Abstract
Two overlapping quantitative trait loci (QTLs) for clot stability, Hmtb8 and Hmtb9, were identified on mouse chromosome 17 in an F2 intercross derived from C57BL/6J (B6) and B6-Chr17A/J (B6-Chr17) mouse strains. The intervals were in synteny with a QTL for thrombotic susceptibility on chromosome 18 in a human study, and there were 23 homologs between mouse and human. The objective of this study was to determine whether any of these genes in the syntenic region are likely candidates as modifiers for clot stability. Seven genes, Twsg1, Zfp161, Dlgap1, Ralbp1, Myom1, Rab31, and Emilin2, of the 23 genes with single nucleotide polymorphisms (SNPs) in the mRNA-UTR had differential expression in B6 and A/J mice. Dlgap1, Ralbp1, Myom1, and Emilin2 also had nonsynonymous SNPs. In addition, two other genes had nonsynonymous SNPs, Lama1 and Ndc80. Of these nine candidate genes, Emilin2 was selected for further analysis since other EMILIN (Elastin Microfibril Interface Located Protein) proteins have known functions in vascular structure and coagulation. Differences were found between B6 and A/J mice in vessel wall architecture and EMILIN2 protein in plasma, carotid vessel wall, and thrombi formed after ferric chloride injury. In B6-Chr17A/J mice both clot stability and Emilin2 mRNA expression were higher compared to those in B6 and A/J mice, suggesting the exposure of epistatic interactions. Although other homologous genes in the QTL region cannot be ruled out as causative genes, further investigation of Emilin2 as a candidate gene for thrombosis susceptibility is warranted.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-010-9274-6) contains supplementary material, which is available to authorized users.
Introduction
Thrombosis occurs as a consequence of vascular injury due to an imbalance between the pathways that regulate thrombus formation and/or dissolution and is the major fatal event of cardiovascular diseases (Voetsch and Loscalzo 2004). Great strides have been made in the diagnosis and treatment of thrombosis in the last decade. However, strategies to prevent thrombosis have lagged far behind partly because of the contribution of multiple and as yet undefined genetic factors that lead to thrombotic risk. Moreover, it remains unclear how genetic background influences the function of molecules and pathways known to regulate thrombus formation and lysis, thereby contributing to the risk of thrombotic disease (Ginsburg 2005).
To investigate genetic differences in thrombosis, we identified two inbred mouse strains, B6 and A/J, that have marked differences in a tail bleeding/rebleeding assay and response to FeCl3-induced carotid injury (Hoover-Plow et al. 2006). In the tail bleeding/rebleeding assay, an established hemostasis and thrombosis reporter assay, there was no difference in the first bleeding time between the B6 and the A/J mice. Interestingly, the time for the second bleeding to occur was increased in A/J mice compared to B6 mice by nearly threefold, indicating enhanced clot stability (Hoover-Plow et al. 2006) in the A/J mice. A marked difference of twofold in arterial thrombus occlusion time was found between the B6 and the A/J mice in the FeCl3 carotid artery injury model.
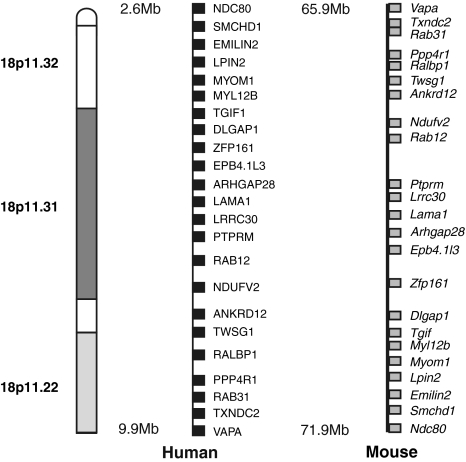
Surveying a panel of 21 chromosome substitution strains (CSS) in which individual A/J chromosomes were replaced in a B6 background, three chromosomes were identified with modifier genes for clot stability (Hoover-Plow et al. 2006): Hmtb1 (chromosome 5), Hmtb2, (chromosome 11), and Hmtb3 (chromosome 17). Quantitative trait locus (QTL) analysis was performed in F2 mice (B6 × B6-Chr5, B6-Chr11, and B6-Chr17 strains) and QTL mapping led to identification of a significant locus on chromosome 5 (Hmtb4), a suggestive locus on chromosome 11 (Hmtb5), and two suggestive overlapping QTLs on mouse chromosome 17 (Hmtb8, 9). The mouse QTL intervals on chromosome 17 are in synteny with a previously identified QTL for thrombosis risk on human chromosome 18 (Soria et al. 2003). The syntenic region contains 23 homologous genes and the purpose of this study was to determine whether any of these genes in the syntenic region are likely candidates as modifiers for clot stability.
Differential expression in B6 and A/J mice in seven of the homologous genes with SNPs in the regulatory regions was confirmed. One of the genes expressed in the cardiovascular system, Emilin2, was differentially expressed in lung of B6, A/J, and B6-Chr17 mice and in liver and bone marrow of B6 and A/J mice. Sequence analysis of Emilin2 identified novel SNPs in the coding and promoter regions between B6 and A/J that could alter expression and/or protein–protein interactions. In B6-Chr17 mice clot stability and Emilin2 expression was higher than in A/J mice, suggesting that there is a modifier gene from another chromosome. EMILIN2 protein was found in platelets, aorta, macrophages, plasma, vessel wall, and thrombi. EMILIN2 may regulate thrombosis by maintaining vessel architecture and/or clot lysis susceptibility. Our data support further investigation of Emilin2 as a promising candidate modifier gene for thrombosis risk.
Methods
Mice
The inbred strains B6 (#000664) and A/J (#000646) were obtained from the Jackson Laboratory (Bar Harbor, ME) at 6 weeks of age. B6-Chr17 mice with A/J chromosome 17 in a B6 background, previously described (Nadeau et al. 2000; Singer et al. 2004), were provided by Dr. Nadeau or by Jackson Laboratory (#004395) and housed and bred at the Biological Resource Unit at the Cleveland Clinic Lerner Research Institute. Experiments were carried out on mice between 7 and 9 weeks of age. All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic.
RT-PCR and real-time PCR
Total RNA was purified from the tissues and the cells of A/J and B6 mice using the RNeasy Mini Kit (Qiagen, Valencia, CA), treated with TURBO DNase (Ambion, Austin, TX), and reverse transcribed into cDNA using SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturers’ instructions. RT-PCR was performed using 50 ng of cDNA from A/J and B6 mice as templates. The PCR product was analyzed on 1 or 1.2% agarose gel and recovered using the QIAquick Gel Extraction Kit (Qiagen). Sequencing (ABI 3730xl, Applied Biosystems, Foster City, CA) was performed at the Genomic Core of Cleveland Clinic. Negative controls that cannot undergo reverse transcription were included in the cDNA synthesis step. Real-time PCR was performed using an iCycler iQ (Bio-Rad Laboratories, Hercules, CA). Each amplification reaction contained 20 ng of cDNA, 300 nM of each primer, 25 μl of 2 × power SYBR® Green Master Mix (Applied Biosystems, Warrington, UK), and 0.5 μl of UNG (Applied Biosystems) added to prevent carryovers. Samples were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Operon, Huntsville, AL). The comparative cycle threshold method was used to analyze the data. RNA was isolated from five to six mice for each group and analyzed in triplicate. All the primers are listed in Supplementary Table 1.
PCR for cloning promoters and noncoding regions
Genomic DNA was isolated from hearts of A/J and B6 mice using a DNA Mini Kit (Qiagen). In order to minimize errors, Expand High Fidelity PCR system (Roche, Indianapolis, IN) was used for PCR. The 25-μl reaction system contained 100 ng of DNA template and 200 nM of each primer (listed in Supplementary Table 1). The estimated error with this method is approximately 3/105 bp. As above, the PCR product was analyzed on agarose gel, recovered, and sequenced by the Genomic Core at the Cleveland Clinic. The cloning and sequencing was repeated twice and the same sequencing results were found.
Generation of EMILIN2 anti-peptide antibody
A polyclonal antibody (E185) for mouse EMILIN2 was generated in rabbits. Since EMILIN2 has a high homology to EMILIN1, a peptide was synthesized from residues 829–843 located in the proline-rich domain specific to EMILIN2. To increase the specificity of the antibody, the rabbit antiserum was subjected to affinity purification using the peptide. To test the specificity of the antibody, HEK293 cells were transfected with the construct E2-N1 that contains EMILIN2-EGFP fusion gene and construct PR-N1 that harbors the PR domain–EGFP fusion gene. The vector N1 that contains only the EGFP gene was used as negative control. Constructs containing EMILIN2 protein domains, namely, Collagen-like C1q domains that do not interact with the antibody, and EGFP fusion genes, were also used as negative controls. Using the purified antibody, Western blot detected a single band at approximately 150 kDa, which is the predicted molecular weight for the EMILIN2-EGFP fusion protein from the E2-N1 construct and approximately 50-kDa band for the PR-EGFP fusion protein. In contrast, no band was detected from the cells transfected with vector alone or the Col-EGFP and C1q-EGFP constructs. Using an antibody against EGFP, the same 150-kDa single band was detected from cells transfected with E2-N1 and a 50-kDa band was detected for PR-EGFP. On the other hand, only one band around 30 kDa (EGFP) was detected from cells transfected with N1 vector. The EGFP antibody also detected the domain-EGFP fusion proteins with the predicted molecular weight for each construct; this indicates that E185 binds specifically to the PR region of EMILIN2.
Western blotting
The mice were perfused with PBS and aortas were harvested, homogenized, and lysed in RIPA buffer (R0278, Sigma, St. Louis, MO) with protease inhibitors (P8340, Sigma). Platelets were gel filtration purified and lysed with RIPA buffer with protease inhibitors. Plasma was collected by centrifuging citrated blood at 1000 g for 10 min and mixed with 2 × Laemmli sample buffer (Bio-Rad). Peritoneal lavage was collected 72 h after treatment, with 0.5 ml of 4% thioglycollate and peritoneal macrophages were harvested, washed with PBS, and suspended in RIPA buffer. About 95% of the cells were macrophages, identified by Wright’s staining, and the viability was greater than 95%, as assessed by trypan blue staining. Protein concentration was measured with the Protein Assay (Bio-Rad). For Western blots, 10 μg of protein was loaded in each lane for SDS-PAGE gels, which were transferred to PVDF membranes. The membranes were incubated with rabbit anti-EMILIN2 (E185), goat anti-EMILIN2 (Q-16, sc-51356, Santa Cruz Biotechnology, Santa Cruz, CA) and EMILIN1 (sc-27569, Y-13, Santa Cruz) antibodies, and HRP-conjugated anti-rabbit or anti-goat secondary antibodies. Protein bands were detected using the ECL detection system (GE Healthcare, Buckinghamshire, UK). A protein loading control, β-actin, was detected with antibody (A5441, Sigma).
FeCl3 carotid injury and immunohistochemical staining
To induce thrombus formation in the carotid artery, a ferric chloride (FeCl3) model of vessel injury was employed as previously described (Hoover-Plow et al. 2006). The flow probe (model 0.5PSB, Transonic Systems, Ithaca, NY) was in place from baseline measurements to several minutes after the stable occlusion had been reached or stopped at 30 min if it had not occluded. Blood flow was recorded every 10 s (model TS420, Transonic Systems). After cardiac perfusion, the carotid arteries were harvested after occlusion, immediately embedded into OCT (Tissue-Tek, Torrance, CA), and frozen. The frozen carotid arteries were sectioned at 10 μm using a cryostat (Leica CM1850, Leica Microsystems, Nassloch, Germany), fixed with acetone at 4°C for 10 min, then blocked with normal serum. EMILIN2 was detected with E185 or Q-16 antibodies and P-selectin with rabbit anti-CD62P antibody (BD Biosciences, San Diego, CA). The sections were then incubated with 1:1000 diluted biotinylated appropriate secondary antibodies (PK-6101, PK-6105 Vectastain ABC Kit, Vector Laboratories, Burlingame, CA), and proteins were visualized with alkaline phosphatase substrate. For quantification analysis, six sections of each carotid and six mice from each group were measured and analyzed using Image-Pro Plus (Media Cybernetics, Silver Spring, MD).
Statistical analysis
Values are presented as mean ± SEM. Statistical differences were assayed by a t test or ANOVA with Newman-Keuls post-test.
Results
Clot stability is enhanced in A/J and B6-Chr17 mice compared to B6 mice
Previously we reported prolonged clot stability times in the tail bleeding/rebleeding assay and rapid occlusion time after FeCl3-induced carotid injury in A/J mice compared to B6 mice (Hoover-Plow et al. 2006). A tail bleeding/rebleeding assay suitable for large-scale screening was used to screen a panel of CSS mice. We identified that the B6-Chr17 mice (the strain containing A/J Chr 17 in the B6 background) had similar clot stability times as those of the A/J strain, suggesting that Chr 17 contains a gene or genes responsible for this phenotypic variation. We tested this phenotypic variation using another model, the FeCl3-induced carotid injury in B6-Chr17, A/J, or B6 mice, to validate the results from the bleeding/rebleeding assays. In the B6-Chr17 mice, the occlusion time, reflecting clot formation, was similar to the B6 times. Measuring the blood flow at 0, 2, 4, and 24 h following administration of FeCl3 tested the patency of the carotids after injury. In both A/J and B6 strains, carotids remained occluded at 2 h. At 4 h, 44% of the vessels were open in the B6 compared to only 22% in the A/J and 0% in the B6-Chr17 (Table 1). At 24 h, 90% of the B6 mice had open carotids while only 64% of A/J and 0% of B6-Chr17 mice had open carotids. Blood flow at 24 h was significantly less in the B6-Chr17 mice than both B6 (P < 0.01) or A/J (P < 0.05) mice. These results suggest that Chr 17 contains candidate genes responsible for the thrombotic susceptibility variation in A/J and B6 mice and that the candidate gene(s) regulate the clot stability/clot lysis rather than clot formation. In addition, the significant difference in patency observed with B6-Chr17 or A/J mice suggests that genes from the B6 background on other chromosomes interact with the causative genes on Chr 17.
Table 1.
Patency in B6, A/J, and B6-Chr17 mice in FeC13-induced vascular injury
| Open percentage | Mean blood flow (ml/min) | |
|---|---|---|
| B6 | Baseline | 0.48 ± 0.08 |
| A/J | Baseline | 0.47 ± 0.07 |
| 2 h | ||
| B6 | 0% (0/5) | 0.05 ± 0.01 |
| A/J | 0% (0/5) | 0.04 ± 0.01 |
| 4 h | ||
| B6 | 44% (4/9) | 0.22 ± 0.06 |
| A/J | 22% (2/9) | 0.16 ± 0.02 |
| B6-Chr17 | 0% (0/4) | 0.06 ± 0.01 |
| 24 h | ||
| B6 | 90% (9/10) | 0.45 ± 0.08* |
| A/J | 64% (7/11) | 0.31 ± 0.08** |
| B6-Chr17 | 0% (0/6) | 0.05 ± 0.01 |
When blood flow is less than 0.1 ml/min, it is assumed that the carotid is closed. Mouse numbers with open carotids/total mouse numbers are in parentheses. Symbols indicate a difference between B6-Chr 17 and B6 (* P < 0.01) and A/J (** P < 0.05)
Expression of homologous genes
The syntenic region for the mouse and human QTLs spanned 5.9 Mb and contained 23 homologs retrieved from the Ensembl Genome Browser (Fig. 1). To test the expression of the candidate genes, real-time PCR was performed and mRNA levels of the 23 genes were analyzed from the liver of A/J and B6 mice (Table 2). Three of the genes were not expressed in the liver: Txndc2, Lrrc30, and Myom1. Expression of six of the genes, Rab12, Zfp161, Emilin2, Twsg1, Ankrd12, and Dlgap1 (indicated in bold in Table 2), was significantly higher (1.6-2.7-fold) in A/J than in B6, suggesting the possibility that these six genes could be candidate genes. The increased expression of EMILIN2 is not indicative of a systematic error. GAPDH was used as the endogenous control for every sample in every analysis, and GAPDH expression levels were similar in the two strains. One possible explanation for the increase in expression in A/J is that A/J has higher gene copy numbers in this region which may alter expression (Cutler et al. 2007; Graubert et al. 2007; Henrichsen et al. 2009; She et al. 2008). In addition, the microarray data from Henrichsen et al. (Microarray data, C. N. Henrichsen et al., Record GSE10744, released in 2009 from Gene Express Omnibus) found differential expression in B6 and A/J in lung for Ralbp1, Twsg1, Ptprm, and Ebp4.1l3 and in heart for Rab31, Ptprm, Myom1, and Emilin2. Thus, of the 23 genes in the homologous region, 11 had differential expression in B6 and A/J mice.
Fig. 1.
Genes in the mouse QTL region and the human syntenic region were determined using Ensembl Genome Browser. Symbols of the genes are indicated. Names of the genes can be found in Table 1
Table 2.
Expression analysis of the homologous genes in the syntenic QTL interval by real-time PCR
| Symbol | Name | Fold change | P value |
|---|---|---|---|
| Vapa | VAMP-associated protein A | 1.28 | 0.19 |
| Txndc 2 | Thioredoxin domain containing 2 | ND | – |
| Rab31 | Member RAS oncogene family | 1.13 | 0.45 |
| Ppp4r1 | Protein phosphatase, regulatory subunit 1 | 1.24 | 0.43 |
| Ralbp1 | ralA binding protein 1 | 1.45 | 0.27 |
| Twsg1 | Twisted gastrulation homolog 1 (drosophila) | 1.63 | 0.001 |
| Ankrd12 | Ankyrin repeat domain 12 | 2.26 | 0.0025 |
| Ndufv2 | NADH dehydrogenase flavoprotein 2 | 1.39 | 0.19 |
| Rab12 | RAB12, member RAS oncogene family | 2.15 | 0.01 |
| Ptprm | Protein tyrosine phosphatase receptor M | 1.25 | 0.31 |
| Lrrc30 | Leucine-rich repeat containing 30 | ND | – |
| Lama1 | Laminin, alpha 1 | 0.71 | 0.89 |
| Arhgap28 | Rho GTPase activating protein 28 | 1.69 | 0.29 |
| Epb4.1l3 | Erythrocyte protein band 4.1-like 3 | 1.31 | 0.33 |
| Zfp161 | Zinc finger protein 161 | 2.74 | 0.01 |
| Dlgap1 | Large (drosophila) homolog-associated protein 1 | 1.96 | 0.035 |
| Tgif | TG interacting factor | 1.04 | 0.83 |
| Myl12b | Myosin light chain 12 B regulatory | 1.86 | 0.10 |
| Myom1 | Myomesin 1 | ND | – |
| Lpin2 | Lipin2 | 1.79 | 0.12 |
| Emilin2 | Elastin microfibril interfacer 2 | 1.58 | 0.005 |
| Smchd1 | SMC hinge domain containing 1 | 1.28 | 0.25 |
| Ndc80 | Kinetochore complex component | 1.41 | 0.26 |
Fold changes of mRNA expression of the genes in A/J compared to B6 and the P values are indicated. n = 6. Genes with significant difference between A/J and B6 are shown in bold
ND not detected
SNPs in the homologous genes
SNPs in the homologous genes were retrieved from the MGI dbSNP Build 128 and the recently available Sanger database of A/J chromosome 17 (Table 3). In addition to the coding region, mRNA-UTR (mRNA-untranslated region), introns, and SNPs 2 kb upstream and downstream of the genes are recorded in the MGI database. The SNPs identified from the MGI database were also identified in the Sanger database. The read depth (number of paired reads mapping Chr17_36/length of Chr17) (Sudbery et al. 2009) of the SNPs in the Sanger database was high, 7-22. In many genes, due to the thorough sequencing coverage of the A/J Chr 17, additional SNPs were identified, especially in the introns of the genes in the Sanger database. Two genes, Txndc2 and Rab12, did not contain any SNPs in A/J or B6 (Table 3). Five genes, Vapa, Ankrd12, Ndufv2, Arhgap28, and Ptprm, had no nonsynonymous SNPs in the coding regions or SNPs in the mRNA-UTR or upstream or downstream noncoding regions (Table 3). Therefore, these seven genes are not likely to be causative genes.
Table 3.
Mouse genes on Chr 17 homologous to genes in human QTL on Chr 18
| Symbol | ID (MGI) | Start | Coding region | Noncoding regions | ||||
|---|---|---|---|---|---|---|---|---|
| Synonymous | Nonsynonymous | Upstream | Downstream | mRNA-UTR | Intron | |||
| Vapa | 1353561 | 65929396 | 1 (1) | |||||
| Txndc2 | Txndc2 | 65986854 | ||||||
| Rab31^ | 1914603 | 66001069 | 1 | 234 (54) | ||||
| Ppp4r1^ | 1917601 | 66169485 | 1 (1) | (5) | (4) | 1 (1) | 135 (38) | |
| Ralbp1#^ | 108466 | 66197781 | 3 (3) | 1 | (5) | (4) | 3 (1) | 101 (39) |
| Twsg1^ | 2137520 | 66272406 | 3 (3) | 15 (11) | ||||
| Ankrd12 | 1914357 | 66316841 | 7 (4) | |||||
| Ndufv2 | 1920150 | 66428135 | 5 (3) | |||||
| Rab12 | 894284 | 66843852 | ||||||
| Ptprm | 102694 | 67016883 | 1 (1) | 2428 (647) | ||||
| Lrrc30 | 2685172 | 67981021 | (1) | |||||
| Lama1# | 99892 | 68046605 | 9 (1) | 5 (1) | 242 (21) | |||
| Arhgap28 | 2147003 | 68192053 | 1 | |||||
| Epb4.1l3 | 103008 | 69506150 | (2) | 14 (11) | ||||
| Zfp161^ | 1195345 | 69732390 | (1) | 2 | 1 | |||
| Dlgap1#^ | 1346065 | 70318413 | 1 (1) | 1 | 5 | 2428 (216) | ||
| Tgif^ | 1194497 | 71193551 | (1) | 2 (1) | 2 (1) | |||
| Myl12b | 107494 | 71323260 | (3) | |||||
| Myom1#^ | 1341430 | 71368861 | 8 | 5 (1) | (1) | 3 | 417 (119) | |
| Lpin2^ | 1891341 | 71531900 | 2 (1) | (2) | 6 (6) | 88 (48) | ||
| Emilin2#^ | 2389136 | 71601516 | 11 (8) | 12 (3) | (3) | 4 | 375 (82) | |
| Smchd1^ | 1921605 | 71693829 | 1 (1) | (5) | 2 | 284 (125) | ||
| Ndc80#^ | 1914302 | 71845442 | 4 (3) | 1 (1) | 2 (1) | 86 (55) | ||
SNPs were retrieved from the Wellcome Trust Sanger Institute Mouse Genome Project and the MGI dbSNP Build 128 (in parenthesis). MGI IDs are indicated. The start site of the gene on mouse chromosome 17 was retrieved from Ensembl annotation of the NCBI Build 37. Synonymous SNPs are SNPs in the coding region with no amino acid change. Nonsynonymous SNPs are SNPs with an amino acid change. Upstream is the region 2 kb from the 5′ coordinate. Downstream is the region 2 kb downstream from the 3′ coordinate. mRNA-UTR indicates SNPs in the mRNA-untranslated region. Items in the Symbol column with no symbol means that there are no SNPs in the nonsynonymous coding regions or SNPs in the mRNA-UTR; # = nonsynonymous SNPs in the coding region; ^ = SNPs in the mRNA UTR. Genes with differential expression in B6 and A/J mice are indicated in bold
Six genes, Ralbp1, Lama1, Dlgap1, Myom1, Emilin2, and Ndc80, had nonsynonymous SNPs in the coding region. Twelve genes, Rab31, Ppp4r1, Ralbp1, Twsg1, Zfp161, Dlgap1,Tgif, Myom1, Lpin2, Emilin2, Smchd1, and Ndc80, had SNPs in the mRNA-UTR. Eleven genes had SNPs in the 2-kb noncoding regions: Ppp4r1, Ralbp1, Lrrc30, Epb4.1l3, Zfp161, Tgif, Myl12b, Myom1, Lpin2, Emilin2, and Smchd1. Although not determined in this study, regions outside the 2-kb region could also alter the expression of genes. Thus, 16 of the 23 homologous genes had SNPs in the coding region or mRNA-UTR and could be candidate genes. Four of these genes, Twsg1, Zfp161, Dlgap1, and Emilin2, with SNPs in the mRNA-UTR, also had differential expression in liver of B6 and A/J mice, suggesting that these four genes have great potential to be candidate genes. In addition, Ralbp1, Myom1, and Rab31 were reported to be differentially expressed in lung or heart (Microarray data, C. N. Henrichsen et al., Record GSE10744, released in 2009 from Gene Express Omnibus). Thus, for 7 of the 16 genes with SNPs in the regulatory regions, differential expression was confirmed. Two genes, Lama1 and Ndc80, had nonsynonymous SNPs. These nine genes (7 with SNPs in regulatory regions and differential expression plus 2 genes with nonsynonymous SNPs) were considered to be candidate genes and their known functions and distribution were evaluated.
Of the known functions and distribution of the genes and gene products identified above with SNPs in the mRNA-UTR and/or nonsynonymous SNPs, Emilin2 seemed the most likely candidate gene. Because EMILIN2 and other EMILIN proteins are associated with the cardiovascular system (Christian et al. 2001; Hayward et al. 1993, 2004; Lembertas et al. 1997; Milanetto et al. 2008), we selected Emilin2 for further analysis. In addition to differential expression in B6 and A/J mice, Emilin2 had nonsynonymous SNPs in the coding region and SNPs in the potential regulatory regions, based on the MGI and Sanger databases. Because Emilin2 could be a causative gene in this QTL, examination of the expression and polymorphisms was undertaken with B6, A/J, or B6-Chr17 mouse strains.
Expression of Emilin2 in lung, bone marrow cells, heart, and aorta
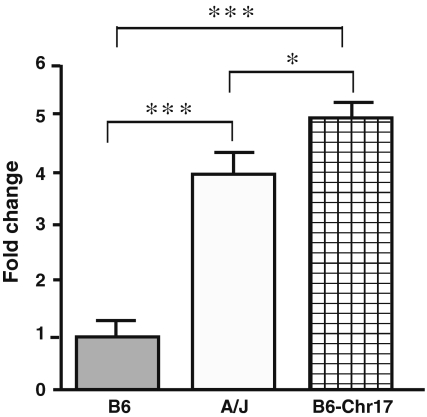
The mRNA expression of Emilin2 was determined by quantitative real-time PCR in lung, heart, aorta, and bone marrow cells. There was a 4.3-fold increase in Emilin2 mRNA expression in the lungs of A/J mice compared to B6 mice (P = 0.005) (Table 4). Expression of Emilin2 in the lungs of B6-Chr17 or A/J mice was 4.8-fold (P = 0.0004) or 3.8-fold (P = 0.004) higher, respectively, compared to B6 mice (Fig. 2). There was no significant difference in expression of Emilin2 in heart and aorta tissue between the two strains (Table 4). In the GEO microarray data, Emilin2 in the lung was 1.4-fold higher in A/J than in B6, which is consistent with increased Emilin2 expression in A/J in our data. However, because of the standard deviation in A/J mice (n = 3) in the microarray data, there was no significant difference between A/J and B6. In the heart, Emilin2 was 60% lower in A/J than in B6 mice in the microarray data, and in our study Emilin2 expression was 44% lower in A/J than in B6 mice. In our data, because of high standard deviation, there was no significant difference between A/J and B6 expression. However, the tendency of reduced expression of Emilin2 in the heart of A/J mice is consistent in both analyses. The microarray data were from only three mice, aged from 11 to 14 weeks, in each group; our data were from six (lung) or five (heart) mice, aged from 6 to 8 weeks, in each group.
Table 4.
EMILIN2 expression by quantitative real-time PCR in tissue
| Tissue | B6 | A/J | P value | Fold change of A/J from B6 | Fold change from liver B6 | Fold change from liver A/J |
|---|---|---|---|---|---|---|
| Liver | 0.50 ± 0.05 | 0.79 ± 0.06 | 0.005 | 1.58 | 1 | 1 |
| Lung | 1.50 ± 0.65 | 6.45 ± 1.42 | 0.01 | 4.30 | 3.00 | 8.16 |
| Heart | 1.93 ± 0.32 | 1.08 ± 0.17 | ns | 0.56 | 3.86 | 1.37 |
| Aorta | 2.60 ± 0.49 | 3.44 ± 0.96 | ns | 1.32 | 5.20 | 4.35 |
| Bone marrow | 32.5 ± 8.06 | 65.2 ± 10.58 | 0.03 | 2.01 | 65.00 | 82.53 |
B6, A/J: samples were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Operon, Huntsville, AL). The comparative cycle threshold method was used to analyze the data. RNA was isolated from five or six mice for each group and analyzed in triplicate. All the primers are listed in Supplementary Table 1
Fig. 2.
Expression of Emilin2 in lung from A/J, B6, and B6-Chr17, n = 5. GAPDH from the same cDNA samples used as endogenous control. The mRNA levels are presented as fold change relative to B6 lung. Statistical difference was determined by ANOVA with Newman-Keuls post-test. *** P < 0.001; * P < 0.05
Since Doliana et al. (2001) demonstrated a robust expression of Emilin2 mRNA in peripheral leukocytes, bone marrow cells were tested for Emilin2 mRNA expression. Emilin2 expression was twofold higher (P = 0.04) in bone marrow cells from A/J than from B6 mice (Table 4). In addition, the expression level of Emilin2 was over tenfold higher in bone marrow cells than in other tissues from the same strain. Bone marrow has hematopoietic cells, including red cells and platelets important for blood clotting. Differences in expression of Emilin2 in tissue and bone marrow between B6, A/J, and B6-Chr17 along with prolonged clot stability in B6-Chr17 suggest that Emilin2 may be a candidate gene for thrombosis susceptibility.
Nucleotide polymorphisms in Emilin2
To verify the SNPs identified in the databases, the protein sequence of Emilin2 was determined for B6 and A/J mice. Emilin2 cDNAs were cloned from A/J or B6 mice by RT-PCR and sequenced in duplicate. The sequences for each strain (B6 GenBank ID GQ872460, A/J GenBank ID GQ872457) were identical from two experiments. The B6 Emilin2 sequence GQ872460 is identical to C57BL/6 Emilin2 sequence NM_145158 from the NCBI GenBank. The predicted amino acid sequences identified 11 amino acid polymorphisms between the two strains (Table 5). Three amino acids, 571, 662, and 834, exist in the SNP database (MGI dbSNP Build 128), while the Sanger database reported 12 nonsynonymous SNPs. There was one discrepancy with the Sanger database, where two SNPs are listed at adjacent positions, 71624555 and 71624556, with respect to our sequencing. Our sequencing revealed that the two SNPs are within one codon and give rise to one amino acid change. Two serine (B6)-proline (A/J) substitutions were detected at residues 136 and 162, and three threonine (B6)-alanine (A/J) substitutions were detected at 483, 917, and 925. There was one valine (B6)-threonine (A/J) substitution at 172 and one glutamine (B6)-arginine (A/J) substitution at 834. The above seven amino acid changes have the potential to alter the charges or structures of the protein. The two serine (B6) with proline (A/J) substitutions at residues 136 and 162 are predicted to have post-translational modifications (Lee et al. 2006), and the threonine (B6) with alanine (A/J) substitution at position 917 is predicted to be a glycosylation site (Lee et al. 2006). Other amino acid polymorphisms, including leucine (B6)-phenylalanine (A/J), serine (B6)-threonine (A/J), valine (B6)-glycine (A/J), and alanine (B6)-valine (A/J), may have less or no effect on the protein functions. Among the 11 nonsynonymous SNPs in the coding region, six amino acids are not conserved in mammals (Supplementary Fig. 1). Two amino acids, Ser162 and Leu182, are conserved in B6 and other mammals but are changed to proline and phenylalanine in A/J. In addition, two other amino acids, proline 136 and alanine 917, are conserved in A/J and other mammals but are substituted by serine and threonine in B6. B6 mice and other mammals have conserved amino acids (threonine and serine) at position 925, which is substituted to alanine in A/J mice. These five amino acid substitutions at the conserved sites have a potential to confer structural and functional differences of the protein in A/J versus B6 mice.
Table 5.
Polymorphisms in Emilin2 between A/J and B6
| Amino acid polymorphisms | SNPs in promoter | ||||
|---|---|---|---|---|---|
| Position | B6 | A/J | Position | B6 | A/J |
| 136a | Serine | Proline | −131 | C | G |
| 162a | Serine | Proline | −246 | C | T |
| 172 | Valine | Threonine | −882 | C | T |
| 182 | Leucine | Phenylalanine | −1312 | G | A |
| 258 | Serine | Threonine | −1313 | Del | C |
| 483 | Threonine | Alanine | −1314 | Del | A |
| 571 | Valine | Glycine | −1315 | Del | C |
| 662 | Alanine | Valine | −1316 | Del | A |
| 834 | Glutamine | Arginine | |||
| 917a | Threonine | Alanine | SNPs in 5′-noncoding region | ||
| 925 | Threonine | Alanine | 156 | G | del |
| 171 | Del | G | |||
In the coding region, the amino acid polymorphisms are indicated. The positions of amino acids are numbered from the starting amino acid. In the promoter and 5′-noncoding regions, the nucleotide polymorphisms are indicated. In the promoter, the nucleotide before the putative transcription start site is numbered as −1. In the 5′-noncoding region, the positions are numbered from the putative transcription start site (as 1)
aPredicted sites for post-translational modification
To determine whether there are nucleotide polymorphisms in the upstream and noncoding regions in Emilin2, the 1.5-kb upstream region and the 5′- and 3′-noncoding mRNA regions were cloned from A/J or B6 mouse strains (Table 5). No polymorphisms were detected in the 3′-noncoding mRNA region. In the 5′-noncoding region, single nucleotide deletions were found in A/J and B6 at two locations (Table 5). A total of eight SNPs were detected in the 1.5-kb putative promoter region, including two substitutions in the proximal region and two substitutions and four deletions (in B6) in the distal region. These SNPs in the promoter and 5′-noncoding region may explain the differential expression of Emilin2 in certain tissues.
Function of Emilin2 in thrombosis
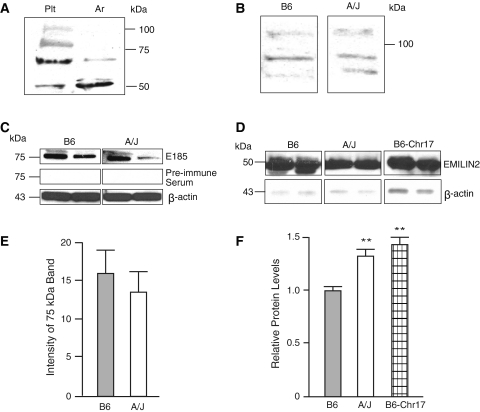
EMILIN2 was considered to potentially play a direct role in the formation or stability of the thrombus after injury. Multimerin1 (Hayward et al. 1993), an EMILIN protein, is found as a complex with coagulation factor V in platelet α-granules; it is released to the platelet surface upon platelet activation and binds to platelets and endothelial cells. A deficiency of Multimerin1 and α-synuclein, a protein that inhibits α-granule release from platelets, had impaired platelet adhesion and thrombin formation that was corrected with Multimerin treatment (Reheman et al. 2010). Whether EMILIN2 plays such a role in thrombosis is not known. EMILIN2 protein was determined by immunoblotting with an anti-peptide antibody. EMILIN2 was detected in platelets, macrophages, and plasma (Fig. 3). Full-length EMILIN2 was detected in platelets, but only fragments of EMILIN2 were detected in aorta. In addition, fragments of EMILIN2 were also detected in macrophages and plasma (Fig. 3c, d). In macrophages, a single band at 75 kDa was detected and a single band at 50 kDa was identified in plasma. The specificity (data not shown) of the EMILIN2 antibody (E185) was verified by inhibition with the antigen peptide, identification of the full-length protein and fragments with a commercially available EMILIN2 antibody, and lack of cross-reactivity with EMILIN1 antibodies. The peptide antigen inhibited the full-length protein as well as the fragments, and an antibody to EMILIN1 did not cross-react with the full-length, or fragments of, EMILIN2. The commercially available antibody detected a pattern of EMILIN2 in the tissues similar to that of the E185 antibody. These results suggested that E185 was specific for EMILIN2 and that the fragments were not artifacts. Although we did not detect a difference in B6 and A/J platelet EMILIN2 in unstimulated platelets or macrophages (Fig. 3b, c, e), plasma EMILIN2 was significantly higher in A/J and B6-Chr17 than in B6 mice (Fig. 3d, f), suggesting a potential role for EMILIN2 in clot stability. EMILIN2 was also identified in the thrombus induced by ferric chloride injury.
Fig. 3.
a–d Western blot of EMILIN2 with antibody E185, from tissue homogenates. a Platelets (Plt) and aorta (Ar) from B6 mice. b Platelets from B6 and A/J mice. c Peritoneal macrophages from B6 and A/J mice. d Plasma from B6, A/J, and B6-Chr17 mice. e Density of 75-kDa bands from Western blot of EMILIN2 protein in peritoneal macrophages from B6 and A/J relative to β-actin. Values are mean ± SEM of four mice per strain. f Density of 50-kDa bands from Western blot of EMILIN2 in plasma. Values are mean ± SEM of six mice per strain. Statistical differences were determined by a t test or ANOVA and Newman-Kuels post-test. ** P < 0 .01
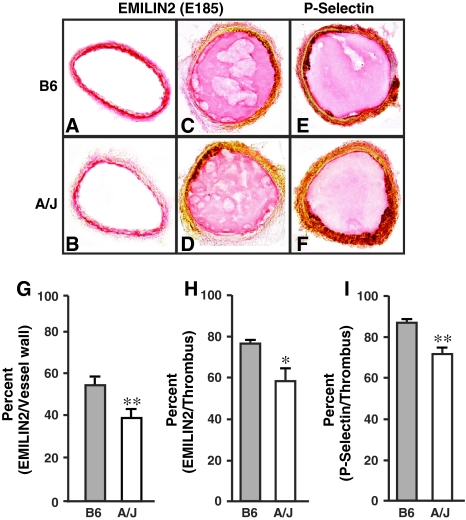
Since EMILIN2 was found in the platelets, macrophages, and plasma, it was assessed in the thrombi after ferric chloride injury. EMILIN2 was found in the vessel wall in the uninjured carotid (Fig. 4a, b) and in the thrombi after injury (Fig. 4c, d). Quantification of the immunostaining indicated reduced EMILIN2 in sections from A/J mice. Identification of platelets (Fig. 4e, f) with a P-selectin antibody suggested a similar pattern of distribution of EMILIN2 in B6 and A/J mice. EMILIN2 was significantly lower in A/J than in B6 mice in the vessel wall and thrombus. Because EMILIN2 is higher in plasma but lower in the vessel wall and thrombi in A/J mice, EMILIN2 in A/J may have defects in binding.
Fig. 4.
a-d Sections immunostained with EMILIN2 antibody E185. a, b Uninjured carotid. c, d Sections immediately after occlusion. e, f Sections immunostained for P-selectin with antibody. Magnification × 100 (a-e). g-i Quantification of immunostaining. The values are mean ± SEM of the percent of EMILIN2/vessel wall (g), EMILIN2/thrombus (h), and P-selectin/thrombus (i) of six mice per strain. Six sections were analyzed for each mouse using Image-Pro Plus software. Statistical differences were determined by t test or ANOVA and Newman-Kuels post-test. * P < 0.05, ** P < 0.01
A second possible role for EMILIN2 in thrombosis is that it is critical for maintenance of the vessel wall architecture. EMILIN1 deficiency causes disruptions of the endothelial cell layer and interferes with elastogenesis (Zanetti et al. 2004), disrupting the elastic lamina of the vessel wall. Collagen was measured in the vessel wall and in the tail of B6 and A/J mice. Because there was a twofold difference in clot stability in the tail bleeding/rebleeding assay in A/J mice than in B6 mice, collagen (Table 6) was assessed in the tail and was found to be 27% higher (P < 0.05) in A/J mice than in B6 mice (Hoover-Plow et al. 2006). The percent of collagen was significantly higher by 45% (P < 0.001) in the arteries of A/J mice than of B6 mice (Table 6). Whether the accumulation of collagen in the A/J mice alters thrombosis and is due to differences in EMILIN2 between the B6 and A/J mice is unclear. The internal and external elastic lamina was evaluated in injured and uninjured carotids (Table 5). Unlike the B6 mice, in the A/J mice there was no change in the elastic lamina after FeCl3-induced injury. In addition, EMILIN2 was identified in the vessel wall in uninjured carotids (Fig. 4). These results suggest that there are functional differences in the two strains that could be related to the differences in the vessel wall ECM composition of the A/J and B6 mice.
Table 6.
Extracellular matrix in vessels of B6 and A/J mice
| B6 | A/J | |
|---|---|---|
| Collagen (%) | ||
| Artery collagen (%) | 33 ± 5 (10) | 48 ± 3** (11) |
| Vein collagen (%) | 47 ± 6 (6) | 57 ± 4 (11) |
| Tail collagen (%) | 33 ± 4 (5) | 42 ± 4* (4) |
| Carotid injury (ratio injured to uninjured) | ||
| Internal elastic lamina | 1.48 ± 0.02 (6) | 1.05 ± 0.07** (6) |
| External elastic lamina | 1.36 ± 0.03 (6) | 0.96 ± 0.05** (6) |
Values are the mean ± SEM. Number of mice indicated in parenthesis
*P < 0.05; **P < 0.001 determined by t test
Summary
The prolonged clot stability in the bleeding/rebleeding assay identified previously in B6-Chr17 mice was confirmed in a direct thrombotic assay of FeCl3-induced carotid injury. In B6 mice only 10% of the carotids remained occluded at 24 h, while in the B6-Chr17 mice 100% remained occluded. Expression analysis of the 23 homologous genes in the syntenic region of QTLs for thrombosis risk in mice and humans revealed 7 genes that had SNPs in the regulatory regions and altered expression in tissues: Rab31, Ralbp1, Twsg1, Zfp161, Dlgap1, Myom1, and Emilin2. Two other genes had nonsynonymous SNPs without increased expression, Lama1 and Ndc80. These nine genes are the most likely candidate genes. EMILIN2 is a member of the EMILIN proteins found in the cardiovascular system and was selected for further analysis. This study found high mRNA expression of Emilin2 in bone marrow compared to other tissues. Sequencing of Emilin2 in B6 or A/J mice identified SNPs in both coding and potential regulatory regions. In addition, EMILIN2 protein differential expression was identified in plasma, the vessel wall, and thrombi, suggesting a potential direct role in clot stability. Furthermore, EMILIN2 in the vessel wall and altered composition and architecture in B6 and A/J vessel walls implicates a second potential role of EMILIN2 in the response to vascular injury.
Discussion
The two QTLs Hmtb8 and Hmtb9, which regulate clot stability, were previously identified on mouse Chr 17 (Sa et al. 2008). These two loci map to a syntenic region that contains a QTL controlling thrombosis susceptibility in humans (Soria et al. 2003). There are several genetic differences that can cause variation in the expression of a gene: (1) Genes have nonsynonymous SNPs that change amino acids in the coding region, (2) genes have SNPs in the regulatory regions, or (3) the gene products differ in their expression levels (Glazier and Nadeau 2002; Wang et al. 2005). In addition to identification of splice variants (Marcucci and Romano 2008; Zavolan and van Nimwegen 2006), recent studies have suggested that copy number variability (Cutler et al. 2007; Graubert et al. 2007; Henrichsen et al. 2009; She et al. 2008) and microRNA regulation (Prasad et al. 2009) can cause differences in gene and gene product expression. This study focused on the identification of SNPs in the homologous genes between B6 and A/J mice.
There were two genes that had nonsynonymous SNPs: Lama1 and Ndc80. Lama1 is a basement membrane component and is limited primarily to epithelium in both embryonic development and in adults (Ekblom et al. 2003). Lama1 deficiency in mice is embryonically lethal (Alpy et al. 2005). Ndc80 is involved in the interface between centromeres and spindle microtubules, and it’s overexpression is associated with tumors (Diaz-Rodriguez et al. 2008). Seven genes that contained SNPs were identified in mRNA-UTR and had confirmed differential expression in B6 and A/J mice: Twsg1, Zfp161, Dlgap1, Ralbp1, Myom1, Rab31, and Emilin2. Mammalian Tswg1 is associated with salivary gland epithelial development. Twsg1-deficient mice have craniofacial and skeletal structure defects and more than half of the deficient mice exhibit dwarfism and fail to thrive (MacKenzie et al. 2009; Melnick et al. 2006). Zfp161 is expressed in neural tissue, is involved in transcription regulation (Numoto et al. 1993), and is linked to a gene associated with an inheritable developmental disorder, holoprosencephaly (Sobek-Klocke et al. 1997). Dlgap1 is expressed in the brain and neural tissues and functions in synaptic transmission and cell-cell signaling (Bouwman et al. 2006; Nguyen et al. 2005). Ralbp1 has GTPase activity, and homozygous and heterozygous mice have increased sensitivity to irradiation, oxidative stress, and impaired glutathione homeostasis (Singhal et al. 2008). Myom1 is a structural constituent of cytoskeleton and is involved in muscle contraction (Price and Gomer 1993; Steiner et al. 1999). Rab31 participates in Golgi vesicle transport and GTP binding (Rodriguez-Gabin et al. 2009). It is not apparent that these eight genes discussed above could be causative genes for thrombosis modifiers.
Of the nine candidate genes, Emilin2 seems to be the most likely candidate for the causative gene for thrombosis. EMILIN2 is one of the EMILIN (Elastin Microfibril Interface Located Protein) proteins, which are a group of extracellular matrix glycoproteins (Colombatti et al. 2000). This family comprises at least four proteins, including EMILIN1, EMILIN2, Multimerin1, and Multimerin2, with the major site of expression in the cardiovascular system, including the vascular wall and platelets (Christian et al. 2001; Hayward et al. 1993; Zacchigna et al. 2006). EMILIN1 deficiency causes disruptions of the endothelial cell layer and interruptions of the elastic lamellae of large vessels (Zanetti et al. 2004). Other extracellular proteins, including elastin (Milewicz et al. 2000), fibulins (Argraves et al. 2003), fibrillar proteins (Robinson and Godfrey 2000), thrombospondins (Stenina et al. 2007), and collagens (Gould et al. 2005; Malfait et al. 2006), important for vascular wall architecture, are required for recovery from injury and prevention of vascular disease. In addition, ECM proteins, including fibulin1 (Argraves et al. 2003), thrombospondins (Stenina et al. 2007), and collagen (Gould et al. 2005; Malfait et al. 2006), are found in plasma and have been used as biomarkers for cardiovascular disease.
In this study we report that EMILIN2 may function in two ways to regulate thrombosis: (1) as a component of the vascular wall and critical for maintaining the vessel architecture and response to injury, and (2) as a component of the thrombus facilitating clot lysis. A recent study reports downregulation of EMILIN2 in brain arteriovenous malformations (Sasahara et al. 2007). Furthermore, a number of proteomic and microarray studies of human tissues have identified EMILIN2 in extracellular fluids (plasma, amniotic, seminal, and synovial), stem cells (hematopoietic, mesenchymal, and osteoblasts), stimulated endometrial stromal cells, and cancer cells (lymphocytic leukemia, hepatic, colorectal, and ovarian cancer). In addition, microarray data (T. C. Somervaille et al., Record GSE13693) released in 2009 from GEO analyzed gene expression in bone marrow cell populations in B6 mice. Analysis of the data revealed that EMILIN2 was significantly increased in mature neutrophils compared to undifferentiated hematopoietic cells (3.6-38-fold, P < 0.001). These studies suggest that EMILIN2 plays important physiological and pathological roles. The identification of EMILIN2 in plasma in humans as well as in mice supports the hypothesis that EMILIN2 could play a role in thrombosis.
Emilin2 mRNA expression has been reported in spleen and uterus (Braghetta et al. 2004); fetal heart, lung, placenta, and peripheral leukocytes (Doliana et al. 2001); cochlear basement membrane (Amma et al. 2003); and optic tissues (Scavello et al. 2005). Emilin2 is expressed in the vasculature and heart in zebrafish during embryonic development and in adults (Milanetto et al. 2008). In this study, both nonsynonymous SNPs and expression differences for Emilin2 were observed in A/J and B6 mice. Furthermore, the amino acid changes are the same in A/J mice and in another strain, 129/svJ (see Supplementary Fig. 2 and Table 2), which also has prolonged clot stability. For the QTLs on chromosome 17, from 66 to 66.6 Mb in the syntenic region, 98% of the SNPs of 129 and B6 mice are identical, but from 66.6 to 71.9 Mb (including Emilin2), 99% of the SNPs of A/J and 129 mice are identical. In the QTL Hmtb4 on chromosome 5, SNPs in some regions such as 100.3-102.5 Mb (99%) and 105-107.4 Mb (94%), B6 and 129 are similar. On the other hand, A/J and 129 are similar (99%) in the region from 107.6 to 110 Mb, and B6 and A/J are similar (99%) in the region from 103.3 to 104.5 Mb. Because of the complexity feature of this trait, it is hard to hypothesize how the interactions happen, but we do not exclude epistatic interactions among loci. The known functions of other EMILIN proteins, the presence of nonsynonymous SNPs in Emilin2, and expression differences in A/J and B6 from the current study strongly suggest further investigation of Emilin2 as a candidate gene underlying thrombosis susceptibility.
In this study, prolonged clot stability in A/J and B6-Chr17, detected in a bleeding/rebleeding assay, was validated using an assay of direct thrombosis formation and stability. The prolonged clot stability in A/J and B6-Chr17 mice corresponded well to the elevated mRNA expression levels of Emilin2 in A/J and B6-Chr17 mice. These data suggest that the quantitative expression difference of Emilin2 may also be responsible for the phenotypic difference between A/J and B6 mice. In addition, the difference in the clot stability and in the expression of Emilin2 between A/J and B6-Chr17 mice suggests that the causative genes on Chr 17 have epistatic interactions with genes on other chromosomes. These findings are consistent with our previous hypothesis that there were interacting genes on Chrs 5 and 17 (Hoover-Plow et al. 2006; Sa et al. 2008). Our previous studies revealed that the homosomic B6-Chr5 or B6-Chr17 mice had prolonged clot stability as did the A/J parental strain, but the heterosomic mice (B6-Chr5 × B6) F1 or (B6-Chr17 × B6) F1 had values no different than B6 mice. In contrast, the F1 mice from a cross of B6-Chr5 × B6-Chr17 restored the prolonged clot stability to the elevated consomic values (Hoover-Plow et al. 2006). Likewise, other studies (Shavit et al. 2009) have reported several loci that modify the von Willibrand factor that is required for platelet aggregation, and Hasstedt et al. (1998) suggested that there was a modifier gene for protein C that increases thromboembolism risk.
In conclusion, this study identifies nine candidate genes on mouse chromosome 17 for thrombosis susceptibility. Of these candidate genes, Emilin2 is a promising candidate gene that warrants further investigation. Emilin2 is expressed in liver, heart, lung, aorta, and most prominently in bone marrow. Emilin2 is differentially expressed in liver, lung, and bone marrow in B6 and A/J mice. B6 and A/J mice have SNPs in Emilin2 in the coding and regulatory regions. In this study we report that EMILIN2 may function to regulate thrombosis as a component of the vascular wall and is critical for maintaining the vessel architecture in response to injury, and/or as a component of the thrombus-facilitating clot lysis. The identification of novel thrombotic susceptibility genes and their homology in humans may uncover new diagnostic and therapeutic factors of thrombotic risk.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Robin Lewis and Subha Raghunathan for assistance with preparation of the manuscript. This study was supported by grants from NIH [HL65204, HL078701 (JHP), and RR12305 (JHN)]. There are no competing financial interests for any of the authors.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alpy F, Jivkov I, Sorokin L, Klein A, Arnold C, et al. Generation of a conditionally null allele of the laminin alpha1 gene. Genesis. 2005;43:59–70. doi: 10.1002/gene.20154. [DOI] [PubMed] [Google Scholar]
- Amma LL, Goodyear R, Faris JS, Jones I, Ng L, et al. An emilin family extracellular matrix protein identified in the cochlear basilar membrane. Mol Cell Neurosci. 2003;23:460–472. doi: 10.1016/S1044-7431(03)00075-7. [DOI] [PubMed] [Google Scholar]
- Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;4:1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman J, Spijker S, Schut D, Wachtler B, Ylstra B, et al. Reduced expression of neuropeptide genes in a genome-wide screen of a secretion-deficient mouse. J Neurochem. 2006;99:84–96. doi: 10.1111/j.1471-4159.2006.04041.x. [DOI] [PubMed] [Google Scholar]
- Braghetta P, Ferrari A, De Gemmis P, Zanetti M, Volpin D, et al. Overlapping, complementary and site-specific expression pattern of genes of the EMILIN/Multimerin family. Matrix Biol. 2004;22:549–556. doi: 10.1016/j.matbio.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Christian S, Ahorn H, Novatchkova M, Garin-Chesa P, Park JE, et al. Molecular cloning and characterization of EndoGlyx-1, an EMILIN-like multisubunit glycoprotein of vascular endothelium. J Biol Chem. 2001;276:48588–48595. doi: 10.1074/jbc.M009604200. [DOI] [PubMed] [Google Scholar]
- Colombatti A, Doliana R, Bot S, Canton A, Mongiat M, et al. The EMILIN protein family. Matrix Biol. 2000;19:289–301. doi: 10.1016/S0945-053X(00)00074-3. [DOI] [PubMed] [Google Scholar]
- Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doliana R, Bot S, Mungiguerra G, Canton A, Cilli SP, et al. Isolation and characterization of EMILIN-2, a new component of the growing EMILINs family and a member of the EMI domain-containing superfamily. J Biol Chem. 2001;276:12003–12011. doi: 10.1074/jbc.M011591200. [DOI] [PubMed] [Google Scholar]
- Ekblom P, Lonai P, Talts JF. Expression and biological role of laminin-1. Matrix Biol. 2003;22:35–47. doi: 10.1016/S0945-053X(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Ginsburg D. Identifying novel genetic determinants of hemostatic balance. J Thromb Haemost. 2005;3:1561–1568. doi: 10.1111/j.1538-7836.2005.01461.x. [DOI] [PubMed] [Google Scholar]
- Glazier A, Nadeau JH. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, et al. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308:1167–1171. doi: 10.1126/science.1109418. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasstedt SJ, Bovill EG, Callas PW, Long GL. An unknown genetic defect increases venous thrombosis risk, through interaction with protein C deficiency. Am J Hum Genet. 1998;63:569–576. doi: 10.1086/301947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward CP, Bainton DF, Smith JW, Horsewood P, Stead RH, et al. Multimerin is found in the alpha-granules of resting platelets and is synthesized by a megakaryocytic cell line. J Clin Invest. 1993;91:2630–2639. doi: 10.1172/JCI116502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward CP, Fuller N, Zheng S, Adam F, Jeimy SB, et al. Human platelets contain forms of factor V in disulfide-linkage with multimerin. Thromb Haemost. 2004;92:1349–1357. doi: 10.1160/TH03-02-0123. [DOI] [PubMed] [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- Hoover-Plow J, Shchurin A, Hart E, Sha J, Hill AE, et al. Genetic background determines response to hemostasis and thrombosis. BMC Blood Disord. 2006;6:6. doi: 10.1186/1471-2326-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, et al. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembertas AV, Perusse L, Chagnon YC, Fisler JS, Warden CH, et al. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest. 1997;100:1240–1247. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie B, Wolff R, Lowe N, Billington CJ, Jr, Peterson A, et al. Twisted gastrulation limits apoptosis in the distal region of the mandibular arch in mice. Dev Biol. 2009;328:13–23. doi: 10.1016/j.ydbio.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait F, Symoens S, Coucke P, Nunes L, De Almeida S. Total absence of the alpha2(I) chain of collagen type I causes a rare form of Ehlers-Danlos syndrome with hypermobility and propensity to cardiac valvular problems. J Med Genet. 2006;43:e36. doi: 10.1136/jmg.2005.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci R, Romano M. Thrombopoietin and its splicing variants: structure and functions in thrombopoiesis and beyond. Biochim Biophys Acta. 2008;1782:427–432. doi: 10.1016/j.bbadis.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Melnick M, Petryk A, Abichaker G, Witcher D, Person AD, et al. Embryonic salivary gland dysmorphogenesis in twisted gastrulation deficient mice. Arch Oral Biol. 2006;51:433–438. doi: 10.1016/j.archoralbio.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanetto M, Tiso N, Braghetta P, Volpin D, Argenton F, et al. Emilin genes are duplicated and dynamically expressed during zebrafish embryonic development. Dev Dyn. 2008;237:222–232. doi: 10.1002/dvdy.21402. [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Urban Z, Boyd C. Genetic disorders of the elastic fiber system. Matrix Biol. 2000;19:471–480. doi: 10.1016/S0945-053X(00)00099-8. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Singer JB, Matin A, Lander ES. Analysing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Nguyen MM, Rivera C, Griep AE. Localization of PDZ domain containing proteins Discs Large-1 and Scribble in the mouse eye. Mol Vis. 2005;11:1183–1199. [PubMed] [Google Scholar]
- Numoto M, Niwa O, Kaplan J, Wong KK, Merrell K, et al. Transcriptional repressor ZF5 identifies a new conserved domain in zinc finger proteins. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SV, Duan ZH, Gupta MK, Surampudi VS, Volinia S, et al. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J Biol Chem. 2009;284:27487–27499. doi: 10.1074/jbc.M109.036541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Price MG, Gomer RH. Skelemin, a cytoskeletal M-disc periphery protein, contains motifs of adhesion/recognition and intermediate filament proteins. J Biol Chem. 1993;268:21800–21810. [PubMed] [Google Scholar]
- Reheman A, Tasneem S, Ni H, Hayward CP. Mice with deleted multimerin 1 and alpha-synuclein genes have impaired platelet adhesion and impaired thrombus formation that is corrected by multimerin 1. Thromb Res. 2010;125(5):e177–e183. doi: 10.1016/j.thromres.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Godfrey M. The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet. 2000;37:9–25. doi: 10.1136/jmg.37.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabin AG, Yin X, Si Q, Larocca JN. Transport of mannose-6-phosphate receptors from the trans-Golgi network to endosomes requires Rab31. Exp Cell Res. 2009;315:2215–2230. doi: 10.1016/j.yexcr.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa Q, Hart E, Hill AE, Nadeau JH, Hoover-Plow JL. Quantitative trait locus analysis for hemostasis and thrombosis. Mamm Genome. 2008;19:406–412. doi: 10.1007/s00335-008-9122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara A, Kasuya H, Akagawa H, Ujiie H, Kubo O, et al. Increased expression of ephrin A1 in brain arteriovenous malformation: DNA microarray analysis. Neurosurg Rev. 2007;30:299–305. doi: 10.1007/s10143-007-0087-3. [DOI] [PubMed] [Google Scholar]
- Scavello GS, Jr, Paluru PC, Zhou J, White PS, Rappaport EF, et al. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- Shavit JA, Manichaikul A, Lemmerhirt HL, Broman KW, Ginsburg D. Modifiers of von Willebrand factor identified by natural variation in inbred strains of mice. Blood. 2009;114:5368–5374. doi: 10.1182/blood-2009-07-233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Burrage LC, Olszens K, Song J, et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Singhal SS, Yadav S, Singhal J, Sahu M, Sehrawat A, et al. Diminished drug transport and augmented radiation sensitivity caused by loss of RLIP76. FEBS Lett. 2008;582:3408–3414. doi: 10.1016/j.febslet.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobek-Klocke I, Disque-Kochem C, Ronsiek M, Klocke R, Jockusch H, et al. The human gene ZFP161 on 18p11.21-pter encodes a putative c-myc repressor and is homologous to murine Zfp161 (Chr 17) and Zfp161-rs1 (X Chr) Genomics. 1997;43:156–164. doi: 10.1006/geno.1997.4784. [DOI] [PubMed] [Google Scholar]
- Soria JM, Almasy L, Souto JC, Buil A, Martinez-Sanchez E, et al. A new locus on chromosome 18 that influences normal variation in activated protein C resistance phenotype and factor VIII activity and its relation to thrombosis susceptibility. Blood. 2003;101:163–167. doi: 10.1182/blood-2002-06-1792. [DOI] [PubMed] [Google Scholar]
- Steiner F, Weber K, Furst DO. M band proteins myomesin and skelemin are encoded by the same gene: analysis of its organization and expression. Genomics. 1999;56:78–89. doi: 10.1006/geno.1998.5682. [DOI] [PubMed] [Google Scholar]
- Stenina OI, Topol EJ, Plow EF. Thrombospondins, their polymorphisms, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2007;27:1886–1894. doi: 10.1161/ATVBAHA.107.141713. [DOI] [PubMed] [Google Scholar]
- Sudbery I, Stalker J, Simpson JT, Keane T, Rust AG, et al. Deep short-read sequencing of chromosome 17 from the mouse strains A/J and CAST/Ei identifies significant germline variation and candidate genes that regulate liver triglyceride levels. Genome Biol. 2009;10:R112. doi: 10.1186/gb-2009-10-10-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voetsch B, Loscalzo J. Genetic determinants of arterial thrombosis. Arterioscler Thromb Vasc Biol. 2004;24:216–229. doi: 10.1161/01.ATV.0000107402.79771.fc. [DOI] [PubMed] [Google Scholar]
- Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchigna L, Vecchione C, Notte A, Cordenonsi M, Dupont S, et al. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell. 2006;124:929–942. doi: 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zanetti M, Braghetta P, Sabatelli P, Mura I, Doliana R, et al. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Mol Cell Biol. 2004;24:638–650. doi: 10.1128/MCB.24.2.638-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan M, van N Nimwegen E. The types and prevalence of alternative splice forms. Curr Opin Struct Biol. 2006;16:362–367. doi: 10.1016/j.sbi.2006.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.