Abstract
Many ultraviolet-A (UVA)-induced biochemical and physiological changes are valid as biomarkers using aquatic species for detection of the degree of stress. Changes in the concentration and activities of enzymes, such as glucose-6-phosphate dehyderogenase (G6PDH), lactate dehyderogenase (LDH), DNA damage and lipid peroxidation (LPO), can be used as biomarkers to identify possible environmental contamination in fish. This study aimed to investigate the impact of UVA on the activity of the selected enzymes, DNA damage and LPO during early developmental stages of the African catfish Clarias gariepinus. Embryo hemogenates were used for measurements of G6PDH, LDH, DNA damage and LPO concentrations and activities spectrophotometrically at 37°C. The normal ontogenetic variations in enzyme activities, DNA damage and LPO of the early developmental stages (24–168 h-PFS; hours-post fertilization stage) were studied. There was a significant decrease in the activity of G6PDH till 120 h-PFS. Then after 120 h-PFS, the activity of such enzymes insignificantly increased toward higher stages. The LDH activity was recorded with a pattern of decrease till 96 h-PFS, followed by a significant increase toward 168 h-PFS. The polynomial pattern of variations in DNA damage and LPO was also evident. The patterns of the enzyme activities, corresponding DNA damage and LPO of the early ontogenetic stages under the influence of three different UVA doses (15, 30 and 60 min), were recorded. The pattern of variations in G6PDH activity in UVA-induced groups was similar to that of the control group with variation in the magnitude of such activity. In all treated groups, LDH activity decreased till 96 h-PFS, then increased till 168 h-PFS. Within each of the embryonic stages, the increase in UVA led to a significant increase in DNA damage. A significant increase in lipid peroxidation under UVA doses was recorded. The variability in number and molecular weight of proteins under exposure to UVA was evident, reflecting some of the genetic and transcriptional changes during exposure and development.
Keywords: UVA, G6PDH, LDH, DNA damage, LPO, African catfish
Introduction
About three percent of the sun's electromagnetic output is emitted as ultraviolet (UV) radiation, but only a fraction reaches the surface of the Earth (Williamson 1995; Brian 2002). Exposure to UV radiation whether of solar or artificial origin that is pointless in aquatic environments carries potential risks to animals and plants, especially those inhabiting shallow water (Williamson et al. 1997; Rozema et al. 2002), through biomolecules, which literally exert the function of internal UV-photon absorbers (Obermüller et al. 2007). Such radiation may have negative effects on the aquatic ecosystems, resulting in decreased biomass productivity, including fish yields (Castro-Pérez 2004). Whether elevated UV levels are a critical environmental threat as only part of the many changes affecting our environment is an important question with no current clear answer (Weatherhead et al. 2007).
Ultraviolet radiation comprises three distinct spectra according to wavelength: UVA (320–400 nm), UVB (290–320), and UVC (200–290 nm) (Kevin 1994). UVA (320–400 nm) constitutes the large majority of solar radiation (Douki et al. 2003). UVA radiation is slightly affected by ozone levels (Weatherhead et al. 1997; WHO 2003) with intensity more constant than UVB during the day and throughout the years (Singh et al. 2006). UVA radiation is able to reach the Earth’s surface and scatters rapidly in water with biologically useful amounts to at least 100-m depth in clear aquatic environments (Losey and Hydes 1998).
Many negative impacts of UVA radiation have been recorded, including skin aging, eye damage, physiological defects, retarded development and immunosuppresssion (Setlow et al. 1993; Setlow and Woodhead 1994; Winckler and Fidhiany 1996; Salo et al. 2000; Clydesdale et al. 2001; WHO 2003; Gallagher and Lee 2006; Dong et al. 2007). UVA has been implicated along with UVB in the development of skin cancers in animals (Gallagher and Lee 2006). Setlow et al. (1993) and Setlow and Woodhead (1994) reported that UVA radiation can induce malignant melanoma in the Xiphophosus fish model. It also induced a behavioral and physiological response with altered and highly variable respiratory intensity. UVA photodestruction of the haem-group in haem-containing enzymes causes direct loss of function and physiological disorder (Gantchev and van Lier 1995; Obermüller et al. 2007). Many other harmful UVA-induced effects were recorded, including reduced hatching rate, increased mortality rate, malformation in embryonic stages, reduced metabolic rate of growing fish, oxidative DNA damage and immunological characteristics (Winckler and Fidhiany 1996; Petersen et al. 2000; Salo et al. 2000; Douki et al. 2003; Merwald et al. 2005; Dong et al. 2007). Furthermore, UVA can have harmful indirect effects, such as the enhanced toxicity of environmental contaminants like polycyclic aromatic hydrocarbons or synergism with pathogens (McCloskey and Oris 1993; Ankley et al. 1994; Mallakin et al. 1999; Little et al. 2000). Such enhanced toxicity of those relatively non-toxic environmental contaminants comes through their UV-induced structural changes (photomodification) or through photosensitization, the mechanisms caused by activation of chemicals bioaccumulated in tissues.
Many of the UVA-induced biochemical and physiological changes are valid as biomarkers using aquatic species for detection of the degree of stress. Changes in the concentrations and activities of enzymes such as LDH and G6PDH (Gaboriau et al. 1993; Tsubai and Matsuo 2002; Armeni et al. 2004; Osman et al. 2007a), DNA damage (Alapetite et al. 1996; Jochen et al. 1996; Yeh et al. 2005) and lipid peroxidation (Morlière et al. 1991; Punnonen et al. 1991; Annie et al. 1993) could be used as biomarkers to identify possible environmental contamination in fishes. So, these biomarkers, in addition to some UVA-induced morphological and histopathological changes, were used in the present work to evaluate the effects of radiation on the early developmental stages (endogenous feeding) of Clarias gariepinus (Burchell 1822), which was considered by many authors, including Degroot (1987), Volckaert et al. (1994), Nguyen and Janssen (2002) and Osman et al. (2007a), as an excellent model for toxicological studies, in addition to its oligotrophic habitats (ponds of 0.5–1.5 m), which are easily penetrated by UV radiation causing detrimental effects. The present findings were also compared and discussed with other types of stress, especially those induced by heavy metals on the larval stages of C. gariepinus (Osman et al. 2007a, b) and other fish species (Nguyen and Janssen 2002; Mekkawy and Lashein 2003).
Materials and methods
Gamete collection
Mature African catfish, C. gariepinus (weight 900–1,500 g) were collected from the River Nile at Assuit, Egypt and transported to the Fish Laboratory, Zoology Department, Assuit University. The criteria applied for the selection of spawners were those described by De Graaf and Janssen (1996). The catfish specimens were kept in 100-l glass tanks to be acclimatized for a 2-week period at 27–29°C, pH = 7.56, with dissolved oxygen 88–94% saturation. The photoperiod was a 12-h light to 12-h dark cycle, and the catfish specimens were fed on a commercial pellet diet (3% of the body weight/day).
For collection of semen, males were anaesthetized with 200 mg/l Ms_222 (tricaine methane sulfonate, Crescent Research Chemicals, Phoenix, AZ) buffered with 800 mg/l sodium bicarbonate, and one of the testes was removed surgically. Alternatively, the fish were killed, and the whole gonads were removed. Blood was cleaned from the testes with surgical towels. The sperms from the testes were pressed through a mesh fabric into a sterile dry petri dish and used directly for dry fertilization. For collection of eggs, ovulation was artificially induced. Females were injected intra-peritoneally with pellets (gonadotropin-releasing hormone analog, GnRHa, D-Ala6, Pro9 Net) containing 2.5–3.0 mg of water-soluble dopamine antagonist metoclopramide (Interfish Ltd., Hungary) dissolved in 0.65% NaCl. One pellet was used per kilogram body weight. About 10–11 h after injection, the fish were stripped, and the eggs were collected in clean dry plastic containers; this was considered dry fertilization.
Experimental setup and sampling
The fertilized eggs were incubated in dechlorinized tap water (pH = 7.56, dissolved oxygen 88–94% saturation, temperature 27–29°C; photoperiod 12:12 light:dark). Exposure started 24 h after fertilization (24 h-PFS) then at intervals, 48, 72, 96, 120, 144 and 168 h PFS. Fertilized eggs were divided into four groups: one control and three groups irradiated once for 15, 30 and 60 min according to Kligman et al. (1983) and Häkkinen and Oikari (2004). Exposure took place in 12 petri dishes (14 cm in diameter and 2.5 cm high) with 3 petri dish replicates for each group. Sampling was done 1 h after UVA-exposure. Four embryos were collected at each sampling point and fixed at −80°C for subsequent measurements. The hatching process started at 22 h post-fertilization.
UVR-A source
The embryos were exposed to UVA (ULTRA-VIOLET Products, Inc. San Cabrial, CA, model UVL-56) using a 6-W self-ballasted long-wave lamp (366 nm) with input voltage 220 V, 60 HZ. The UVA source was fitted at 10 cm above the Petri dish bottom (water level was 2 cm).
Measurements of enzyme activities
Because of the small size of the embryos, whole body homogenates were used for the measurements of the enzyme activities (Osman et al. 2007a). Homogenization of the whole animal is a reliable method that provides a reasonable index of total enzyme activity in individuals whose body mass is predominantly composed of muscle tissue (Berges and Ballantyne 1991; Lemos et al. 2003). The embryos were pulverized under liquid nitrogen, and ~100 mg of ground tissue powder was added to five volumes of buffer [50 mM Tris, pH 7.4,1 Mm ethylene diamine tetra-acetic acid (EDTA) and 2 Mm Mg Cl2]. Tissue was homogenized briefly with an Ultra-T URRAX (temperature was maintained at 4°C during homogenization). The homogenate was centrifuged for 15 min at 10,000g and 4°C, and the supernatant was used for the enzyme activity assay. Activities were determined in the supernatant with a spectrophotometer (Micro Lab 200 Vital Scientific, Dieren, The Netherlands) at a wavelength of 340 nm and at 37°C using kits, Stanbio LDH (UV-Rate) procedure no. 0940 USA for the quantitative determination of lactate dehydrogenase (Kachmar and Moss 1976) and RANDOX Laboratories Ltd., PD410, UK BT294QY, for the quantitative determination of glucose-6-phosphate dehydrogenase (Kornberg 1955). The calculation of the catalytic activity content for the selected enzymes was in accordance with the recommendations of the French Society of Clinical Biology (Societe Francaise de Biologie Clinique 1982). The principle of the assay consists of the kinetic determination of LDH activity, based on the rate of NADH oxidation. We determined the oxidation rate, which is directly proportional to LDH activity, by measuring the decrease in absorbance at 340 nm. To standardize the expressions of enzyme activity, the units of enzyme activity have been defined as the quantity of enzyme that catalyzes the reaction of 1 mmol of substrate per minute. The catalytic concentration was expressed as U/l.
Isoenzymes
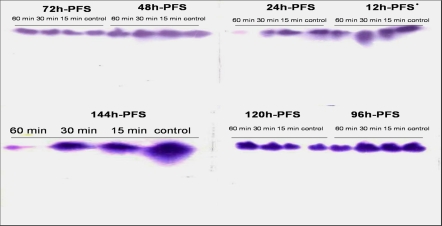
For electrophoresis, adopting the method used by Partington and Mills (l988), ten specimen samples of the control and exposed groups were homogenized by hand using a glass rod. The homogenates were centrifuged (−4°C) at 12,000 rpm for l5 min. The extractions were electrophoresed according to the procedures mentioned in Helena Laboratories Publications (l984) for detection of LDH using cellulose acetate plates. Electra HR buffer, Tittan iso vis cellulose acetate plates and LDH reagent were used for electrophoresis at 300 V (constant voltage) and 10-min duration time at 4°C. The electrophoretic isoenzyme patterns (Plate 1) were analyzed and graphed by Gel-Pro Analyzer Package V3.1 for Windows XP/NT (Media Cybernetica 1993–1997).
Plate 1.
Representatives of the electrophoretic patterns of C. gariepnius embryo LDH enzyme in different larval stages of C. gariepinus under three UVA doses in comparison with the control. h-PFS hours post fertilization stages
Protein analysis by polyacrylamide gel electrophoresis (SDS–PAGE)
Embryos (~0.1 g fresh weight) of each treatment in addition to control were suspended in 1.0 ml lysing buffer, heated at 100°C for 5 min., centrifuged at 10,000 rpm for 30 min, and 50 μl of each extracted protein treatment was used for protein analysis using SDS–PAGE according to Laemmli (1970) in the first dimension. The low-molecular weight standards (Pierce, USA) were run concurrently, and the protein molecular mass was determined using Gel-Pro Analyzer package V3.1 for Windows XP/NT (Media Cybernetica 1993–1997).
Lipid peroxidation and total protein measurements
Total protein contents were determined according to the Biuret method (Gornall et al. 1949) using bovine serum albumin (E. Merck-Darmstadt, Germany) as a standard. Lipid peroxidation (LPO) in the embryos was determined by the procedure of Utley et al. (1967). The absorbance of each aliquot was measured at 535 nm. The rate of lipid peroxidation was expressed as nmol of thiobarbituric acid reactive substance (TBARS) formed per hour per milligram of protein using a molar extinction coefficient of 1.56 M−1 cm−1 (Buege and Aust 1978).
DNA fragmentation measurement
DNA fragmentation was determined according to the procedure of Kurita-Ochiai et al. (1999) using a spectrophotometer (Micro Lab 200 Vital Scientific, Dieren, The Netherlands) at 575 or 600 nm against a reagent blank. In brief, 100 mg of tissue (10% W/V) was added to 1 ml of buffer (102 mg Tris + 29 mg EDTA + 200 μl triton in 100 ml distilled water). Then the mixture was incubated in ice for 10 min and centrifuged at 8,000 rpm for 10 min. The supernatant was used for the measurement of fragmented DNA while the pellet was used for the determination of intact DNA. For the measurement of fragmented DNA, 200 μl of supernatant was added to 200 μl of trichloroacetic acid (TCA). The total volume was centrifuged at 8,000 rpm for 10 min. After centrifugation, 50 μl supernatant or standard was added to 1 ml of diphenylamine reagent, boiled for 10 min in water bath and then cooled on ice. For the determination of intact DNA, the pellet was added to 1 ml buffer (102 mg Tris + 29 mg EDTA + 200 mg SDS in 100 ml distilled water). The mixture was heated in a water bath at 40°C and centrifuged for 10 min at 5,000 rpm; 200 μl of supernatant was added to 200 μl of (TCA). The total volume was centrifuged at 5,000 rpm for 10 min. After centrifugation, 50 μl of supernatant was added to 1 ml of diphenylamine reagent, boiled for 10 min in a water bath and cooled on ice. The developing blue color was measured at 575 or 600 nm against a blank (diphenylamine solution). The percentage of fragmented DNA was estimated by the following formula: percentage of fragmented DNA = fragmented DNA/(fragmented + intact DNA) × 100.
Comet assay
The comet assay was performed according to a modified protocol based on Jarvis and Knowles (2003). Embryos were macerated with forceps in 25 μl Ca2+, Mg2+-free phosphate buffer saline. A volume of 50 μl 1% low melting point (37°C) agarose was added. The total volume (75 μl) was layered onto a frosted microscope slide previously coated with 1% normal melting point agarose. The slide was incubated at 4°C for 15 min to allow solidification and was subsequently coated with an additional layer of 1% low melting point agarose. After solidification at 4°C for 20 min, the embedded cells were lysed in lysing buffer (2.5 M NaCl, 100 mM NaEDTA, 10 mM Tris base, pH 10, 1% Triton X-100, 10% DMSO) at 4°C for 120 min. After 30 min incubation in electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH ≥13) electrophoresis was carried out at 20 V and 300 mA for 30 min. Subsequently, neutralization was performed in three washing steps in 0.4 M Tris–HCl (pH 7.5). The slides were fixed in 100% ethanol (5 min), rehydrated in ultra-pure water (5 min) and stained with ethidium bromide solution (20 μg ml−1) for 10 min, followed by a final washing step in ultra-pure water. Analysis was performed with a Zeiss Axioplan2 fluorescence microscope (200×) and a digital 3 CCD color video camera (Sony, AVT-Horn) using the TriTek Comet image analysis software.
Statistical analysis
The basic statistics, means, standard errors and ranges were estimated. The patterns of variation due to developmental stages and UVA doses and their interaction were studied by two-way analysis of variance using the SPSS package (SPSS 1998) at the 0.05 significance level. Levene’s test of equality of error variance of the dependent variables was applied with rejection of the null hypothesis for raw, log-transformed and SQRT-transformed data. So, the homogeneity of variance was assumed for raw data. The model considered was: intercept + age + treatment + age treatment (adjusted R 2 was 0.998, 0.999, 0.999 and 0.997 for G6PDH, LDH, DNA and LOP, respectively). A further design (age + treatment + age treatment) exhibited a similar significant pattern of variation for the main effect of age and treatment factors and their interaction (P < 0.0001) (adjusted R 2 was 0.999, 1, 0.999 and 0.999 for G6PDH, LDH, DNA and LOP, respectively). The pattern of variation was also recorded by one-way analysis of variance, revealing significant difference within the developmental stages and treatments (P < 0.0001); no homogeneity of variance, even with log and SQRT transformations, was recorded. The Tukey-HSD test was considered for multiple comparisons. Moreover, the Dunnett T test was applied, measuring the control against other treatments in each developmental stage.
Results
Biochemical characteristics
The ontogenetic changes of the activities of G6PDH, LDH, the corresponding DNA damage and LPO/TP ratio during normogenesis were firstly studied as requirements before elucidation of the effects of different UVA doses on these enzymes, DNA and LPO/TP ratio.
The normal ontogenetic variations in enzyme activities, DNA damage and LPO/TP ratio
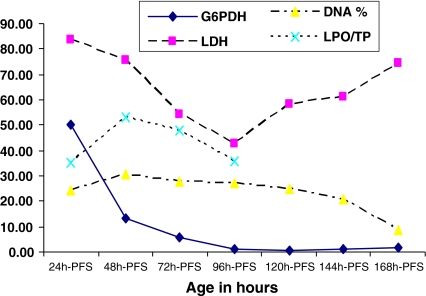
The activities of G6PDH, LDH and the associated DNA damage and lipid LPO/TP ratio showed variability during the early developmental stages (24–168 h-PFS) under normal conditions (Tables 1, 2, 3, 4; Fig. 1). The pattern of the ontogenetic variations exhibited a significant (P < 0.05) exponential decrease in the activities of G6PDH till 120 h-PFS. After 120 h-PFS, the activities of G6PDH increased insignificantly toward higher stages (P > 0.05). A polynomial pattern of variation (6th order, R 2 = 0.9808, P < 0.05) in the LDH activities was recorded with a pattern of decrease till 96 h-PFS, followed by a significant increase toward 168 h-PFS. The polynomial pattern of variation in DNA damage and LPO/TP ratio was also evident. DNA damage across embryonic stages was insignificantly correlated with that of LDH (R = −0.29, P > 0.05) and G6PDH (R = 0.18, P > 0.05) activities. DNA damage increased till 72 h-PFS, then decreased weakly till 168 h-PFS. No significant correlation between LDH activity and LPO/TP ratio (R = 0.02, P > 0.05) was recorded.
Table 1.
Effect of different doses of UVA (366 nm) on the activity of G6PDH (mean ± SE) during early developmental stages of the African catfish Clarias gariepinus
| Embryonic stage (h-PFS)a | Control | 15-min exposure | 30-min exposure | 60-min exposure |
|---|---|---|---|---|
| 24 | 50.15 ± 0.58 (49.16–51.18)a (A) | 31.96 ± 0.89 (30.99–33.74)a (B) | 22.36 ± 0.78 (21.12–23.81)a (C) | 21.91 ± 0.94 (20.02–22.92)a (C) |
| 48 | 13.37 ± 0.25 (12.96–13.84)b (A) | 6.20 ± 0.23 (5.88–6.66)b (B) | 3.45 ± 0.25 (3.04–3.91)b (C) | 3.12 ± 0.08 (2.99–3.27)b (C) |
| 72 | 5.53 ± 0.29 (5.00–6.01)c (A) | 5.25 ± 0.19 (4.99–5.64)b (A) | 6.04 ± 0.08 (5.89–6.18)c (A) | 4.08 ± 0.09 (3.94–4.25)b (B) |
| 96 | 0.98 ± 0.02 (0.94–1.02)de (A) | 1.12 ± 0.06 (1.00–1.22)cd (AB) | 1.38 ± 0.10 (1.18–1.52)de (B) | 0.44 ± 0.02 (0.39–0.48)c (C) |
| 120 | 0.48 ± 0.01 (0.46–0.50)d (A) | 0.54 ± 0.03 (0.48–0.60)c (A) | 0.84 ± 0.03 (0.80–0.90)e (B) | 0.32 ± 0.03 (0.28–0.38)c (C) |
| 144 | 1.00 ± 0.05 (0.92–1.12)de (A) | 1.29 ± 0.09 (1.13–1.44)cd (A) | 1.82 ± 0.10 (1.66–2.00)de (B) | 0.48 ± 0.04 (0.42–0.54)c (C) |
| 168 | 1.79 ± 0.09 (1.62–1.92)e (A) | 2.47 ± 0.12 (2.23–2.64)d (B) | 2.91 ± 0.10 (2.79–3.11)bd (C) | 0.65 ± 0.05 (0.59–0.74)c (D) |
Values are in units per liter
aEmbryonic stages showing similar lower case letters are insignificant within the doses at 0.05 levels (vertical comparison). Stages showing similar capital letters are insignificant within the embryonic stages at 0.05 levels (horizontal comparison)
bThe hatching process started 22 h after fertilization
Table 2.
Effect of different doses of UVA (366 nm) on the activity of LDH (mean ± SE) during early developmental stages of the African catfish Clarias gariepinus
| Embryonic stage (h-PFS)a | Control | 15-min exposure | 30-min exposure | 60-min exposure |
|---|---|---|---|---|
| 24 | 83.5363 ± 0.3 (83.33–83.96)a (A) | 85.9035 ± 1.3(83.24–87.24)a (A) | 94.1692 ± 2.4 (91.77–98.97)a (B) | 109.1475 ± 0.24 (108.67–109.39)a (C) |
| 48 | 75.64 ± 0.95 (74.69–77.57)b (A) | 75.57 ± 0.48 (74.6–76.06)b (A) | 91.99 ± 0.14 (91.73–92.13)a (B) | 118.0 ± 2.28 (113.52–120.98)b (C) |
| 72 | 54.50 ± 1.156 (53.35–56.82)c (A) | 57.64 ± 0.62 (57.02–58.88)c (AB) | 60.40 ± 0.28 (60.13–60.97)b (B) | 80.98 ± 1.51 (79.47–84.01)c (C) |
| 96 | 42.69 ± 0.57 (42.12–43.85)d (A) | 44.58 ± 44.58 (44.28–45.21)d (A) | 47.84 ± 0.091(47.76–48.03)c (B) | 57.37 ± 1.0864 (55.2–58.46)d (C) |
| 120 | 57.98 ± 0.58 (56.82–58.57)e (A) | 62.91 ± 0.18 (62.74–63.28)e (B) | 70.28 ± 0.31 (69.97–70.91)d (C) | 82.46 ± 0.20 (82.05–82.67)c (D) |
| 144 | 61.03 ± 0.14 (60.89–61.33)f (A) | 67.39 ± 0.15 (67.09–67.54)f (B) | 77.85 ± 0.74 (77.11–79.34)e (C) | 118.53 ± 0.47 (117.59–119.01)b (D) |
| 168 | 74.62 ± 0.56 (74.07–75.75)b (A) | 89.85 ± 0.3 (89.25–90.16)g (B) | 172.15 ± 3.14 (169.0–178.44)f (C) | 179.83 ± 2.65 (177.18–185.15)e (C) |
Values are in units per liter
aEmbryonic stages showing similar lower case letters are insignificant within the doses at 0.05 levels (vertical comparison). Stages showing similar capital letters are insignificant within the embryonic stages at 0.05 level (horizontal comparison)
bThe hatching process started 22 h after fertilization
Table 3.
Effect of different doses of UVA (366 nm) on the lipid peroxidation/total protein ratio (mean ± SE) during early developmental stages of the African catfish Clarias gariepinus
| Embryonic stage (h-PFS)a | Control | 15-min exposure | 30-min exposure | 60-min exposure |
|---|---|---|---|---|
| 24 | 26.6224 ± 1.5067 (23.61–8.23)a (A) | 29.5016 ± 1.1457 (27.93–31.73)a (B) | 16.9504 ± 1.7815 (13.83–20)a (C) | 10.608 ± 0.3532 (10.06–1.27)a (D) |
| 48 | 22.0188 ± 1.3983 (20.23–4.77)b (A) | 88.4724 ± 11.9643 (67.65–9.09)b (B) | 57.2116 ± 4.083 (49.43–3.24)b (BC) | 17.336 ± 0.8358 (15.67–18.3)b (C) |
| 72 | 21.3726 ± 0.8808 (19.83–22.89)c (A) | 32.9534 ± 4.6292 (24.06–39.64)b (A) | 13.3229 ± 0.579 (12.7–14.48)c (B) | 10.1434 ± 0.2584 (9.74–10.63)c (B) |
| 96 | 13.0878 ± 0.796 (11.76–14.52)a (A) | 41.4606 ± 3.5011 (34.48–45.45)a (B) | 11.3997 ± 0.8625 (9.7–12.5)d (B) | 5.4423 ± 0.2161 (5.07–5.81)c (B) |
Values are in nmol/mg
aEmbryonic stages showing similar lower case letters are insignificant within the doses at 0.05 level (vertical comparison). Stages showing similar capital letters are insignificant within the embryonic stages at 0.05 levels (horizontal comparison)
bThe hatching process started 22 h after fertilization
Table 4.
Effect of different doses of UVA (366 nm) on the percentage of DNA damage (%) (mean ± SE) during early developmental stages of the African catfish Clarias gariepinus
| Embryonic stage (h-PFS)a | Control | 15-min exposure | 30-min exposure | 60-min exposure |
|---|---|---|---|---|
| 12 | 17.3387 ± 0.2619 (16.67–8.02)a (A) | 20.4602 ± 0.3091 (19.67–21.27)b (A) | 30.9084 ± 0.4669 (29.71–32.13)c (A) | 32.7286 ± 0.4944 (31.46–34.02)d (A) |
| 24 | 24.2003 ± 0.3656 (23.27–5.15)a (B) | 31.6064 ± 0.4775 (30.39–32.85)b (B) | 40.0791 ± 0.6055 (38.53–41.66)c (B) | 50.7535 ± 0.7667 (48.79–52.75)d (B) |
| 48 | 30.3513 ± 0.4585 (29.18–1.55)a (C) | 36.7898 ± 0.5558 (35.37–38.24)b (C) | 50.8409 ± 0.768 (48.88–52.84)c (C) | 70.2654 ± 1.0615 (67.55–73.03)d (C) |
| 72 | 27.6144 ± 0.4172 (26.55–28.7)a (D) | 31.1355 ± 0.4704 (29.93–2.36)b (B) | 44.9846 ± 0.6796 (43.25–46.76)c (D) | 47.6534 ± 0.7199 (45.81–49.53)d (B) |
| 96 | 27.1865 ± 0.4107 (26.14–28.26)a (D) | 33.7149 ± 0.5093 (32.41–5.04)b (D) | 34.1033 ± 0.5152 (32.79–35.45)b (E) | 40.8259 ± 0.6167 (39.25–42.43)c (D) |
| 120 | 24.684 ± 0.3729 (23.73–25.66)a (B) | 25.4957 ± 0.3852 (24.51–26.5)a (E) | 36.2016 ± 0.5469 (34.8–37.63)b (E) | 38.1875 ± 0.5769 (36.71–39.69)c (D) |
| 144 | 20.6185 ± 0.3115 (19.82–21.43)a (E) | 31.6064 ± 0.4775 (30.39–2.85)b (B) | 39.1149 ± 0.5909 (37.6–40.66)c (B) | 41.0052 ± 0.6195 (39.42–42.62)c (D) |
| 168 | 8.6416 ± 0.1305 (8.31–8.98)a (F) | 22.695 ± 0.3428 (21.82–23.59)b (F) | 23.371 ± 0.3531 (22.47–24.29)b (F) | 30.6274 ± 0.4627 (21.82–23.59)c (A) |
aEmbryonic stages showing similar lower case letters are insignificant within the doses at the 0.05 level (vertical comparison). Stages showing similar capital letters are insignificant within the embryonic stages at the 0.05 level (horizontal comparison)
bThe hatching process started 22 h after fertilization
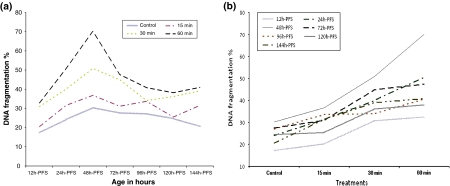
Fig. 1.
Patterns of G6PDH, LDH, DNA and LPO/TP during the different developmental stages of C. gariepinus under normal conditions
The enzyme activities and the corresponding DNA damage and LPO/TP ratio after exposure to UVA in comparison with normogenesis
The patterns of the enzyme activities and corresponding DNA damage and LPO/TP ratio of the different early ontogenetic stages under the influence of three different UVA doses (15, 30 and 60 min) in comparison with the control are revealed (Tables 1, 2, 3, 4). Different patterns of enzyme activities and the corresponding DNA damage and LPO/TP ratio were recorded within each embryonic stage due to exposure to different doses of UVA. Moreover, in addition to their significant main effects (P < 0.0001), the interaction between UVA doses and the embryonic stages was significant (P < 0.0001) in all cases.
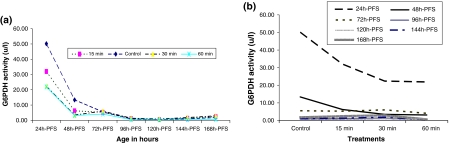
In the three different UVA-treated groups, the activities of G6PDH decreased significantly with developmental progress in a polynomial manner of different order (P < 0.001). In 24 and 48 h-PFS the G6PDH activity decreased significantly (P < 0.001) with increasing UVA dose. In the other treated stages significant fluctuations in G6PDH activity were evident (Table 1) (Fig. 2a, b). The pattern of variations in G6PDH activity in UVA-induced groups was similar to that of the control group with variation in the magnitude of such activity. Significant interaction between treatment and age was evident (P < 0.0001, R 2 = 0.998). No significant correlation between G6PDH activities and DNA damage or LPO/TP ratio in the treated groups (P > 0.05) was recorded, except for 48 h-PFS (R = 0.99, P < 0.01).
Fig. 2.
Effect of UVA irradiation on G6PDH activity during the different developmental stages of C. gariepinus within stages (a) and between treatments (b). h-PFS hours post fertilization stages
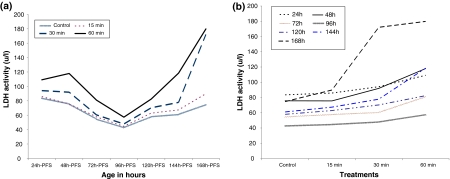
The significant six-ordered polynomial pattern of LDH activity appearing in the control group was evident in the treated ones with variability in the magnitudes. Within each early embryonic stage, the LDH activity increased significantly with the increasing of UVA. In all treated groups, LDH synthesis decreased till 96 h-PFS, then increased (P < 0.0001) toward the next stages (Table 2; Fig. 3a, b). Significant interaction between UVA doses and early embryonic stages was evident (P < 0.0001, R 2 = 1).
Fig. 3.
Effect of UVA irradiation on LDH activity during the different developmental stages of C. gariepinus within stages (a) and between treatments (b). h-PFS hours post fertilization stages
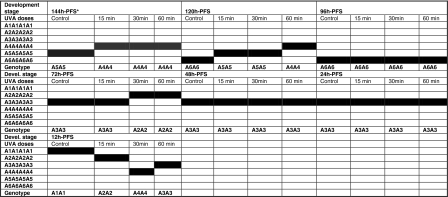
The electrophoretic pattern of LDH (Plate 1) referred to the fact that UVA exposure led to activation and inactivation of LDH alleles through different developmental stages in comparison with controls; a genetic switching process of alleles of LDH locus was identified (Table 5) to adapt the UVA exposure.
Table 5.
Different LDH alleles and genotypes identified in different larval stages of C. gariepinus under three UVA doses in comparison with the control
* h-PFS hours post fertilization stages
DNA damage fluctuated under UVA exposure (P < 0.0001) with two peaks, one at 72 h-PFS in all treatments. Within each of the embryonic stages, the UVA increase led to a significant increase in a linear trend in DNA damage (P < 0.0001) (Table 4; Fig. 4a, b). The significant interaction between UVA exposure and embryonic stages was evident (P < 0.0001). No significant correlation between DNA damage and LDH or G6PDH of the treated groups was recorded. DNA damage was significantly correlated (P < 0.0001) with the LPO/TP ratio in 15- and 60-min UVA doses (R = 0.76 and 0.78 respectively).
Fig. 4.
Effect of UVA irradiation on DNA fragmentation % during the different developmental stages of C. gariepinus within stages (a) and between treatments (b). h-PFS hours post fertilization stages
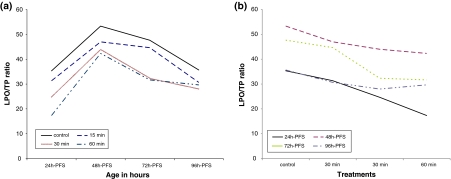
Significantly increased lipid peroxidation (LPO) under UVA stress was recorded in the early larval stages (endogenous feeding) of C. gariepinus (P < 0.001). Such LPO was found to be correlated with DNA damage (P < 0.01). The corresponding total protein exhibited fluctuation with high values in 96 h-PFS. Interaction between UVA doses and larval age was highly significant for LPO and TP. LPO/TP ratio reflected another pattern of variations. A decreased linear trend in the LPO/TP ratio was evident in the early embryonic stages (24–96 h-PFS) with the increase of UVA doses (Table 3) (Fig. 5a, b). In each treated group, an increase in LPO/TP ratio followed by a decrease was evident (P < 0.0001), a situation recorded in the control group with different magnitudes. Except for 24 and 72 h-PFS, insignificant negative correlations were recorded. In the 15- and 60-min UVA doses the LPO/TP ratio was significantly correlated with DNA damage (R = 0.76 and R = 0.78 respectively) in comparison with the control and 30-min UVA doses.
Fig. 5.
Effect of UVA irradiation on LPO/TP ratio during the different developmental stages of C. gariepinus within stages (a) and between treatments (b). h-PFS hours post fertilization stages
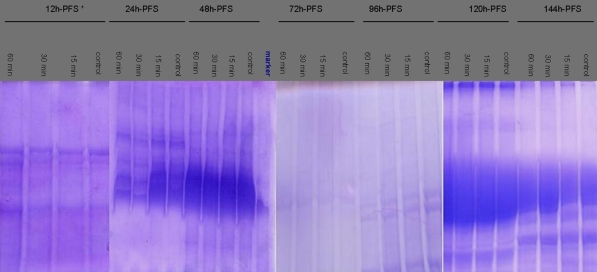
In addition to the aforementioned biomarkers identified under UVA stress, the protein fractions (Fig. 6; Tables 6, 7) were identified in terms of their molecular weights and percentage.
Fig. 6.
Protein fractions identified in different larval stages of C. gariepinus under different doses of UVA (366 nm) radiation in comparison with control. * h-PFS hours post fertilization stages
Table 6.
Molecular weight (in kDa) identification of protein fractions in different larval stages of C. gariepinus under different doses of UVA (366 nm) radiations in comparison with control
| Stage | 12 h-PFSa | 24 h-PFS | 48 h-PFS | 72 h-PFS | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lanes | Marker | Control | 15 Min | 30 Min | 60 Min | Control | 15 Min | 30 Min | 60 Min | Control | 15 Min | 30 Min | 60 Min | Control | 15 Min | 30 Min | 60 Min |
| Protein fractions | Molecular weight | ||||||||||||||||
| r1 | 239.1 | 240.9 | |||||||||||||||
| r2 | 223 | 221.2 | 224.8 | 221.2 | 224.8 | 231.1 | 231.9 | 226.6 | 223.9 | 223 | 223.9 | 227.5 | |||||
| r3 | 216.7 | 208.6 | 212.2 | 214.9 | |||||||||||||
| r4 | 196.1 | 195.2 | 188.9 | ||||||||||||||
| r5 | 168.3 | 167.4 | |||||||||||||||
| r6 | 154.8 | ||||||||||||||||
| r7 | 136 | 132.4 | |||||||||||||||
| r8 | 125.3 | 123.5 | 127 | ||||||||||||||
| r9 | 110 | 108.2 | 114.4 | 106.3 | 113.6 | 111.8 | 110.9 | 109.1 | |||||||||
| r10 | 98.1 | 101.8 | 94.4 | 97.2 | 102.7 | ||||||||||||
| r11 | 93.5 | 88 | 83.4 | ||||||||||||||
| r12 | 81.6 | 80.5 | |||||||||||||||
| r13 | 70.7 | 72.9 | 73.9 | 73.9 | |||||||||||||
| r14 | 62 | 57.7 | 68.5 | 63.1 | 67.5 | 62 | |||||||||||
| r15 | 46.8 | 45.9 | 46.3 | 54.4 | 53.3 | 54.4 | 50.1 | 48.9 | |||||||||
| r16 | 45.7 | 44.7 | 43 | 44.9 | 43.3 | 45.7 | |||||||||||
| r17 | 38.8 | 39.6 | 40.6 | ||||||||||||||
| r18 | 38.5 | 36.9 | |||||||||||||||
| r19 | 31.8 | 34.7 | 34.7 | 33.7 | 34.2 | 34.7 | 34.7 | 34.5 | 33 | ||||||||
| r20 | |||||||||||||||||
| r21 | 24.8 | ||||||||||||||||
| r22 | |||||||||||||||||
| r23 | |||||||||||||||||
| r24 | 18.9 | 18.7 | |||||||||||||||
| r25 | 16.5 | 14.9 | 14.5 | 16.1 | 15.7 | ||||||||||||
| r26 | 11.9 | 11.6 | |||||||||||||||
| Stage | 96 h-PFS | 120 h-PFS | 144 h-PFS | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lanes | Marker | Control | 15 Min | 30 Min | 60 Min | Control | 15 Min | 30 Min | 60 Min | Control | 15 Min | 30 Min | 60 Min | ||||
| Protein fractions | Molecular weight | ||||||||||||||||
| r1 | 244.5 | ||||||||||||||||
| r2 | 223 | ||||||||||||||||
| r3 | 205.9 | 207.7 | 207.7 | 206.9 | 209.5 | 214.9 | 207.8 | 210.4 | |||||||||
| r4 | |||||||||||||||||
| r5 | 165.6 | 168.3 | 170.9 | 169.2 | |||||||||||||
| r6 | 152.2 | 149.5 | 152.1 | 147.7 | |||||||||||||
| r7 | 141.4 | 140.5 | 143.2 | 137.8 | |||||||||||||
| r8 | |||||||||||||||||
| r9 | 110 | 105.4 | 114.5 | 114.5 | 112.7 | ||||||||||||
| r10 | 98.1 | ||||||||||||||||
| r11 | 88 | 83.4 | |||||||||||||||
| r12 | 81.6 | ||||||||||||||||
| r13 | |||||||||||||||||
| r14 | 66.4 | 64.2 | 65.3 | 58.8 | |||||||||||||
| r15 | 46.8 | 56.6 | 55.5 | 47.9 | 46.3 | ||||||||||||
| r16 | 42.5 | 43.3 | 43.1 | ||||||||||||||
| r17 | 38.8 | 40.9 | 41.4 | 41.2 | 40.1 | 40.9 | |||||||||||
| r18 | 35.3 | 35.8 | |||||||||||||||
| r19 | 31.8 | 34.5 | 32.9 | ||||||||||||||
| r20 | 28.1 | 28.8 | 27.5 | 27.5 | 28.8 | 30.8 | 27.8 | ||||||||||
| r21 | 24.8 | 24.8 | 24.8 | 25.8 | 25.5 | 25.5 | |||||||||||
| r22 | 23 | 22.4 | 22.8 | 22.2 | 24 | 23 | 23 | ||||||||||
| r23 | 20.1 | 19.7 | |||||||||||||||
| r24 | 17.1 | 17.1 | 19.1 | 19.5 | 18.1 | 18.3 | |||||||||||
| r25 | 16.5 | 16.3 | 16.7 | 15.7 | 15.71 | ||||||||||||
| r26 | 11.9 | 11.9 | 12.2 | 12.9 | 12.9 | ||||||||||||
a h-PFS hours post fertilization stages
Table 7.
Protein fractions (in percent) identified in different larval stages of C. gariepinus under different doses of UVA (366 nm) radiation in comparison with control
| Stage | 12 h-PFSa | 24 h-PFS | 48 h-PFS | 72 h-PFS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lanes | Control | 15 min | 30 min | 60 min | Control | 15 min | 30 min | 60 min | Control | 15 min | 30 min | 60 min | Control | 15 min | 30 min | 60 min |
| Protein fractions | % | |||||||||||||||
| r1 | 32.9 | 14.1 | 11 | 58 | ||||||||||||
| r2 | 42.2 | 48.3 | 301 | 35.1 | 37 | 40.1 | 25 | 44.6 | 11 | 29.5 | ||||||
| r3 | 25.3 | 20 | 10 | 7.2 | 13.8 | |||||||||||
| r4 | 6.2 | 9.8 | ||||||||||||||
| r5 | 11.1 | |||||||||||||||
| r6 | ||||||||||||||||
| r7 | 15.1 | 28.3 | ||||||||||||||
| r8 | 20.4 | 30.4 | 29.2 | 23.7 | 21 | |||||||||||
| r9 | 2 | 38 | 29.4 | 34.1 | 11.6 | |||||||||||
| r10 | 4.2 | 2.8 | 18.4 | 10.8 | 37.2 | |||||||||||
| r11 | 2.4 | 29.4 | 22.5 | 9.4 | ||||||||||||
| r12 | 3.5 | 18.9 | ||||||||||||||
| r13 | 1.9 | 2.9 | 42 | 46.9 | ||||||||||||
| r14 | 8.3 | 3.4 | 20.5 | 13.4 | 20.2 | |||||||||||
| r15 | 10.2 | 11.1 | 14.1 | 16.6 | 5.6 | 5.4 | 8.1 | 24.7 | ||||||||
| r16 | 2.4 | 11.3 | 8.9 | 12 | 12.2 | |||||||||||
| r17 | 12.6 | 7.5 | 8.4 | |||||||||||||
| r18 | 6.8 | 16.2 | ||||||||||||||
| r19 | 23 | 13.9 | 25.8 | 19.2 | 22.8 | 40.6 | 37.2 | 12 | ||||||||
| r20 | ||||||||||||||||
| r21 | ||||||||||||||||
| r22 | ||||||||||||||||
| r23 | ||||||||||||||||
| r24 | 13.1 | 11 | ||||||||||||||
| r25 | 6.2 | 3 | 7.4 | 5.4 | ||||||||||||
| r26 | 1 | |||||||||||||||
| Stage | 96 h-PFS | 120 h-PFS | 144 h-PFS | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lanes | Control | 15 min | 30 min | 60 min | Control | 15 min | 30 min | 60 min | Control | 15 min | 30 min | 60 min | ||||
| Protein fractions | % | |||||||||||||||
| r1 | 15.2 | |||||||||||||||
| r2 | ||||||||||||||||
| r3 | 25.7 | 30.3 | 30.4 | 27.9 | 29.1 | 32.5 | 34.5 | 27.3 | ||||||||
| r4 | ||||||||||||||||
| r5 | 10.3 | 5.8 | 16.2 | 27.9 | ||||||||||||
| r6 | 3.8 | 4.3 | 1.7 | 5.4 | ||||||||||||
| r7 | 5.4 | 4.9 | 3.6 | 3.7 | ||||||||||||
| r8 | ||||||||||||||||
| r9 | 18.2 | 15.9 | 11.6 | 6.8 | ||||||||||||
| r10 | 5.8 | |||||||||||||||
| r11 | 31.1 | 32.8 | ||||||||||||||
| r12 | ||||||||||||||||
| r13 | ||||||||||||||||
| r14 | 11.5 | 12 | 10.9 | 9.7 | ||||||||||||
| r15 | 36.8 | 33 | 45 | 46.3 | ||||||||||||
| r16 | 7.5 | 7 | 44.1 | |||||||||||||
| r17 | 5.9 | 42.6 | 5.5 | 3.4 | 6.2 | 0 | ||||||||||
| r18 | 5.7 | 1.2 | ||||||||||||||
| r19 | 1.2 | 0 | ||||||||||||||
| r20 | 9.6 | 7.9 | 7.8 | 9.5 | 5.1 | 2.9 | 49.8 | |||||||||
| r21 | 2.1 | 6.1 | 12 | 9.2 | 40 | |||||||||||
| r22 | 6.8 | 6 | 8.1 | 8.8 | 5.2 | 5.2 | 5.5 | |||||||||
| r23 | 19.8 | 18.2 | ||||||||||||||
| r24 | 9.9 | 6.5 | 17.7 | 20.9 | 9.6 | 10.6 | ||||||||||
| r25 | 11.6 | 7.7 | 12.9 | 13.4 | ||||||||||||
| r26 | 7.7 | 6.1 | 8.3 | 7.2 | ||||||||||||
a h-PFS hours post fertilization stages
Discussion
Except for the reports of Mekkawy and Lashein (2003) and Osman et al. (2007a), there is no literature available concerning the pattern of ontogenetic metabolic enzyme activities in early developmental stages of fish before hatching and before exogenous feeding. However, ontogenetic variations during development after the start of exogenous feeding have been reported (Somero and Childress 1985; Clarke et al. 1992; Segner and Verreth 1995).
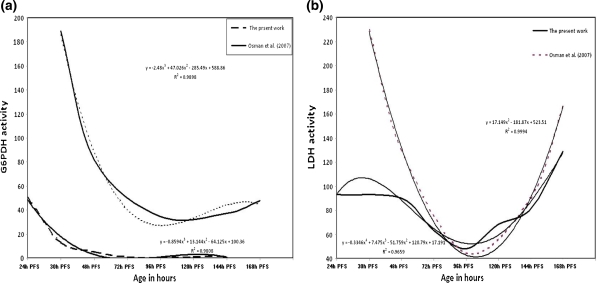
The ontogenetic variations in two metabolic enzyme systems, LDH and G6PDH, of some endogenous-feeding embryonic stages of Ctenopharyngodon idellus and C. gariepinus were studied by Mekkawy and Lashein (2003) and Osman et al. (2007a). These authors discussed the progress of activation and inactivation of the maternal and zygotic genes through those early developmental prehatching stages. The switching time (i.e., blastula, gastrula and organogenesis) from maternal switching time to zygotic LDH and G6PDH-genes was determined. In the present work the enzymatic activities of G6PDH and LDH and the corresponding DNA damage and LPO exhibited variability during the early embryonic stages (24–168 h-PFS), a period of post-hatching intervals with endogenous feeding. The pattern of variations in G6PDH activity exhibited a significant decrease till 120 h-PFS, followed by a relative overall insignificant increase, a polynomial trend (R 2 = 0.999) with a relatively low level of decrease (Fig. 7a). A similar polynomial trend in G6PDH decrease followed by a significant increase was recorded by Osman et al. (2007a) in the same species; the rate of decrease in Osman et al. (2007a) was best described by a power functional equation and by a polynomial equation in the present work (Fig. 7a). The differences in patterns and magnitudes of G6PDH activities in the present work and that of Osman et al. (2007a) may be due to water temperature (29 and 27°C, respectively), species strains and enzyme activity measurement conditions. The decrease of G6PDH activities till 96 h-PFS (Osman et al. 2007a) or 120 h-PFS (present work) of C. graiepinus led to the conclusion that the embryo in its early embryonic stages depended on the maternal enzymes. A similar conclusion can be postulated for LDH activities (Fig. 7b); after such stages, the zygotic genes began to work as expressed by the increasing activities of these enzymes (Mekkawy and Lashein 2003). A similar utilization of the maternal G6PDH and LDH enzyme stores and their subsequent degradation before zygotic translation of mRNA was postulated by Mekkawy and Lashein (2003) for grass carp, Ctenopharyngodon idellus. Those authors, on a genetic basis, concluded that the zygotic genes of the C. idellus embryo appear to be inactive up to the process of organogenesis. In C. gariepinus there is still such inactivation up to 96 h-PFS, during which time the major switching process between maternal and zygotic gene activation occurs. This means the utilization, still, of maternal enzyme mRNA for a long period, in comparison with C. idellus (Mekkawy and Lashein 2003) and other fish species in which maternal proteins are synthesized up to the blastula stages, or even to gastrulation.
Fig. 7.
The activities of G6PDH and LDH in the different developmental stages of C. gariepinus in control embryos in comparison with Osman et al. (2007a). a G6PDH, b LDH. PFS post fertilization stage
The increase in G6PDH and LDH activities from 96, 120 h-PFS onwards, before the onset of exogenous feeding (at 196 h-PFS of C. gariepinus) (Osman et al. 2007a), indicates the early substantial capacity for NADPH generation based on switching on zygotic gene mechanisms (Segner and Verreth 1995). Segner and Verreth (1995) referred to such a capacity existing from the onset of exogenous feeding onwards, as evidenced by the increase of G6PDH activity with age. Many authors referred to the increase of LDH and G6PDH enzyme activities with increasing larval age of different fish species (Pelletier et al. 1995; Nathanailides 1996). However, Nathanailides (1996) referred to the decrease of LDH activity during the development of Tilapia larvae.
Maintenance of genome integrity during development is critical. So, can one detect DNA damage and LPO during normogensis of C. graiepinus? In the present work, DNA damage and LPO have been detected as evidenced by the fact that cell division in the developing embryo leads to more exposure of DNA to unknown agents and oxidative stress; the genome transcriptionally becomes active. The DNA damage and LPO have insignificant correlation with LDH and G6PDH activities (P > 0.05). Moreover, an insignificant relatively high correlation between DNA damage and LPO was evident (R = 0.73).
The UVA-exposed embryos exhibited a pattern of enzyme activities similar to that of the normal embryos, but with a significant variability in the magnitude. However, within the embryonic stage in comparison with the control, two patterns of enzyme activities were identified, a sharp decreased polynomial pattern (order 2) in 24 and 48 h-PFS and a polynomial one (order 3) with a peak in 30-min dose in the remainder of embryonic stages (72–168 h-PFS). These findings referred to UVA-induced inactivation of G6PDH, especially in the early larval stages, which have higher G6PDH activities in comparison with later ones. A few studies referred to such UV-induced inactivation of G6PDH (Dovrat and Weinreb 1995; Thomas et al. 2001; Lee et al. 2002; Andrew and Jacob 2003; Estey et al. 2007). Other studies recorded an increase in such enzyme activities under UVA radiation, emphasizing its antioxidant role (Tsubai and Matsuo 2002). A similar increase in G6PDH activity under pollutant stress was recorded (Wu and Lam 1997; Stephensen et al. 2000; Pandey et al. 2003; Rosety-Rodriguez et al. 2005; Osman et al. 2007a). The high activity of G6PDH was due to the increased production of NADH for the detoxification process (Stephensen et al. 2000; Osman et al. 2007a). In the present work, the detoxification process or antioxidant role of G6PDH was absent and may be related to the increased DNA damage through the growing developmental stages and increased doses of UVA radiation. Such damage may lead to a decreased rate of transcription with an insignificant contribution of regulatory mechanisms acting at post-transcriptional levels.
LDH is generally associated with cellular metabolic activity (Osman et al. 2007a). Such activity is inhibited under stress, especially after exposure to heavy metals (Singh and Sharma 1998; Almeida et al. 2001; Elumalai et al. 2002; Osman et al. 2007a). Inhibition of the enzyme activity may be due to the formation of an enzyme-inhibition complex (Singh and Sharma 1998; Osman et al. 2007a), to ion imbalance or to intracellular action of metal subsequent to initial plasma membrane damage (Sastry and Gupta 1980). Some authors (Dovrat and Weinreb 1995; Thomas et al. 2001; Lee et al. 2002; Andrew and Jacob 2003; Estey et al. 2007) referred to the inactivation of LDH activity under UVA radiation, whereas other recorded an increase in UVA-induced enzyme activity (Dovrat and Weinreb 1995; Thomas et al. 2001; Lee et al. 2002; Andrew and Jacob 2003; Estey et al. 2007). Does UVA radiation disrupt the balance between the overall stability and flexibility of LDH? Coquelle et al. (2007) discussed such a balance and amino acid substitutions in thermal environments. The presence of different isoenzymes of LDH and switching between the activation and inactivation process of their alleles during development and under stress (Mekkawy and Lashein 2003) facilitate the balance process between enzyme stability and flexibility. UVA-induced LDH electrophoretic patterns of C. graepinus in the present work in comparison with the control through the developmental stages emphasized that in these findings UVA radiation did not change the polynomial pattern of LDH activity, but affected the magnitude with increases of UVA doses, especially in 168 h-PFS. Such an increased response of LDH activity to UVA referred to a responsible degree of cell membrane damage, to its protective role against oxidative damage and to the increase of anaerobic activity. The UVA-induced damage in membrane properties may be related to the lipid peroxidation process (Gaboriau et al. 1993). Fu et al. (2000) postulated that UVA increased the release of the plasma enzyme LDH and lipid peroxidation, but decreased the activity of glutathione peroxidase. In the present work LDH release as a sign of cell membrane damage and its decreased fluidity (Legrand et al. 1992; Gaboriau et al. 1993) was not correlated with lipid peroxidation under stress (P > 0.05).
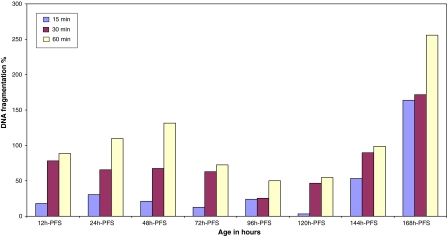
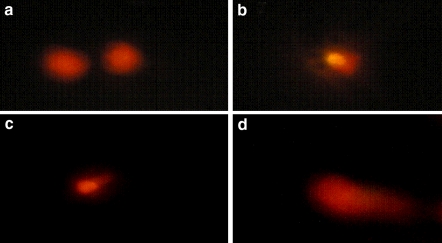
The adverse effect of UV radiation on DNA was considered by many authors (Armstrong et al. 2002; Sinha and Häder 2002; Browman et al. 2003; Ciereszko et al. 2005; Kino and Sugiyama 2005). The absolute and relative amounts of DNA damage vary according to the specific wavelength of radiation (Kielbassa et al. 1997) in addition to their doses, age and species. While UV radiation, particularly UVA, can assist in repairing DNA damage, the primary effect of UV appears to be damaging to both DNA and individual cells (Weatherhead and Stevermer 2001). So, does the DNA damage recorded in the UVA-treated fish represent a primary effect of UVA radiation? What are the limits of repairing and damaging DNA? The range of differences in DNA damage in comparison with the control was 3.3 (120 h-PFS)–163.8% (168 h-PFS), 25.44 (96 h-PFS)–171.7% (168 h-PFS) and 50.2 (96 h-PFS)–255.8% (168 h-PFS) for 15, 30 and 60 min of exposure, respectively (Fig. 8). These findings may be considered as limits of no damage repair. So, one can conclude that the acute exposure to UVA inhibits the developed repair or tolerance mechanisms that counteract the DNA damage caused by UV photoreactivation with the help of the enzyme photolyase, which may be severely suppressed under this acute exposure. DNA damage is due to UV-induced impairment of SOD and catalase, two important components of the antioxidants defense system. The UVA-induced changes recorded in DNA of C. gariepinus contradicted the statement of Sinha and Häder (2002) that UVA wavelengths are less efficient in inducing DNA damage. These authors interpreted their conclusion by the assumption that UVA is not absorbed by native DNA, but it still produces secondary photoreactions of existing DNA photoproducts and damages DNA via indirect photosensitizing reactions. McFadzen et al. (2000) stated that the primary products of UV radiation are rapidly formed free radicals producing biological effects lasting from minutes to years. DNA is the main target for UV radiation-induced damage. Preliminarily, the DNA damage in the present work was confirmed by Comet Assay technique (Fig. 9), suggesting further study using such a technique (Osman et al. 2008).
Fig. 8.
DNA fragmentation % in the different developmental stages of C. gariepinus during exposure to different doses of UVA in comparison with control. h-PFS hours post fertilization stage
Fig. 9.
Grade of DNA damage (representative stages) of embryos of C. gariepinus exposed to UVA. a Control, undamaged nucleus of 144 h-PFS; b 15 min, low damage nucleus of 120 h-PFS; c 30 min, median damaged nucleus of 96 h-PFS; d 60 min, highly damaged nucleus of 72 h-PFS. h-PFS hours post fertilization stage
The increased DNA damage in the UVA-exposed fish may be due to the deficiencies of natural processes, such as the chromatin package and abortive apoptosis during development and cell division. Petersen et al. (2000) also reported that UVA irradiation increased the intracellular levels of hydrogen peroxide (H2O2) and caused oxidative DNA damage, single strand breaks and alkali-lobile sites. Douki et al. (2003) stated that bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage from solar UVA radiation. UVA-induced DNA damage of C. gariepinus endogenous feeding larvae was found to be positively correlated with LPO. These results were emphasized by many authors (Armstrong et al. 2002; Sinha and Häder 2002; Browman et al. 2003; Ciereszko et al. 2005; Kino and Sugiyama 2005) referring to the oxidative nature of DNA damage.
In conclusion, this work showed the destructive effects of ultraviolet-A radiation (366 nm) on the biochemical characteristics of the early developmental stages of African catfish, C. gariepinus.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alapetite C, Wachter T, Sage E, Moustacchi E. Use of alkaline comet assay to detect DNA repair deficiencies in human fibroblasts exposed to UVC, UVB, UVA and gamma-rays. Int J Radiat Biol. 1996;69:359–369. doi: 10.1080/095530096145922. [DOI] [PubMed] [Google Scholar]
- Almeida JA, Novelli ELB, Silva MD, Alves R (2001) Environmental cadmium exposure and metabolic responses of the Nile tilapia, Oreochromis niloticus. Environ Pollut 114:169–175 [DOI] [PubMed]
- Andrew TEH, Jacob GS (2003) Epithelial activity of hexokinase and glucose-6-phosphate dehydrogenase in cultured bovine lenses recovering from pharmaceutical-induced optical damage. Mol Vis 9:594–600 [PubMed]
- Ankley GT, Collyard SA, Monson PD, Kosian PA. Influence of ultraviolet light on the toxicity of sediment contaminated with polycyclic aromatic hydrocarbons. Environ Conta Toxicol. 1994;13:1791–1796. [Google Scholar]
- Annie M, Marquis I, Gaboriau F, Santus R, Dubertret L, Morlière P. Ultraviolet A-induced lipid peroxidation and antioxidant defense systems in cultured human skin fibroblasts. J Invest Dermatol. 1993;100:692–698. doi: 10.1111/1523-1747.ep12472352. [DOI] [PubMed] [Google Scholar]
- Armeni T, Damiani E, Battino M, Greci L, Principato G. Lack of in vitro protection by a common sunscreen ingredient on UVA-induced cytotoxicity in keratinocytes. Toxic. 2004;203:165–178. doi: 10.1016/j.tox.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Armstrong TN, Reimschuessel R, Bradly BP (2002) DNA damage, histological changes and DNA repair in larval Japanese medaka (Oryzias latipes) exposed to ultraviolet-B radiation. Aquat Toxicol 58:1–14 [DOI] [PubMed]
- Berges JA, Ballantyne JS (1991) Size scaling of whole body maximal enzyme activities in aquatic crustaceans. Can J Fish Aquac Sci 48:2385–2394
- Brian LD. Sources and measurements of ultraviolet radiation. Methods. 2002;28(1):4–13. doi: 10.1016/S1046-2023(02)00204-9. [DOI] [PubMed] [Google Scholar]
- Browman HI, Vetter RD, Flodriguez CA, Cullen JJ, Davis RF, Lynn E, Pierre J (2003) Ultraviolet (280–400 nm)-induced DNA damage in the eggs and larvae of Calanus finmarchicus G. (Copepoda) and atlantic cod (Gadus morhua). Photochem Photobiol 77(4):397–404 [DOI] [PubMed]
- Buege JA, Aust SD. Microsomal lipid peroxidation. Methods Enzymol. 1978;52:302–310. doi: 10.1016/S0076-6879(78)52032-6. [DOI] [PubMed] [Google Scholar]
- Castro-Pérez CA (2004) Effects of ultraviolet radiation exposure on the swimming performance and hematological parameters of tambaqui, colossoma macropomum. International Congress on the Biology of Fish, Tropical Hotel Resort, Manaus Brazil, August: 1–5
- Ciereszko A, Tobie DW, Konrad D (2005) Analysis of DNA damage in sea lamprey (Petromyzon marinus) spermatozoa by UV, hydrogen peroxide, and the toxic antbisazir. Aquat Toxicol 73(2):128–138 [DOI] [PubMed]
- Clarke ME, Calvi C, Domeier M, Edmonds M, Walsh PJ (1992) Effects of nutrition and temperature on metabolic enzyme activities in larval and juvenile red drum, Sciaenops ocellatus and lane snapper, Lutjanus synagris. Marine Biol 112:31–36
- Clydesdale GJ, Dandie GW, Muller HK. Ultraviolet light induced injury: Immunological and inflammatory effects. Immunol Cell Biol. 2001;79(6):547–568. doi: 10.1046/j.1440-1711.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Coquelle N, Fioravanti E, Weik M, Vellieux F, Madern D (2007) Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments. J Mol Biol 374:547–562 [DOI] [PubMed]
- De Graaf GJ, Janssen H (1996) Artificial reproduction and pond rearing of the African catfish Clarias gariepinus in sub-Saharan Africa. FAO, Fish Tech Pap 362:1–73
- Degroot SJ. Culture of Clarias species. Aquaculture. 1987;63:1–36. doi: 10.1016/0044-8486(87)90056-1. [DOI] [Google Scholar]
- Dong Q, Svoboda K, Tiersch TR, Monroe WT. Photobiological effects of UVA and UVB light in zebrafish embryos: evidence for a competent photorepair system. J Photochem Photob B Biol. 2007;88:137–146. doi: 10.1016/j.jphotobiol.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochem. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- Dovrat A, Weinreb O (1995) Recovery of lens optics and epithelial enzymes after ultraviolet A radiation. Invest Ophthalmol Vis Sci 36:12 [PubMed]
- Elumalai M, Antunes C, Guilhermino L (2002) Effects of single metals and their mixtures on selected enzymes of Carcinus maenas. Water Air Soil Pollut 141:273–280
- Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V (2007) Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem 282(7):4382–4392 [DOI] [PubMed]
- Fu Y, Jin X, Wei S, Lin H, Kacew S (2000) Ultraviolet radiation and reactive oxygen generation as inducers of keratinocyte apoptosis: protective role of tea polyphenols. J Toxicol Environ Health 61(3):177–188 [DOI] [PubMed]
- Gaboriau F, Morliére P, Marquis I, Moysan A, Géze M, Dubertret L. Membrane damage induced in cultured human skin fibroblasts by UVA irradiation. Photochem Photobiol. 1993;58:515–520. doi: 10.1111/j.1751-1097.1993.tb04924.x. [DOI] [PubMed] [Google Scholar]
- Gallagher RP, Lee TK. Adverse effects of ultraviolet radiation: a brief review. Prog Biophys Mol Biol. 2006;92:119–131. doi: 10.1016/j.pbiomolbio.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Gantchev TG, van Lier JE. Catalase inactivation following photosensitization with tetrasulfonated metallophtalocyanines. Photochem Photobiol. 1995;62:123–134. doi: 10.1111/j.1751-1097.1995.tb05248.x. [DOI] [PubMed] [Google Scholar]
- Gornall AC, Bardawill CJ, David MM (1949) Determination of serum proteins by means of the Biuret reaction. J Biol Chem 177:751–766 [PubMed]
- Häkkinen J, Oikari A (2004) A field methodology to study effects of UV radiation on fish larvae. Water Res 38:2891–2897 [DOI] [PubMed]
- Jarvis RB, Knowles JF (2003) DNA damage in zebrafish larvae induced by exposure to low-dose rate gamma-radiation: detection by the alkaline comet assay. Mutat Res Genet Toxicol Environ Mutagen 541:63–69 [DOI] [PubMed]
- Jochen G, Lidia L, Hubert T, Ute R, Claus U. Isolation, sequencing and overproduction of the single-stranded DNA binding protein from Pseudomonas aeruginosa PAO. Gene. 1996;182:137–143. doi: 10.1016/S0378-1119(96)00535-5. [DOI] [PubMed] [Google Scholar]
- Kachmar JF, Moss DW ( 1976) In: Tietz NW (ed) Fundamentals of clinical chemistry, 2nd edn. WB Saunders, Philadelphia, p 652
- Kevin RA. Impact of ozone depletion on phytoplankton growth in the Southern Ocean: large-scale spatial and temporal variability. Mar Ecolo Prog. 1994;114:1–12. doi: 10.3354/meps114001. [DOI] [Google Scholar]
- Kielbassa C, Len R, Bernd E (1997) Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis 18:811–816 [DOI] [PubMed]
- Kino K, Sugiyama H (2005) UVR-induced G–C to C–G transversions from oxidative DNA damage. Mutat Res 571:33–42 [DOI] [PubMed]
- Kligman LH, Akin FJ, Kligman AM (1983) Sunscreens promote repair of ultraviolet radiation-induced dermal damage. J Investig Dermatol 81:98–102 [DOI] [PubMed]
- Kornberg A (1955) Lactic dehydrogenase of muscle. In: Lowestein JM (ed) Methods in enzymology. Academic Press, New York, pp 441–442
- Kurita-Ochiai T, Fukushima K, Ochiai K. Lipopolysaccharide stimulates butyric acid-induced apoptosis in human peripheral blood mononuclear cells. Infect Immun. 1999;67:22–29. doi: 10.1128/iai.67.1.22-29.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227: 680–685 [DOI] [PubMed]
- Lee SM, Koh H, Park D, Song BJ, Huh T, Park J (2002) Cytosolic NADP+-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med 32(11):1185–1196 [DOI] [PubMed]
- Legrand C, Bour JM, Jacob C, Capiaumont J, Martial J, Marc A, Wudtke M, Kretzmer G, Demangel C, Duval D, Hache J (1992) Lactate dehydrogenase (LDH) activity of the number of dead cells in the medium of cultured eukaryotic cells as marker. J Biotechnol 25:231–243 [DOI] [PubMed]
- Lemos D, Salomon M, Gomes V, Phan V, Buchholz E (2003) Citrate synthase and pyruvate kinase activities during early life stages of the shrimp Farfantepenaeus paulensis (Crustacea, Decapoda, Penaeidae): effects of development and temperature. Comp Biochem Physiol B 135:707–719 [DOI] [PubMed]
- Little EE, Cleveland L, Hurtubise R, Barron MG. Assessment of the photoenhanced toxicity of a weathered petroleum to the tidewater silverside. Environ Toxicol Chem. 2000;19:926–932. doi: 10.1002/etc.5620190420. [DOI] [Google Scholar]
- Losey GS, Hydes D. The UV visual world of fishes. J Fish Biol. 1998;54:921–945. doi: 10.1111/j.1095-8649.1999.tb00848.x. [DOI] [Google Scholar]
- Mallakin A, McConkey BJ, Miao G, Mckibben B, Snieckus V, Dixon DG, Greenberg BM. Impacts of structural photomodification on the toxicity of environmental contaminants: anthracene photooxidation products. Ecotoxic Envir saf. 1999;43:204–212. doi: 10.1006/eesa.1999.1764. [DOI] [PubMed] [Google Scholar]
- McCloskey JT, Oris JT. Effect of anthracene and solar ultraviolet radiation exposure on gill ATPase and selected hematologic measurements in the bluegill sunfish (Lepomis macrochirus) Aquat Toxicol. 1993;24:207–218. doi: 10.1016/0166-445X(93)90072-9. [DOI] [Google Scholar]
- McFadzen I, Baynes S, Hallam J, Beesley A, Lowe D (2000) Histopathology of the skin of UV-B irradiated sole (Solea solea) and turbot (Scophthalmus maximus) larvae. Marine Environ Res 50:273–277 [DOI] [PubMed]
- Mekkawy IAA, Lashein FE (2003) The effect of lead and cadmium on LDH and G-6-PDH isozyme patterns exhibited during the early embryonic development of the teleost fish, Ctenopharyngodon idellus with emphasis on the corresponding morphological variations. The Big Fish Bang, the proceeding of the 26th Annual Larval Fish Conference (LFC2002), 22-26 July, 2002, Bergen, Norway, edited by: Howard I. Browman and Anne Berit Skiftesvik
- Merwald H, Klosner G, Kokesch C, Der-Petrossian M, Honigsmann H, Trautinger F. UVA-induced oxidative damage and cytotoxicity depend on the mode of exposure. J Photochem Photob B Biol. 2005;79(3):197–207. doi: 10.1016/j.jphotobiol.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Morlière P, Annie M, René S, Gabriele H, Jean-Claude M, Louis D. UVA-induced lipid peroxidation in cultured human fibroblasts. BBA Lipids Lipid Metab. 1991;1084(3):261–268. doi: 10.1016/0005-2760(91)90068-S. [DOI] [PubMed] [Google Scholar]
- Nathanailides C (1996) Metabolic specialization of muscle during development in cold water and warm water fish species exposed to different thermal conditions. Can J Fish Aquat Sci 53:2147–2155
- Nguyen LTH, Janssen CR. Embryo-larval toxicity tests with the African catfish (Clarias gariepinus): comparative sensitivity of endpoints. Arch Environ Contam Toxicol. 2002;42:256–262. doi: 10.1007/s00244-001-0007-4. [DOI] [PubMed] [Google Scholar]
- Obermüller B, Puntarrulo S, Abele D. UV-tolerance and instantaneous physiological stress responses of two Antarctic amphipod species Gondogeneia antarctica and Djerboa furcipes during exposure to UV radiation. Mar Environ Res. 2007;64:267–285. doi: 10.1016/j.marenvres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Osman AG, Mekkawy IAA, Verreth J, Kirschbaum F. Effects of lead nitrate on the activity of metabolic enzymes during early developmental stages of the African catfish, Clarias gariepinus (Burchell, 1822) Fish Physiol Biochem. 2007;33:1–13. doi: 10.1007/s10695-006-9111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman AG, Wuertz S, Mekkawy IAA, Exner H, Kirschbaum F. Lead induced malformations in embryos of the African catfish Clarias gariepinus (Burchell, 1822) Environ Toxicol. 2007;22(4):375–389. doi: 10.1002/tox.20272. [DOI] [PubMed] [Google Scholar]
- Osman AG, Mekkawy IAA, Verreth J, Wuertz S, Kloas W, Kirschbaum F. Monitoring of DNA breakage in embryonic stages of the African catfish Clarias gariepinus (Burchell, 1822) after exposure to lead nitrate using alkaline comet assay. Environ Toxicol. 2008;23:679–687. doi: 10.1002/tox.20373. [DOI] [PubMed] [Google Scholar]
- Pandey S, Parvez S, Sayeed I, Haque R, Bin-Hafeez B, Raisuddin S (2003) Biomarkers of oxidative stress: a comparative study of river Yamuna fish Wallago attu (Bl. & Schn.). Sci Total Environ 309:105–115 [DOI] [PubMed]
- Pelletier D, Blier PU, Dutil JD, Guderley H (1995) How should enzyme activities be used in fish growth studies? J Exp Biol 198:1493–1497 [DOI] [PubMed]
- Petersen AB, Gniadecki R, Vicanova J, Thorn T, Wulf HC. Hydrogen peroxide is responsible for UVA-induced DNA damage measured by alkaline comet assay in HaCaT keratinocytes. J photo photob B Biology. 2000;59:123–131. doi: 10.1016/S1011-1344(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Punnonen K, Puntala A, Ansen CT, Ahotupa M. UVB irradiation induces lipid peroxidation and reduced antioxidant enzyme activities in human keratinocytes in vitro. Acta Dermato-venerologica. 1991;1:239–273. [PubMed] [Google Scholar]
- Rosety-Rodriguez M, Ordonez F, Rosety I, Rosety J, Rosery M (2005) Erythrocyte antioxidant enzymes of gilthead as early-warning bio-indicators of oxidative stress induced by malathion. Haematology 8:237–240
- Rozema J, Van Geel B, Bjorn LO, Lean J, Madronich S. Toward solving the UV puzzle. Science. 2002;296:1621–1622. doi: 10.1126/science.1070024. [DOI] [PubMed] [Google Scholar]
- Salo H, Jokinen E, Markkula S, Aaltonen T, Penttilä H. Comparative effects of UVA and UVB irradiation on the immune system of fish. J photo photob B. Biology. 2000;56:154–162. doi: 10.1016/S1011-1344(00)00072-5. [DOI] [PubMed] [Google Scholar]
- Sastry KV, Gupta PK (1980) Alterations in the activities of a few dehydrogenases in the digestive system of 2 teleost fishes exposed to lead nitrate. Ecotoxicol Environ Saf 4:232–239 [DOI] [PubMed]
- Segner H, Verreth J (1995) Metabolic enzyme activities in larvae of the African catfish, Clarias gariepinus—changes in relation to age and nutrition. Fish Physiol Biochem 14:385–398 [DOI] [PubMed]
- Setlow RB, Woodhead AD. Temporal changes in the incidence of malignant melanoma: explanation from action spectra. Mutat Res. 1994;307:365–374. doi: 10.1016/0027-5107(94)90310-7. [DOI] [PubMed] [Google Scholar]
- Setlow RB, Gist E, Thompson K, woodhead Avril D. Wavelengths effective in induction of malignant melanoma. Genetics. 1993;90:6666–6670. doi: 10.1073/pnas.90.14.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Sharma B (1998) Carbofuran-induced biochemical changes in Clarias batrachus. Pest Sci 53:285–290
- Singh SS, Pankaj k, Ashwani k (2006) Ultraviolet radiation stress: molecular and physiological adaptations in trees in A biotic stress tolerance in plants Springer etherlands editer. doi:10.1007/14020-4389
- Sinha RP, Häder D (2002) UV-induced DNA damage and repair: a review. Photochem Photobiol Sci 1:225–236 [DOI] [PubMed]
- Societe Francaise de Biologie Clinique Enzymology commission. Recommendations. Ann Biol Clin (Paris) 1982;40:87–164. [PubMed] [Google Scholar]
- Somero S, Childress J (1985) Scaling of oxidative and glycolytic enzyme activities in fish muscle. Springer, Berlin
- Stephensen E, Svavarsson J, Sturve J, Ericson G, Adolfsson-Erici M, Förlin L (2000) Biochemical indicators of pollution exposure in shorthorn sculpin (Myoxocephalus scorpius), caught in four harbours on the southwest coast of Iceland. Aquat Toxicol 48:431–442 [DOI] [PubMed]
- Thomas E, Ursula F, Rainhardt O (2001) DNA damage and apoptosis in mononuclear cells from glucose-6-phosphate dehydrogenase-deficient patients (G6PD Aachen variant) after UV irradiation. J Leukoc Biol 69:340–342 [PubMed]
- Tsubai T, Matsuo M. Ultraviolet light-induced changes in the glucose 6-phosphate dehydrogenase activity of porcine corneas. Cornea. 2002;21(5):495–500. doi: 10.1097/00003226-200207000-00011. [DOI] [PubMed] [Google Scholar]
- Utley HC, Bernheim F, Hachslien P (1967) Effects of sulfhydryl reagent on peroxidation in microsome. Arch Biochem Biophys 260:521–531
- Volckaert FAM, Hellemans BA, Galbusera P, Ollevier F, Sekkali B, Belayew A. Replication, expression and fate of foreign DNA during embryonic and larval development of the African catfish Clarias gariepinus. Mol Mar Biol Biotechnol. 1994;3:57–69. [PubMed] [Google Scholar]
- Weatherhead EC, Stevermer A (2001) Ultraviolet radiation. In: Encyclopedia of global environmental change, vol 1. John Wiley and Sons Inc
- Weatherhead EC, George CT, Gregory CR, John EF, John JD, Dongseok C. Analysis of long-term behavior of ultraviolet radiation measured by Robertson-Berger meters at 14 sites in the United States. J Geophys Res. 1997;102(d7):8737–8754. doi: 10.1029/96JD03590. [DOI] [Google Scholar]
- Weatherhead SC, Haniffa M, Lawrence CM. Melanomas arising from naevi and de novo melanomas does origin matter? Briti J derma. 2007;156:72–86. doi: 10.1111/j.1365-2133.2006.07570.x. [DOI] [PubMed] [Google Scholar]
- WHO . Climate change and human health—risks and responses—summary. Geneva: World Health Organization; 2003. [Google Scholar]
- Williamson CE. What role does UV-B radiation play in freshwater ecosystems? Limno oceanog. 1995;40(2):386–392. doi: 10.4319/lo.1995.40.2.0386. [DOI] [Google Scholar]
- Williamson CE, Metgar SL, Lovera PA, Moeller RE. Solar ultraviolet radiation and the spawning habitat of yellow Perch, Perca flavescens. Ecol Appl. 1997;7(3):1017–1023. doi: 10.1890/1051-0761(1997)007[1017:SURATS]2.0.CO;2. [DOI] [Google Scholar]
- Winckler K, Fidhiany L. Significant influence of UVA on the general metabolism in the growing Ciclid fish, Cichlasoma nigrofasciatum. J photochem photob B. Biology. 1996;33:131–135. doi: 10.1016/1011-1344(95)07238-1. [DOI] [PubMed] [Google Scholar]
- Wu RSS, Lam PKS (1997) Glucose-6-phosphate dehydrogenase and lactate dehydrogenase in the green-lipped mussel (Perna viridis). Possible biomarker for hypoxia in the marine environment. Water Res 31:2797–2801
- Yeh S, Wang W, Huang C, Hu M. Pro-oxidative effect of β-carotene and the interaction with flavonoids on UVA-induced DNA strand breaks in mouse fibroblast C3H10T1/2 cells. J Nutr Biochem. 2005;16(12):729–735. doi: 10.1016/j.jnutbio.2005.03.012. [DOI] [PubMed] [Google Scholar]