells in the upper chamber were gently scraped away, and adherent cells present on the lower surface of the insert were stained with Hema-3 and photographed. The number of cells that had migrated to the lower side of the filter was counted under a light microscope with five high-power fields (400x). Experiments were performed in triplicate. The cell migration in response to serum-free DMEM was used as the basal migration rate. In overexpression experiments, the migration of cells transduced with a vector control in response to serum-free DMEM was used as the basal migration rate.
Fibrin zymography
The enzymatic activity and molecular weight of electrophoretically separated forms of uPA were determined in conditioned medium of cells by SDS-PAGE as described previously (19). Briefly, the SDS-PAGE gel contains acrylamide to which purified plasminogen and fibrinogen were substrates before polymerization. After polymerization, equal amounts of proteins in the samples were electrophoresed and the gel was washed and stained as described previously (19).
Reverse transcription-PCR analysis
Cellular RNA was isolated from MSCs using the Qiagen RNeasy kit. RNA (1 μg) was treated with DNase (10 units/μg of RNA for 1 h) and used as a template for the RT reaction (20 μl). The RT reaction mix (Invitrogen) contained 1 μl (10 pm) of primers. The resultant cDNA was then used in PCR reactions and analyzed by gel electrophoresis. We used the following primers for PCR: uPA-sense, 5′-TGC GTC CTG GTC GTG AGC GA-3′, and uPA-antisense, 5′-CTA CAG CGC TGA CAC GCT TG-3′; GAPDH-sense, 5′-CGG AGT CAA CGG ATT TGG TCG TAT-3′, and GAPDH-antisense, 5′-AGC CTT CTC CAT GGT GGT GAA GAC-3′. PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The final extension was at 72°C for 5 min. Quantitative reverse transcription-PCR (qRT-PCR) was done using the iCycler IQ real-time PCR system (Bio-Rad) with the SYBR Green Mastermix as per manufacturer’s instructions. All reactions were performed in triplicate. No reverse transcriptase or no template served as the negative controls.
Nuclear extract preparation and immunoblot analysis
Nuclear extracts were prepared from control and TSA-treated CB-MSC and BM-MSC using a nuclear extraction kit from Panomics, Inc. (Redwood City, CA) as per the manufacturer’s instructions. Equal amounts of nuclear extracts were resolved by SDS-PAGE and then blotted with anti-acetylated histone H3, anti-acetylated histone H4 and histone H3 (Cell Signaling Technology, Beverly, MA). For immunoblot analysis of uPA and uPAR protein expression, 30 μg of total cell lysate were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes by electroblotting. The following antibodies were used: anti-uPA (Biomeda, Foster City, CA), anti-uPAR (American Diagnostics Inc., Greenwich, CT) and anti-GAPDH (Abcam, Cambridge, MA). Horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) were used for detection of immunoreactive proteins by chemiluminescence (Amersham Biosciences).
Chromatin Immunoprecipitation Assay
ChIP assays were performed as per the manufacturer’s instructions (catalog no. 17–295, Upstate Biotechnology, Lake Placid, NY). In brief, cells (1×106 cells/100 mm dish) were fixed by adding formaldehyde at a final concentration of 1% and incubating for 10 min at 37°C. The cells were washed twice with ice-cold phosphate-buffered saline containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A), harvested, and treated with SDS lysis buffer for 10 min on ice. The resulting lysates were sonicated to shear the DNA to fragment lengths below 1000 bp (amplitude 60%, 4 × 10 s, Fisher Sonic Dismembrator 60, Pittsburgh, PA). After pre-clearing the lysates, 4 μg of specific antibodies (anti-acetylated histone H3, anti-acetylated histone H4, anti-HDAC1, anti-HDAC3, and anti-HDAC7, Cell Signaling Technology Inc., Beverly, MA) were used to immunoprecipitate the protein-DNA complexes. Antibody controls were also included for each ChIP assay; no precipitation was observed. The antibody-protein complexes were collected using salmon sperm DNA-protein A-agarose slurry and washed several times as per the manufacturer’s instructions. The immunocomplexes were eluted with 1% SDS and 0.1 M NaHCO3, and the cross-links were reversed by incubation at 65°C for 4 hrs in the presence of 200 nM NaCl. The samples were treated with proteinase K for one hour, and the DNA was purified by phenol/chloroform extraction and ethanol precipitation. The recovered DNA was resuspended in 30 μl of H2O and used as templates for PCR of uPA or β-actin gene promoters. The following primers were used for PCR: uPA promoter-sense, 5′-CAG GTG CAT GGG AGG AAG C-3′, and uPA promoter-antisense, 5′-AGG GGC GGC GCC GGG GCG G-3′; β-actin promoter-sense, 5′-CCA ACG CCA AAA CTC TCC C-3′, and β-actin promoter-antisense, 5′-AGC CAT AAA AGG CAA CTT TCG-3′. Initially, PCR was performed with different numbers of cycles or dilutions of input DNA to determine the linear range of the amplification; all results shown fall within this range. Following 30 cycles of amplification, PCR products were run on 2% agarose gels and analyzed by ethidium bromide staining.
Methylation-specific PCR
Genomic DNA treated with sodium bisulfite was amplified using primers specific to the methylated and unmethylated forms of DNA sequences of interest. We used the following MSP primers: for the unmethylated uPA sequence: 5’-AGT GTT GTG GAA GTA TGT GG -3′ (sense) and 5′-CCA CCA CAA CCC CAC CCA AA -3′ (antisense); for the methylated uPA sequence: 5′-AGC GTT GCG GAA GTA CGC GG -3′ (sense) and 5′-CCG CCG CAG CCC CGC CCA AA -3′ (antisense). After denaturation at 95°C for 5 min, 40 PCR cycles were completed with the bisulfite-treated genomic DNA as a template. 10 μL of the PCR-amplified fragments were loaded onto 2% agarose gels for analysis. Positive controls used for methylation-specific PCR included DNA from PC3 cells as unmethylated DNA control and CpGenome Universal methylated DNA as methylated DNA control (Chemicon International). Negative control MS-PCR reactions were performed using water only as a template.
Immunofluorescence detection
MSCs treated with control and TSA were fixed with 4% paraformaldehyde and incubated with anti-AcH4 (1:500; Cell Signaling Technology, Beverly, MA). After washing, fluorescent secondary antibodies (Santa Cruz Biotechnology) were added at a 1:500 dilution. The cells were again washed three times with PBS, and counterstained with Hoechst. Fluorescent images were acquired using a charge-coupled device RT Slider Spot Camera (Diagnostic Instruments Inc., Sterling Heights, MI) connected to a microscope (Olympus, Melville, NY) and managed by a computer equipped with the spot RT software version 3.5 (Diagnostic Instruments, Sterling Heights, MI).
Densitometry
ImageJ software (National Institutes of Health) was used to quantify band intensities. Data are represented as relative to the intensity of the indicated loading control.
Statistical Analysis
Statistical comparisons were performed using analysis of variance for analysis of significance between different values using GraphPad Prism software (San Diego, CA). Values are expressed as mean ± S.D. from at least three separate experiments, and differences were considered significant at a p value of <0.05.
Supplementary Material
Acknowledgments
We thank Shellee Abraham for preparing the manuscript and Diana Meister and Sushma Jasti for manuscript review.
This research was supported by National Cancer Institute Grant CA75557, CA116708, CA138409, and Caterpillar, Inc., OSF St. Francis, Inc. Peoria, IL (to J.S.R.). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
Author contributions:
S.M.K.P.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; B.G.: data analysis and interpretation, collection and assembly of data; V.R.D.: collection and assembly of data, data analysis and interpretation; C.S.G.: collection and assembly of data, data analysis and interpretation; J.S.R.: conception and design, data analysis and interpretation, financial support, administrative support, final approval of manuscript.
Reference List
- 1.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 2.Buzanska L, Machaj EK, Zablocka B, Pojda Z, Domanska-Janik K. Human cord blood-derived cells attain neuronal and glial features in vitro. J Cell Sci. 2002;115:2131–8. doi: 10.1242/jcs.115.10.2131. [DOI] [PubMed] [Google Scholar]
- 3.Chen N, Hudson JE, Walczak P, et al. Human umbilical cord blood progenitors: the potential of these hematopoietic cells to become neural. Stem Cells. 2005;23:1560–70. doi: 10.1634/stemcells.2004-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramm CM, Sena-Esteves M, Barnett FH, et al. Gene therapy for brain tumors. Brain Pathol. 1995;5:345–81. doi: 10.1111/j.1750-3639.1995.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakamizo A, Marini F, Amano T, et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65:3307–18. doi: 10.1158/0008-5472.CAN-04-1874. [DOI] [PubMed] [Google Scholar]
- 6.Newman MB, Willing AE, Manresa JJ, Sanberg CD, Sanberg PR. Cytokines produced by cultured human umbilical cord blood (HUCB) cells: implications for brain repair. Exp Neurol. 2006;199:201–8. doi: 10.1016/j.expneurol.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Pereboeva L, Komarova S, Mikheeva G, Krasnykh V, Curiel DT. Approaches to utilize mesenchymal progenitor cells as cellular vehicles. Stem Cells. 2003;21:389–404. doi: 10.1634/stemcells.21-4-389. [DOI] [PubMed] [Google Scholar]
- 8.Studeny M, Marini FC, Champlin RE, Zompetta C, Fidler IJ, Andreeff M. Bone marrow-derived mesenchymal stem cells as vehicles for interferon-beta delivery into tumors. Cancer Res. 2002;62:3603–8. [PubMed] [Google Scholar]
- 9.Son BR, Marquez-Curtis LA, Kucia M, et al. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells. 2006;24:1254–64. doi: 10.1634/stemcells.2005-0271. [DOI] [PubMed] [Google Scholar]
- 10.Sordi V, Malosio ML, Marchesi F, et al. Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood. 2005;106:419–27. doi: 10.1182/blood-2004-09-3507. [DOI] [PubMed] [Google Scholar]
- 11.Wynn RF, Hart CA, Corradi-Perini C, et al. A small proportion of mesenchymal stem cells strongly expresses functionally active CXCR4 receptor capable of promoting migration to bone marrow. Blood. 2004;104:2643–5. doi: 10.1182/blood-2004-02-0526. [DOI] [PubMed] [Google Scholar]
- 12.Ziu M, Schmidt NO, Cargioli TG, Aboody KS, Black PM, Carroll RS. Glioma-produced extracellular matrix influences brain tumor tropism of human neural stem cells. J Neurooncol. 2006;79:125–33. doi: 10.1007/s11060-006-9121-5. [DOI] [PubMed] [Google Scholar]
- 13.Jo M, Thomas KS, Somlyo AV, Somlyo AP, Gonias SL. Cooperativity between the Ras-ERK and Rho-Rho kinase pathways in urokinase-type plasminogen activator-stimulated cell migration. J Biol Chem. 2002;277:12479–85. doi: 10.1074/jbc.M111147200. [DOI] [PubMed] [Google Scholar]
- 14.Marks PA, Rifkind RA, Richon VM, Breslow R. Inhibitors of histone deacetylase are potentially effective anticancer agents. Clin Cancer Res. 2001;7:759–60. [PubMed] [Google Scholar]
- 15.Pulukuri SM, Gorantla B, Rao JS. Inhibition of histone deacetylase activity promotes invasion of human cancer cells through activation of urokinase plasminogen activator (uPA) J Biol Chem. 2007;282:35594–603. doi: 10.1074/jbc.M705867200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.De Felice L, Tatarelli C, Mascolo MG, et al. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res. 2005;65:1505–13. doi: 10.1158/0008-5472.CAN-04-3063. [DOI] [PubMed] [Google Scholar]
- 17.Gregory RI, O’Neill LP, Randall TE, et al. Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1 locus in mouse embryonic stem cells. J Biol Chem. 2002;277:11728–34. doi: 10.1074/jbc.M105775200. [DOI] [PubMed] [Google Scholar]
- 18.Pulukuri SM, Rao JS. Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–46. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Pulukuri SM, Gondi CS, Lakka SS, et al. RNA Interference-directed Knockdown of Urokinase Plasminogen Activator and Urokinase Plasminogen Activator Receptor Inhibits Prostate Cancer Cell Invasion, Survival, and Tumorigenicity in Vivo. J Biol Chem. 2005;280:36529–40. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Gutova M, Najbauer J, Frank RT, et al. Urokinase plasminogen activator and urokinase plasminogen activator receptor mediate human stem cell tropism to malignant solid tumors. Stem Cells. 2008;26:1406–13. doi: 10.1634/stemcells.2008-0141. [DOI] [PubMed] [Google Scholar]
- 21.Akashi K. Lineage promiscuity and plasticity in hematopoietic development. Ann N Y Acad Sci. 2005;1044:125–31. doi: 10.1196/annals.1349.016. [DOI] [PubMed] [Google Scholar]
- 22.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci U S A. 2007;104:12371–6. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–9. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- 24.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–25. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 25.Della RF, Criniti V, Della PV, et al. Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Lett. 2001;499:199–204. doi: 10.1016/s0014-5793(01)02539-x. [DOI] [PubMed] [Google Scholar]
- 26.Tan NC, Tan DY, Tan LC. An Unusual Headache: Lemierre’s Syndrome. J Neurol. 2003;250:245–6. doi: 10.1007/s00415-003-0964-9. [DOI] [PubMed] [Google Scholar]
- 27.Davie JR, Spencer VA. Control of histone modifications. J Cell Biochem. 1999;(Suppl 32–33):141–8. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 28.Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–8. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 29.Kargiotis O, Chetty C, Gogineni V, et al. uPA/uPAR downregulation inhibits radiation-induced migration, invasion and angiogenesis in IOMM-Lee meningioma cells and decreases tumor growth in vivo. Int J Oncol. 2008;33:937–47. [PMC free article] [PubMed] [Google Scholar]
- 30.Salvi A, Arici B, De Petro G, Barlati S. Small interfering RNA urokinase silencing inhibits invasion and migration of human hepatocellular carcinoma cells. Mol Cancer Ther. 2004;3:671–8. [PubMed] [Google Scholar]
- 31.Schweinitz A, Steinmetzer T, Banke IJ, et al. Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents. J Biol Chem. 2004;279:33613–22. doi: 10.1074/jbc.M314151200. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto M, Sawaya R, Mohanam S, et al. Expression and localization of urokinase-type plasminogen activator in human astrocytomas in vivo. Cancer Res. 1994;54:3656–61. [PubMed] [Google Scholar]
- 33.Ossowski L, Aguirre-Ghiso JA. Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr Opin Cell Biol. 2000;12:613–20. doi: 10.1016/s0955-0674(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen DH, Hussaini IM, Gonias SL. Binding of urokinase-type plasminogen activator to its receptor in MCF-7 cells activates extracellular signal-regulated kinase 1 and 2 which is required for increased cellular motility. J Biol Chem. 1998;273:8502–7. doi: 10.1074/jbc.273.14.8502. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen DH, Webb DJ, Catling AD, et al. Urokinase-type plasminogen activator stimulates the Ras/extracellular signal-regulated kinase (ERK) signaling pathway and MCF-7 cell migration by a mechanism that requires focal adhesion kinase, Src, and Shc. Rapid dissociation of GRB2/Sps-Shc complex is associated with the transient phosphorylation of ERK in urokinase-treated cells. J Biol Chem. 2000;275:19382–8. doi: 10.1074/jbc.M909575199. [DOI] [PubMed] [Google Scholar]
- 36.Kiyan J, Kiyan R, Haller H, Dumler I. Urokinase-induced signaling in human vascular smooth muscle cells is mediated by PDGFR-beta. EMBO J. 2005;24:1787–97. doi: 10.1038/sj.emboj.7600669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo M, Thomas KS, Marozkina N, et al. Dynamic assembly of the urokinase-type plasminogen activator signaling receptor complex determines the mitogenic activity of urokinase-type plasminogen activator. J Biol Chem. 2005;280:17449–57. doi: 10.1074/jbc.M413141200. [DOI] [PubMed] [Google Scholar]
- 38.Dasari VR, Spomar DG, Li L, Gujrati M, Rao JS, Dinh DH. Umbilical cord blood stem cell mediated downregulation of Fas improves functional recovery of rats after spinal cord injury. Neurochem Res. 2008;33:134–49. doi: 10.1007/s11064-007-9426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanamandra N, Kondraganti S, Gondi CS, et al. Recombinant adeno-associated virus (rAAV) expressing TFPI-2 inhibits invasion, angiogenesis and tumor growth in a human glioblastoma cell line. Int J Cancer. 2005;115:998–1005. doi: 10.1002/ijc.20965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.