Abstract
Chitin is an essential part of the carbohydrate skeleton of the fungal cell wall and is a molecule that is not represented in humans and other vertebrates. Complex regulatory mechanisms enable chitin to be positioned at specific sites throughout the cell cycle to maintain the overall strength of the wall and enable rapid, life-saving modifications to be made under cell wall stress conditions. Chitin has also recently emerged as a significant player in the activation and attenuation of immune responses to fungi and other chitin-containing parasites. This review summarises latest advances in the analysis of chitin synthesis regulation in the context of fungal pathogenesis.
Introduction
Chitin is an essential component of the cell walls and septa of all pathogenic fungi, and occurs in the cyst walls of pathogenic amoebae, the egg-shells and gut lining of parasitic nematodes and the exoskeletons of invertebrate vectors of human disease including mosquitoes, sand flies, ticks and snails. Despite the fact that chitin is only outweighed in abundance in nature by cellulose and is present and essential in so many parasites and pathogens, fundamental information about its biosynthesis and recognition by the immune system is lacking.
Chitin, a β(1,4)-linked homopolymer of N-acetylglucosamine, is a simple polysaccharide that is represented in the cell walls of all fungi studied to date [1,2]. The nascent primary polysaccharide of fungi folds back on itself to form anti-parallel chains, which form intra-chain hydrogen bonds that further stiffen the carbohydrate into immensely strong fibrous microfibrils tougher than any other molecule in nature, and stronger, weight-for-weight, than bone or steel (Figure 1). The 3D network of chitin microfibrils is covalently attached to β(1,3)-glucan — a second load-bearing polysaccharide present in most fungal cell walls [3]. In many species, such as the human pathogen Candida albicans, the major classes of cell wall proteins are attached through a GPI-remnant to β(1,3)-glucan or chitin via a branched β(1,6)-glucan linker [3]. A variable proportion of fungal chitin is synthesised and then deacetylated to chitosan by the action of one or more chitin deacetylases. In C. albicans, less than 5% of chitin is deacetylated to chitosan, while most zygomycetes and the basidomycete Cryptococcus neoformans have more than two thirds deacetylated to chitosan [4,5]. Chitin deacetylation may make the polymer more elastic and protect it from the action of hostile chitinases.
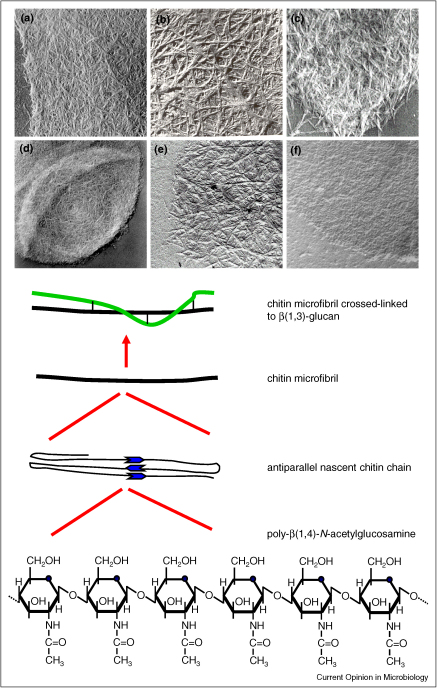
Figure 1.
Chitin structure and diversity in fungi. Chitin is a β(1,4)-homopolymer of N-acetylglucosamine that folds in an anti-parallel manner forming intra-chain hydrogen bonds. Chitin chains are cross-linked covalently to β(1,3)-glucan (green) to form the inner skeleton of most fungi. Examples of shadow cast electron microscopy images of chitin from (a)Neurospora crassa; (b)Coprinus cinereus; (c) chitin–chitosan from Mucor mucedo; and (d)Candida albicans. In (e) and (f), the structure of chitin from C. albicans is shown in a chs3Δ and chs8Δ mutant, respectively, demonstrating that the architecture of chitin is genetically determined [24].
The apparent simplicity of the chitin primary structure belies a complex underlying biosynthetic process. Chitin is synthesised by large families of chitin synthase (CHS) enzymes that fall into seven discernable classes (Table 1). We have chosen to follow the classification proposed by Niño-Vega et al. [6] and Roncero [7], and not that of Chigira et al. [8] and Choquer et al. [9] who reverse classes VI and VII. The functional significance of all CHS classes is not clear and seems to differ in different fungi [1,2]. The roles of individual CHS genes have been investigated principally by analysis of specific CHS deletion strains. Class I enzymes are most readily measured in in vitro biochemical assays, yet they normally make only a minor fraction of cell wall chitin, and mutants lacking class I CHS genes are invariably viable with mild phenotypes under non-stressed conditions [1]. Class II enzymes often are immeasurable in enzyme assays, and make little chitin, but their deletion results in marked deleterious effects on cell viability through effects on vital processes such as primary septum formation [10]. Class IV enzymes often make substantial amounts of wall chitin, but mutants are usually viable although sometimes attenuated in virulence [11]. Class IV, V and VII sequences share some sequence homology, and class V and some class VII proteins also contain myosin-like domains. Class III, V, VI and VII have only been identified in filamentous fungi and some dimorphic fungi and are absent from yeasts like Saccharomyces cerevisiae and C. albicans.
Table 1.
Members of the seven classes of CHS enzymes in various fungia
| Fungusb | I | II | III | IV | V | VI | VII | Total |
|---|---|---|---|---|---|---|---|---|
| S. cerevisiaed | Chs1 | Chs2 | Chs3 | 3 | ||||
| C. albicansd | Chs2 Chs8 |
Chs1 | Chs3 | 4 | ||||
| A. nidulansd | ChsC | ChsA | ChsB ChsF |
ChsD | CsmA | ChsG | CsmB | 8 |
| A. fumigatusd | ChsA | ChsB | ChsC ChsG |
ChsF | ChsE | ChsD | Afu2g13430 | 8 |
| W. dermatitidisd | Chs2 | Chs1 | Chs3 | Chs4 | Chs5 | Chs6 | Chs7 | 7 |
| U. maydise | Chs3 Chs4 |
Chs2 | Chs1 | Chs5 Chs7 |
Mcs1 | Chs6 | 8 | |
| C. neoformanse | Chs6 Chs8 |
Chs6 Chs8 |
Chs2 Chs7 |
Chs1 Chs3 |
Chs5 | Chs4 | 8 | |
| P. blakesleeanusf | Chs5 Chs6 |
Chs1 Chs2 Chs3 Chs4 |
Chs7 Chs8 Chs9 Chs10 |
10c |
Standard genetic nomenclature for S. cerevisiae and C. albicans has been used to designate CHS proteins from all fungi. The enzymes have been assigned to classes based on the classification proposed by Niño-Vega et al. [6] and Roncero [7].
d: ascomycete; e: basidomycete; and f: zygomycete.
34 putative CHSs have been annotated in the P. blakesleeanus genome (http://genome.jgi-psf.org/Phybl2/Phybl2.home.html).
The multiplicity of CHS enzymes suggests that they may have redundant roles in cell wall synthesis. This is the case for some but not all CHS enzymes. It is clear that the expression and activity of CHS is highly regulated both throughout the cell cycle and under conditions of stress, such as in response to potentially lethal challenges to cell integrity imposed by lytic enzymes or antibiotics or oxidants that are generated by the respiratory burst within the phagolysomes of lymphocytes.
Transcriptional regulation of chitin synthesis
In most fungi, chitin and cell wall synthesis occurs at sites of polarised growth. During early bud growth, cell wall material is deposited at the bud tip [12]. A period of isotropic growth occurs in large budded cells where material is deposited over the entire bud surface. Following nuclear division, a repolarisation phase begins where material is directed towards the mother-bud neck to prepare for cytokinesis. In hyphal or filamentous forms, cell extension is a continuous and indefinite process of apical growth [13].
Accordingly, chitin synthesis must be regulated both temporally and spatially in relation to the cell cycle. The S. cerevisiae class II enzyme ScChs2 synthesises the primary septum and ScChs1 of class I acts a repair enzyme that replenishes chitin in the birth/bud scar immediately after cytokinesis [1,14]. A genome-wide analysis of cell cycle regulation at the mRNA level using synchronised S. cerevisiae cultures indicated that expression of ScCHS2 peaked in M-phase and ScCHS1 in M/G1, both at appropriate times for the functions of these enzymes [15]. Transcription during the cell cycle of an opaque C. albicans MTLa FAR1OP strain synchronised after release from α-factor arrest showed that expression of CaCHS1 (orthologous to ScCHS2), CaCHS8 and to a lesser extent CaCHS3 peaked in G2 phase, whereas expression of CaCHS2 was non-periodic [16•].
Disruption of cell wall biosynthetic genes or treatments with cell wall perturbing agents often results in compensatory alterations in the cell wall, including activation of chitin synthesis, in an attempt to maintain cellular integrity (reviewed in [17]). Defects in the cell wall are sensed in S. cerevisiae by transmembrane proteins such as ScWsc1 and ScMid2, or the signalling mucins ScMsb2 and ScHkr1 that activate downstream mitogen-activated protein (MAP) kinase cascades to bring about cell wall remodelling (Figure 2). In S. cerevisiae this so-called ‘cell wall salvage’ or ‘cell wall compensatory’ pathway is mediated primarily through the protein kinase C (PKC) cell integrity MAP kinase cascade and its downstream target the transcription factor ScRlm1 [18]. A second MAP kinase cascade, the high osmolarity glycerol response (HOG) pathway, has also been suggested to play a role in regulating cell wall architecture [19] (Figure 2).
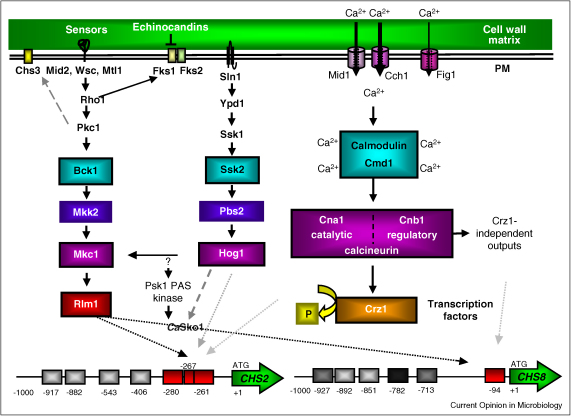
Figure 2.
Signalling pathways that regulate Candida albicansCHS gene expression. The HOG, PKC and the Ca2+/calcineurin signalling pathways regulate chitin synthesis and CHS gene expression [20]. The Rlm1 transcription factor downstream of the PKC MAP kinase cascade controls the expression of a number of cell wall related genes in S. cerevisiae. Putative Rlm1 binding motifs (red boxes) in the promoters of CaCHS2 and CaCHS8 contribute to their cell wall stress-activated regulation [21]. Activation of the calcineurin pathway results in de-phosphorylation of the CaCrz1 transcription factor. CaCrz1 then moves to the nucleus and induces expression of genes with CDREs (calcium-dependent response elements) within their promoter sequences. C. albicans has significant re-wiring of signalling pathways compared to S. cerevisiae, for example, the role of the CaSko1 transcription factor in response to caspofungin is independent of the CaHog1 MAP kinase but involves the CaPsk1 PAS kinase [26]. Potential CDREs and Sko1 binding sites identified in silico upstream of CaCHS2 and CaCHS8 (grey boxes) were not required for regulation of gene expression [21]. Sequences in the first 347 bp and 125 bp of the CaCHS2 and CaCHS8 promoters governed expression through these three signalling pathways [21].
Adapted from [17].
In C. albicans, the PKC and HOG MAP kinase cascades and the Ca2+/calcineurin pathway regulate CHS gene expression and chitin synthesis in response to cell wall stresses [20]. Promoter dissection experiments have defined the regulatory regions of the class I CHS promoter sequences and revealed that C. albicans uses different transcription factors and/or consensus binding sequences to regulate chitin synthesis compared to S. cerevisiae [21] (Figure 2).
Upregulation of chitin synthesis in response to cell wall stress may be clinically relevant. In C. albicans, the transcriptional activation of chitin synthesis through the PKC, HOG and Ca2+/calcineurin pathways confers resistance to the echinocandin class of antifungal drugs [22•], and in Aspergillus fumigatus, calcineurin-mediated elevated transcription of AfCHSA and AfCHSC is required for paradoxical growth in the presence of high concentrations of the echinocandin drug caspofungin [23••].
Post-transcriptional regulation of chitin synthesis
Coordinated synthesis of chitin also requires the localisation of the enzymes to be regulated throughout the cell cycle. Fungal CHS enzymes that are responsible for synthesising cell wall and/or septal chitin tend to be localised to sites of polarised growth. For example, in C. albicans, the class IV enzyme CaChs3 is involved in both cell wall and septum synthesis [11]. It is localised to the tip of growing buds and hyphae and relocates to sites of septum formation before cytokinesis [24]. Similarly, in A. nidulans, the class III enzyme AnChsB appears to function at polarised growth sites and in forming septa during hyphal growth and conidia development [25].
Both the localisation and stability of CHS enzymes can be regulated by phosphorylation. Recent work has shown that protein kinases and the establishment of cell polarity appear to be important for cell wall regulation in C. albicans [26]. In S. cerevisiae, the primary septum is synthesised by ScChs2 [1,14]. Phosphorylation of ScChs2 by ScCdk1 at 4 N-terminal sites retains this enzyme in the Endoplasmic Reticulum (ER) until mitotic exit [27]. Deletion of these phospho-sites resulted in ScChs2 degradation [28], indicating that phosphorylation regulates chitin synthesis by ScChs2 at specific stages of the cell cycle, either by regulating the cellular localisation or stability of the protein. Three of the four CHS enzymes are phosphorylated in C. albicans [29], and phosphorylation of CaChs3 on S139 is required to target this enzyme to sites of polarised growth [30•] (Figure 3).
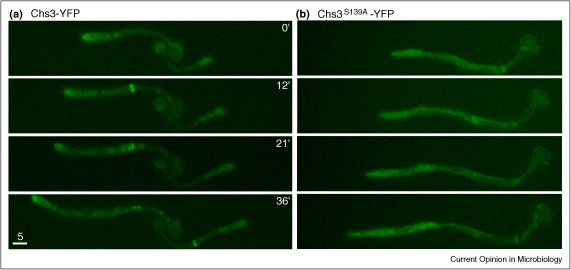
Figure 3.
Phosphorylation of Candida albicans Chs3 on a specific serine residue is required to target the CHS to sites of polarised growth. CaChs3 was tagged with yellow-fluorescent protein (YFP) and the localisation of the CHS in growing hyphae was observed by time-lapse fluorescence microscopy. (a)CaChs3-YFP localises to the tip of the growing hypha and then flashes at the site of septum formation. (b) Mislocalisation of CaChs3 is observed when the serine at position 139 has been mutated to an alanine that cannot be phosphorylated (Chs3S139A-YFP).
Modified from [30•].
The regulation of localisation of the class IV CHS enzyme in S. cerevisiae has been studied extensively and involves a number of different post-transcriptional regulatory mechanisms. Export of ScChs3 from the ER is controlled by the chaperone ScChs7 and transportation from the Golgi to the plasma membrane (PM) occurs in specialised vesicles called chitosomes [31,32]. Generation of the PtdIns(4)P lipid by ScPik1 promotes forward transport of ScChs3 to the PM and subsequent de-phosphorylation by ScSac1 terminates the signal, allowing ScChs3 to remain in the PM and synthesise chitin [33]. ScChs3 is targeted to the bud neck for septum formation at an appropriate time in the cell cycle through interactions with ScChs4, ScBni4, ScGlc7 and the septins [14,34–38].
In addition to mechanisms involving post-translational modifications or protein–protein interactions, some CHSs are hybrid proteins with N-terminal myosin motor-like domains (MMD) and C-terminal CHS domains. These enzymes tend to fall into classes V and VII, and the best characterised examples from human pathogens include WdChs5 (the class V enzyme from Wangiella dermatitidis; [39]), AnCsmA and AnCsmB (class V and VII from A. nidulans; [40,41]). Using their MMD, these enzymes appear to localise themselves to sites of polarised cell wall expansion in an actin-dependent manner [40,42–44]. The MMD may not possess the motor activity of traditional myosins [43], however, in the plant pathogen Ustilago maydis, the myosin-like domain of the class V UmMcs1 enzyme has been shown to be essential for its apical localisation and is involved in retention of UmMcs1 in the apical dome (G Steinberg, personal communication).
Chitin and immune recognition
The immune system has evolved to detect conserved, basic molecular components of microorganisms called pathogen associated molecular patterns (PAMPs). β(1,3)-Glucan is a well studied PAMP that is detected by Dectin-1, a C-type lectin receptor of monocytes and macrophages, and is of major importance in activating a strong, pro-inflammatory response by the innate immune system [45]. In C. albicans, access to the inner cell wall layer containing β-glucan and chitin is normally shielded by the superficial mannans and so immune detection of intact cells is initially focused on mannan-immune receptor interactions [46]. However, β-glucan can be unmasked by exposure to host enzymes, antifungal drugs, heat-treatment and in mannosylation deficient mutants [47]. In a similar way, an outer α(1,3)-glucan layer masks detection of the inner chitin/β-glucan layer in Histoplasma capsulatum [48]. In the case of β-glucan, unmasking has major immunomodulatory consequences. What therefore is the immunological role of chitin, the other highly conserved signature molecule in the inner cell wall?
Recent studies have begun to reveal a complex picture regarding the immunological properties of chitin [49]. Immune responses seem to be highly dependent on the size of the chitin fragments used to stimulate immune cells [50]. Very large (>100 μm) chitin fragments, normally prepared from invertebrate sources, seem to be immunologically inert, while intermediate (40–70 μm) and small chitin (<40 μm) seem capable of activating macrophages and eliciting IL-17, TNF and IL-23 production via a range of pattern recognition receptors (PRRs) [51]. All of these particles are quite large relative to the sizes of fungal cells and the molecular scale of immune receptor–ligand interactions. Also, such experiments have yet to accommodate the fact that the structure of chitin microfibrils varies significantly in different organisms and even in different parts of the cell wall [24] (Figure 1). Size-dependent immune reactivity helps explain the importance of chitin-degrading proteins, such as acidic mammalian chitinase [52] and chitotriosidase [53,54] in allergic immune responses. These enzymes presumably digest chitin into smaller particles, capable of mediating allergic responses. Their ability to digest large chitin fragments results in the accumulation of IL-4 expressing basophils, eosinophils and neutrophils in tissues and induces alternative macrophage activation that is associated with the immunity to certain chitin-containing parasites such as the helminth Nippostrongylus brasiliensis [55••]. Reciprocally, it appears as though certain human proteins sequester chitin to dampen immune responses [56].
Recently, several candidate mediators of chitin mediated immune responses have been identified. RegIIIg (HIP/PAP) is a C-type lectin with an affinity for chitin that is expressed in the neutrophil-like Paneth cells of the small intestine and is induced by certain bacterial peptidoglycans [57]. Peptidoglycans are chemically related to chitin in so far as they are N-acetylglucosamine containing polysaccharides and the two molecules can activate some common cellular responses [58]. FIBCD1 has also been identified as a calcium-dependent acetyl group-binding tetrameric chitin-binding receptor [59••]. The binding between this chitin receptor and acetylated BSA could be inhibited by a variety of acetylated compounds, but not glucosamine or glucose [59••]. The requirement for acetylation suggests that this receptor is unresponsive to chitosan, which is known to be able to activate dendritic cells via a TLR4-dependent mechanism [60]. This is important because fungal pathogens such as the zygomycetous fungi and C. neoformans have substantial amounts of deacetylated chitin in their walls. Therefore further chitin and chitosan immune receptors remain to be identified and the role of fungal chitin in immune recognition requires investigation.
Chitin as a target for anti-fungal chemotherapy
Chitin and chitosan are hallmark polysaccharides that are present in all known fungal pathogens and not in humans. Inhibition of chitin synthesis has therefore been proposed as an attractive target for antifungal therapies. However, no CHS inhibitor has ever progressed into clinical practice [61]. Existing CHS inhibitors such as the nikkomycins and polyoxins are most potent and specific against class I enzymes but are less effective inhibitors of other classes of CHS enzymes, and of fungal growth in vivo [61,62].
The discovery of the essential role of C. albicans CHS1 [10] prompted the screening for class II CHS inhibitors. Roche developed one compound, RO-09-3143, a specific inhibitor of CaChs1 that was fungistatic to C. albicans, but cidal in a chs2Δ mutant background [63]. This highlighted the possibility of compensatory functions for different CHS enzymes and was reinforced by the observation that C. albicans yeast cells stimulated to synthesise chitin by treatment with Ca2+ and Calcofluor White were able to grow and divide by forming a novel salvage septum in the absence the otherwise essential CaCHS1 gene [22•].
A common response to cell wall damage is strengthening of the wall by the production of excess chitin, primarily by the class IV enzymes such as ScChs3 and CaChs3 [20,64]. This compensatory mechanism is triggered when fungi are treated with echinocandin drugs [22•,65,66]. Enhanced chitin levels reduces susceptibility to echinocandin drugs in C. albicans [17,22•], but reciprocally, combinations of CHS and glucan synthase inhibitors are more potent against C. albicans and A. fumigatus than individual drug treatments [22•,67]. These studies highlight the potential of combination therapies that target the synthesis of the two major structural polysaccharides found in most fungi, in achieving fungicidal regimens that would prevent the emergence of resistance mechanisms. In addition, cell wall synthase inhibitors, applied in combination with antagonists of the signalling pathways that regulate synthase expression and activity, may have potential as potent antifungal combination therapies. For example, the calcineurin pathway is important for regulation of chitin synthesis in C. albicans and A. fumigatus as well as the response to echinocandin drugs, and drugs that block the calcineurin pathway act synergistically with the echinocandins [20,22•,67,68].
Conclusions
The primary structure of chitin comprises a single sugar type and a single inter-sugar linkage. None-the-less it is diverse in structure and form and is assembled by different classes of enzymes encoded by families of genes whose expression is regulated in a cell cycle-dependent manner at the transcriptional and post-transcriptional levels. Recent advances in our understanding of how chitin synthesis is regulated and responds to cell wall stress sustains the attraction of this process as a potential antifungal drug target, possibly in combination with inhibitors of β(1,3)-glucan synthesis and/or the cell wall compensatory pathway. The relevance of fungal chitin synthesis in human disease is also evident in emerging research defining the role of this fungal signature molecule in immune recognition mechanisms.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Our work in this area is supported by the Wellcome Trust, EC Marie Curie programme and the BBSRC.
References
- 1.Munro C.A., Gow N.A.R. Chitin synthesis in human pathogenic fungi. Med Mycol. 2001;39 S1:41–53. [PubMed] [Google Scholar]
- 2.Latge J.P. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 3.Klis F.M., Boorsma A., De Groot P.W. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- 4.Baker L.G., Specht C.A., Donlin M.J., Lodge J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 6.Niño-Vega G.A., Carrero L., San-Blas G. Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med Mycol. 2004;42:51–57. doi: 10.1080/1369378031000153811. [DOI] [PubMed] [Google Scholar]
- 7.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 8.Chigira Y., Abe K., Gomi K., Nakajima T. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr Genet. 2002;41:261–267. doi: 10.1007/s00294-002-0305-z. [DOI] [PubMed] [Google Scholar]
- 9.Choquer M., Boccara M., Goncalves I.R., Soulie M.C., Vidal-Cros A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem. 2004;271:2153–2164. doi: 10.1111/j.1432-1033.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 10.Munro C.A., Winter K., Buchan A., Henry K., Becker J.M., Brown A.J., Bulawa C.E., Gow N.A.R. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol Microbiol. 2001;39:1414–1426. doi: 10.1046/j.1365-2958.2001.02347.x. [DOI] [PubMed] [Google Scholar]
- 11.Bulawa C.E., Miller D.W., Henry L.K., Becker J.M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci U S A. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheu Y.J., Barral Y., Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer R., Zekert N., Takeshita N. Polarized growth in fungi-interplay between the cytoskeleton, positional markers and membrane domains. Mol Microbiol. 2008;68:813–826. doi: 10.1111/j.1365-2958.2008.06193.x. [DOI] [PubMed] [Google Scholar]
- 14.Bulawa C.E. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- 15.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Côte P., Hogues H., Whiteway M. Transcriptional analysis of the Candida albicans cell cycle. Mol Biol Cell. 2009;20:3363–3373. doi: 10.1091/mbc.E09-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]; The difficulty in synchronising the cell cycle within a population of C. albicans cells has hampered investigations into cell cycle regulation in this organism. Here, the authors describe a novel system allowing C. albicans cells of particular genetic background to be synchronised using mating pheromone treatment.
- 17.Walker L.A., Gow N.A.R., Munro C.A. Fungal echinocandin resistance. Fungal Genet Biol. 2010;47:117–126. doi: 10.1016/j.fgb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia R., Bermejo C., Grau C., Perez R., Rodriguez-Pena J.M., Francois J., Nombela C., Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- 19.Bermejo C., Rodriguez E., Garcia R., Rodriguez-Pena J.M., Rodriguez de la Concepcion M.L., Rivas C., Arias P., Nombela C., Posas F., Arroyo J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munro C.A., Selvaggini S., de Bruijn I., Walker L., Lenardon M.D., Gerssen B., Milne S., Brown A.J., Gow N.A.R. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenardon M.D., Lesiak I., Munro C.A., Gow N.A.R. Dissection of the Candida albicans class I chitin synthase promoters. Mol Genet Genomics. 2009;281:459–471. doi: 10.1007/s00438-009-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Walker L.A., Munro C.A., de Bruijn I., Lenardon M.D., McKinnon A., Gow N.A.R. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows how chitin is upregulated in response to echinocandin treatment and that concomitant inhibition of β(1,3)-glucan and chitin synthesis acts synergistically in killing C. albicans. The paper also describes how activation of the chitin compensatory response can overcome the otherwise lethal effects of mutations in the normally essential CHS1 gene of C. albicans.
- 23••.Fortwendel J.R., Juvvadi P.R., Perfect B.Z., Rogg L.E., Perfect J.R., Steinbach W.J. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob Agents Chemother. 2010;54:1555–1563. doi: 10.1128/AAC.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; The activity of echinocandin drugs is attenuated when high concentrations of the drug are used to treat C. albicans and A. fumigatus. Work presented in paper complements and extends our understanding of this ‘parodoxical effect’ in C. albicans [20,22•] by elucidating the molecular mechanisms regulating the paradoxical effect in A. fumigatus.
- 24.Lenardon M.D., Whitton R.K., Munro C.A., Marshall D., Gow N.A.R. Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol Microbiol. 2007;66:1164–1173. doi: 10.1111/j.1365-2958.2007.05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda K., Yamada K., Deoka K., Yamashita S., Ohta A., Horiuchi H. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot Cell. 2009;8:945–956. doi: 10.1128/EC.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blankenship J.R., Fanning S., Hamaker J.J., Mitchell A.P. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teh E.M., Chai C.C., Yeong F.M. Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle. 2009;8:2965–2976. [PubMed] [Google Scholar]
- 28.Martinez-Rucobo F.W., Eckhardt-Strelau L., Terwisscha van Scheltinga A.C. Yeast chitin synthase 2 activity is modulated by proteolysis and phosphorylation. Biochem J. 2009;417:547–554. doi: 10.1042/BJ20081475. [DOI] [PubMed] [Google Scholar]
- 29.Beltrao P., Trinidad J.C., Fiedler D., Roguev A., Lim W.A., Shokat K.M., Burlingame A.L., Krogan N.J. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Lenardon M.D., Milne S.A., Mora-Montes H.M., Kaffarnik F.A.R., Peck S.C., Brown A.J.P., Munro C.A., Gow N.A.R. Phosphorylation regulates polarisation of chitin synthesis in Candida albicans. J Cell Sci. 2010 doi: 10.1242/jcs.060210. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates for the first time that phosphorylation of a chitin synthase (CaChs3) is essential for its normal localisation in both yeast and hyphal cells.
- 31.Chuang J.S., Schekman R.W. Differential trafficking and timed localization of two chitin synthase proteins. Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartnicki-Garcia S. Chitosomes: past, present and future. FEMS Yeast Res. 2006;6:957–965. doi: 10.1111/j.1567-1364.2006.00158.x. [DOI] [PubMed] [Google Scholar]
- 33.Schorr M., Then A., Tahirovic S., Hug N., Mayinger P. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr Biol. 2001;11:1421–1426. doi: 10.1016/s0960-9822(01)00449-3. [DOI] [PubMed] [Google Scholar]
- 34.Reyes A., Sanz M., Duran A., Roncero C. Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane. J Cell Sci. 2007;120:1998–2009. doi: 10.1242/jcs.005124. [DOI] [PubMed] [Google Scholar]
- 35.DeMarini D.J., Adams A.E., Fares H., De Virgilio C., Valle G., Chuang J.S., Pringle J.R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozubowski L., Panek H., Rosenthal A., Bloecher A., DeMarini D.J., Tatchell K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol Biol Cell. 2003;14:26–39. doi: 10.1091/mbc.E02-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson J.R., Bharucha J.P., Ceaser S., Salamon J., Richardson C.J., Rivera S.M., Tatchell K. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:3040–3051. doi: 10.1091/mbc.E08-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larson J.R., Kozubowski L., Tatchell K. Changes in Bni4 localization induced by cell stress in Saccharomyces cerevisiae. J Cell Sci. 2010;123:1050–1059. doi: 10.1242/jcs.066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., Kauffman S., Becker J.M., Szaniszlo P.J. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot Cell. 2004;3:40–51. doi: 10.1128/EC.3.1.40-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshita N., Yamashita S., Ohta A., Horiuchi H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol. 2006;59:1380–1394. doi: 10.1111/j.1365-2958.2006.05030.x. [DOI] [PubMed] [Google Scholar]
- 41.Fujiwara M., Horiuchi H., Ohta A., Takagi M. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem Biophys Res Commun. 1997;236:75–78. doi: 10.1006/bbrc.1997.6907. [DOI] [PubMed] [Google Scholar]
- 42.Abramczyk D., Park C., Szaniszlo P.J. Cytolocalization of the class V chitin synthase in the yeast, hyphal and sclerotic morphotypes of Wangiella (Exophiala) dermatitidis. Fungal Genet Biol. 2009;46:28–41. doi: 10.1016/j.fgb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuizaki M., Takeshita N., Ohta A., Horiuchi H. Myosin motor-like domain of the class VI chitin synthase CsmB is essential to its functions in Aspergillus nidulans. Biosci Biotechnol Biochem. 2009;73:1163–1167. doi: 10.1271/bbb.90074. [DOI] [PubMed] [Google Scholar]
- 44.Takeshita N., Ohta A., Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol Biol Cell. 2005;16:1961–1970. doi: 10.1091/mbc.E04-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid D.M., Gow N.A.R., Brown G.D. Pattern recognition: recent insights from Dectin-1. Curr Opin Immunol. 2009;21:30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Netea M.G., Gow N.A.R., Munro C.A., Bates S., Collins C., Ferwerda G., Hobson R.P., Bertram G., Hughes H.B., Jansen T. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Netea M.G., Brown G.D., Kullberg B.J., Gow N.A.R. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 48.Rappleye C.A., Eissenberg L.G., Goldman W.E. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C.G., Da Silva C.A., Lee J.Y., Hartl D., Elias J.A. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol. 2008;20:684–689. doi: 10.1016/j.coi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Da Silva C.A., Chalouni C., Williams A., Hartl D., Lee C.G., Elias J.A. Chitin is a size-dependent regulator of macrophage TNF and IL-10 production. J Immunol. 2009;182:3573–3582. doi: 10.4049/jimmunol.0802113. [DOI] [PubMed] [Google Scholar]
- 51.Da Silva C.A., Hartl D., Liu W., Lee C.G., Elias J.A. TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. J Immunol. 2008;181:4279–4286. doi: 10.4049/jimmunol.181.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z., Zheng T., Homer R.J., Kim Y.K., Chen N.Y., Cohn L., Hamid Q., Elias J.A. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 53.Gordon-Thomson C., Kumari A., Tomkins L., Holford P., Djordjevic J.T., Wright L.C., Sorrell T.C., Moore G.P. Chitotriosidase and gene therapy for fungal infections. Cell Mol Life Sci. 2009;66:1116–1125. doi: 10.1007/s00018-009-8765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee C.G. Chitin, chitinases and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med J. 2009;50:22–30. doi: 10.3349/ymj.2009.50.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Reese T.A., Liang H.E., Tager A.M., Luster A.D., Van Rooijen N., Voehringer D., Locksley R.M. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447:92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors show that chitin activates macrophages through the alternative pathway and induces the accumulation of innate immune cells associated with the allergic response in tissues. Accumulation of these immune cells was abolished in the presence of human chitinase. This is the first report directly linking chitin to allergy.
- 56.de Jonge R., Thomma B.P. Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 2009;17:151–157. doi: 10.1016/j.tim.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Cash H.L., Whitham C.V., Behrendt C.L., Hooper L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X.L., Lee R.T., Fang H.M., Wang Y.M., Li R., Zou H., Zhu Y., Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 59••.Schlosser A., Thomsen T., Moeller J.B., Nielsen O., Tornoe I., Mollenhauer J., Moestrup S.K., Holmskov U. Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J Immunol. 2009;183:3800–3809. doi: 10.4049/jimmunol.0901526. [DOI] [PubMed] [Google Scholar]; This is the first report of a genuine candidate for a chitin receptor. Here, the authors show that FIBCD1 binds acetylated compounds such as chitin with high-affinity in a calcium-dependent manner. This receptor, however, does not recognise deacetylated compounds, leaving the way open for investigations to find additional chitin and chitosan receptors. The full tissue expression profile for this receptor also remains to be determined.
- 60.Villiers C., Chevallet M., Diemer H., Couderc R., Freitas H., Van Dorsselaer A., Marche P.N., Rabilloud T. From secretome analysis to immunology: chitosan induces major alterations in the activation of dendritic cells via a TLR4-dependent mechanism. Mol Cell Proteomics. 2009;8:1252–1264. doi: 10.1074/mcp.M800589-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munro C.A., Gow N.A.R. Chitin biosynthesis as a target for antifungals. In: Dixon G.K., Copping L.G., Hollomon D.W., editors. Antifungal Agents: Discovery and Mode of Action. Bios Scientific; 1995. pp. 161–171. [Google Scholar]
- 62.Gaughran J.P., Lai M.H., Kirsch D.R., Silverman S.J. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J Bacteriol. 1994;176:5857–5860. doi: 10.1128/jb.176.18.5857-5860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sudoh M., Yamazaki T., Masubuchi K., Taniguchi M., Shimma N., Arisawa M., Yamada-Okabe H. Identification of a novel inhibitor specific to the fungal chitin synthase. Inhibition of chitin synthase 1 arrests the cell growth, but inhibition of chitin synthase 1 and 2 is lethal in the pathogenic fungus Candida albicans. J Biol Chem. 2000;275:32901–32905. doi: 10.1074/jbc.M003634200. [DOI] [PubMed] [Google Scholar]
- 64.Carotti C., Ferrario L., Roncero C., Valdivieso M.H., Duran A., Popolo L. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast. 2002;19:1113–1124. doi: 10.1002/yea.905. [DOI] [PubMed] [Google Scholar]
- 65.Reinoso-Martin C., Schuller C., Schuetzer-Muehlbauer M., Kuchler K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot Cell. 2003;2:1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cota J.M., Grabinski J.L., Talbert R.L., Burgess D.S., Rogers P.D., Edlind T.D., Wiederhold N.P. Increases in SLT2 expression and chitin content are associated with incomplete killing of Candida glabrata by caspofungin. Antimicrob Agents Chemother. 2008;52:1144–1146. doi: 10.1128/AAC.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fortwendel J.R., Juvvadi P.R., Pinchai N., Perfect B.Z., Alspaugh J.A., Perfect J.R., Steinbach W.J. Differential effects of inhibiting chitin and 1,3-(beta)-d-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob Agents Chemother. 2009;53:476–482. doi: 10.1128/AAC.01154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steinbach W.J., Reedy J.L., Cramer R.A., Jr., Perfect J.R., Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]