Abstract
AIM: To investigate the gene expression pattern of hepatocyte nuclear factor 6 (HNF6) and other liver-enriched transcription factors in various segments of the human intestine to better understand the differentiation of the gut epithelium.
METHODS: Samples of healthy duodenum and jejunum were obtained from patients with pancreatic cancer whereas ileum and colon was obtained from patients undergoing right or left hemicolectomy or (recto)sigmoid or rectal resection. All surgical specimens were subjected to histopathology. Excised tissue was shock-frozen and analyzed for gene expression of liver-enriched transcription factors by semiquantitative reverse transcription polymerase chain and compared to the human colon carcinoma cell line Caco-2. Protein expression of major liver-enriched transcription factors was determined by Western blotting while the DNA binding of HNF6 was investigated by electromobility shift assays.
RESULTS: The gene expression patterning of liver-enriched transcription factors differed in the various segments of the human intestine with HNF6 gene expression being most abundant in the duodenum (P < 0.05) whereas expression of the zinc finger protein GATA4 and of the HNF6 target gene ALDH3A1 was most abundant in the jejunum (P < 0.05). Likewise, expression of FOXA2 and the splice variants 2 and 4 of HNF4α were most abundantly expressed in the jejunum (P < 0.05). Essentially, expression of transcription factors declined from the duodenum towards the colon with the most abundant expression in the jejunum and less in the ileum. The expression of HNF6 and of genes targeted by this factor, i.e. neurogenin 3 (NGN3) was most abundant in the jejunum followed by the ileum and the colon while DNA binding activity of HNF4α and of NGN3 was confirmed by electromobility shift assays to an optimized probe. Furthermore, Western blotting provided evidence of the expression of several liver-enriched transcription factors in cultures of colon epithelial cells, albeit at different levels.
CONCLUSION: We describe significant local and segmental differences in the expression of liver-enriched transcription factors in the human intestine which impact epithelial cell biology of the gut.
Keywords: Liver-enriched transcription factors, Human intestine, Caco-2, Gene expression
INTRODUCTION
Numerous studies established the pivotal role of liver-enriched transcription factors in organ development and cellular function. These nuclear proteins are known to work in a hierarchical and cooperative network. The timely expression of specific transcription factors is necessary for cellular differentiation[1,2] and in situ hybridization studies of staged embryos demonstrate that hepatocyte nuclear factor 6 (HNF6) and its target gene FOXA2 are expressed in the hepatic diverticulum. More detailed analysis of the developmental expression patterns of HNF6 and FOXA2 provides evidence for their colocalization in intestinal epithelium. The expression patterns of these 2 transcription factors do not overlap in other endoderm-derived tissues[3].
There is growing evidence that the liver-enriched transcription factor plays a role in cancerous diseases of the digestive tract[4], and recent studies from our own laboratory provide evidence for HNF6 and FOXA2 as key regulators in colorectal liver metastases[5].
To better understand the molecular pathology of colorectal liver metastases we, and others, carried out a genome-wide expression analysis[6,7]. Essentially, the genes coding for the liver-enriched transcription factors HNF6, HNF1β and CCAAT enhancer binding protein γ (C/EBPγ) were selectively regulated but protein expression of regulated transcription factors identified unacetylated HNF6 to be a hallmark of colorectal liver metastases[5]. For its proposed interaction with HNF6, expression of FOXA2 and HNF6 was investigated. Notably, FOXA2 was significantly induced in colorectal liver metastases[5]. From the electromobility shift assay, evidence was obtained for HNF6 DNA binding activity to be specifically repressed in nuclear extracts of colorectal liver metastases. Taken collectively, we found HNF6 expression in colorectal liver metastases to be driven by the hepatic environment. Its expression is not observed in healthy colon nor in primary colonic cancer. Thus, HNF6 DNA binding is selectively prevented through lack of posttranslational modification and interaction with FOXA2.
As HNF6 is only expressed in colorectal liver metastases but not in primary colonic cancer or healthy colonic tissue we wished to investigate the regulation of HNF6 and of other liver-enriched transcription factors in different segments of the human intestine thereby providing information on their expression patterning. We therefore mapped HNF6 and other liver-enriched transcription factors in the human duodenum, jejunum, ileum and colon of patients undergoing large intestinal surgery. Regional differences in the expression and regulation of transcription factors might participate in growth of intestinal tumors and may influence metastatic spread. We also investigated regulation of liver-enriched transcription factors in the human colon carcinoma cell line Caco-2 that is considered to be valuable for the study of gut epithelial biology. The expression patterning of liver-enriched transcription factors was investigated as a function of time and confluency. Note, upon confluency the Caco-2 colon carcinoma cell line acquires many of the features of the enterocyte as detailed elsewhere[8,9]. We additionally investigated the DNA binding of HNF6, HNF4α and the HNF6 target neurogenin 3 (NGN3) to well known regulatory DNA sequences to link the DNA binding activity of these transcription factors to gene expression data.
Overall, we report the gene expression pattern of liver-enriched transcription factors in the human intestine and compare these findings with results obtained from the human colon carcinoma cell line, to facilitate the construction of a gene expression map for a better understanding of their regulation in gut biology.
MATERIALS AND METHODS
Ethical approval and patient’s characteristics
Approval for the use of surplus tissue material from elective surgery was obtained from the ethics committee of the Medical School of Hanover, Germany. All patients participating in this study gave written informed consent and were fully aware of the aims of the study. A summary of the patients’ characteristics is given in Table 1. Duodenum and jejunum were obtained from patients mainly with pancreatic cancer whereas ileum and colon was obtained from patients undergoing right or left hemicolectomy, (recto)sigmoid resection or rectal resection. All surgical specimens were subjected to histopathology. Excised tissue was shock-frozen in liquid nitrogen and stored at -80°C until analyzed.
Table 1.
Patient characteristics
| Patient | Sex | Age (yr) | Tumor localization | Tumor type | TNM classification | UICC classification |
| CN2/CP2 | M | 64 | Rectum | Colorectal cancer | pT2 pN0 M0-G2 | I |
| CN3/CP3 | F | 74 | Colon transversum | Colorectal cancer | pT2 pN1 pM1-G2 | IV |
| CN7/CP7 | F | 51 | Colon ascendens | Colorectal cancer | pT3 pN2 M0-G2 | III |
| CN8/CP8 | M | 81 | Colon sigmoideum | Colorectal cancer | pT3 pN0 M0-G3 | II |
| CN9/CP9 | F | 63 | Colon sigmoideum | Colorectal cancer | pT2 pN1 M0-G2 | III |
| CN10/CP10 | M | 81 | Rectum | Colorectal cancer | pT3 pN1 M0-G2 | III |
| CN11/CP11 | F | 49 | Colon ascendens | Colorectal cancer | pT3 pN2 M1-G2 | IV |
| CN15/CP15 | M | 73 | Colon sigmoideum | Colorectal cancer | pT4 pN0 M0-G3 | II |
| CN16/CP16 | M | 72 | Rectum | Colorectal cancer | pT2 pN0 M0-G2 | I |
| CN17/CP17 | M | 44 | Rectum | Colorectal cancer | pT2 pN1 M1-G2 | IV |
| CN18/CP18 | F | 67 | Rectum | Colorectal cancer | pT3 pN1 M1-G2 | IV |
| CN19/CP19 | M | 61 | Rectum | Colorectal cancer | pT2 pN2 M1-G2 | IV |
| CN20/CP20 | M | 56 | Rectum | Colorectal cancer | pT3 pN0 M0-G2 | II |
| CN21/CP21 | M | 61 | Colon sigmoideum | Colorectal cancer | pT3 pN2 M1-G2 | IV |
| P34 CN/CP | F | 61 | Colon sigmoideum | Colorectal cancer | pT4 pN1 M1-G2 | IV |
| P38 CN/CP | M | 67 | Rectum | Colorectal cancer | pT2 pN0 M0-G3 | I |
| Duo P39 | M | 72 | Pancreas | Pancreatic carcinoma | ||
| Duo P44 | F | 66 | Pancreas | Pancreatic cyst | ||
| Duo P47 | F | 57 | Pancreas | Pancreatic carcinoma | ||
| Jej P41 | M | 53 | Pancreas | Pancreatic carcinoma | ||
| Jej P44 | F | 66 | Pancreas | Pancreatic cyst | ||
| Jej P45 | M | 61 | Pancreas | Bile duct cancer | ||
| Ileum P40 | F | 78 | Colon ascendens | Colorectal cancer | ||
| Ileum P43 | M | 78 | Colon ascendens | Colorectal cancer | ||
| Ileum P46 | M | 64 | Colon ascendens | Colorectal cancer |
Cell culture
Expression of liver-enriched transcription factors was compared between colonic epithelium obtained from tissue resection material and Caco-2 cells, which were derived from a Caucasian patient with colonic adenocarcinoma. The Caco-2 cell line was obtained from the European Collection of Cell Cultures (Salisbury, UK) and were cultured as described previously[10].
RNA isolation and cDNA synthesis
RNA was isolated form tissue samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Quality and quantity of isolated RNA were checked by capillary electrophoresis (Bioanalyzer 2100, Agilent Technologies) following the manufacturer’s instructions or by gel electrophoresis. Reverse transcription (RT) used 2 μg total RNA from each sample. RNA and random primer (Promega, Mannheim, Germany) were preheated for 10 min at 70°C and then chilled on ice for 2 min. A total of 5 × RT-avian myeloblastosis virus (AMV) buffer (Promega), dNTP (10 mmol/L), RNAsin, AMV-RT (avian myeloblastosis virus-reverse transcriptase) (all Promega) and DEPC-H2O were added to a final volume of 20 μL. RT was carried out for 60 min at 42°C and was stopped by heating to 95°C for 5 min. The resulting cDNA was frozen at -20°C until additional experimentation.
Semiquantitative RT polymerase chain reaction
Primer design was done with the program Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Cross-reaction of primers with the genes was excluded by comparison of the sequence of interest with a database (Blast 2.2, US National Centre for Biotechnology Information) and all primers used in our study were intron spanning. Polymerase chain reaction (PCR) reactions were undertaken with a 20 μL reaction mixture containing HotStarTaq Master Mix (Qiagen, Hilden, Germany), DEPC, 1 μL cDNA and 1.0 μmol/L concentration of the 3’- and 5’-specific oligomers (synthesized by Invitrogen, Hilten, Germany). PCR reactions were carried out on a thermal cycler (T3, Biometra). Detailed oligonucleotide sequence information and the PCR amplification protocol were published previously[5]. DNA contamination was determined by direct amplification of RNA extracts before conversion to cDNA. Contamination of RNA extracts with genomic DNA was determined by gel electrophoresis and by DNA digest prior to cDNA synthesis. PCR reactions were done within the linear range of amplification, and amplification products were separated using 1.5% agarose gel and stained with ethidium bromide. Gels were photographed on a transilluminator (Kodak Image Station 440), and amplicons were quantified using the Kodak 1D 3.5 network software.
Western blotting experiments
Western immunoblotting was done as follows: total protein (100 μg) or nuclear protein (30 μg) extracts of Caco-2 cell cultures were denaturated at 95°C for 5 min, followed by sodium dodecyl sulphate polyacrylamide gel electrophoresis on 12% polyacrylamide gels, and blotted onto a polyvinylidene difluoride membrane (NEN, Dreieich, Germany) at 350 mA for 2 h in a buffer containing 400 mmol/L glycine and 50 mmol/L Tris (pH 8.3). Non-specific binding sites were blocked with Rotoblock (Roth, Germany) in 1 × TBS buffer. After electroblotting of proteins, membranes were incubated with polyclonal antibodies for HNF1α (Santa Cruz sc6548), FOXA2 (Santa Cruz sc6554), FOXA3 (Santa Cruz sc5360), HNF4α (Santa Cruz sc 6556), and HNF6 (kind gift of Dr. Costa RH, Chicago, Illinois, USA) for 1 h and washed 3 times with 1 × TBS buffer containing 0.1% Tween-20 (Roth, Germany). Subsequently, the membranes were incubated with a 1:5000 diluted anti-α rabbit antibody (Chemicon, Hofheim, Germany) for 1 h at room temperature, followed by 3 successive washes with 1 × TBS buffer containing 0.1% Tween-20 (Roth, Germany). Immunoreactive proteins were visualized with a chemiluminescence reagent kit (NEN, Dreieich, Germany) according to the manufacturer’s instructions, and bands were scanned with the Kodak Image Station CF 440 and analyzed using the Kodak 1D 3.5 imaging software (Eastman Kodak Company, USA).
Electrophoretic mobility shift assay
The procedure for electrophoretic mobility shift assays was adapted from a previously described method[11]. Briefly, 5 μg of Caco-2 nuclear extract were incubated with the binding buffer consisting of 25 mmol/L HEPES (pH 7.6), 5 mmol/L MgCl2, 34 mmol/L KCl, 2 mmol/L DTT, 2 mmol/L Pefablock (Roche Diagnostics GmbH, Mannheim, Germany), 0.5 μL aprotinin (2.2 mg/mL, Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), 50 ng poly (dl-dC) and 80 ng bovine serum albumin (PAA Laboratories GmbH, Cölbe, Germany). The binding reaction was carried out for 20 min on ice, and free DNA and DNA-protein complexes were resolved on a 6% polyacrylamide gel. For supershift studies, a specific HNF6 and/or HNF4α antibody (Santa Cruz Biotechnology Inc., Heidelberg, Germany) was added to the reaction mix 10 min before addition of the labeled probe. In the case of NGN3, no commercial antibody was available. Thus, a competition assay at 100- and 500-fold access of unlabeled oligonucleotide probe specific to NGN3 was used. Gels were blotted to Whatman 3 MM paper, dried under vacuum, exposed to imaging screens (Imaging Screen-K, Bio-Rad Laboratories GmbH, Munich, Germany) for autoradiography overnight at room temperature and analyzed using a phosphor imaging system (Molecular Imager FX pro plus; Bio-Rad Laboratories GmbH, Munich, Germany) and the Quantity One Version 4.2.2 software (Bio-Rad Laboratories GmbH, Munich, Germany).
Statistical analysis
We applied the Wilcoxon signed rank test and the Student t-test to determine significance, with P < 0.05 being statistically significant.
RESULTS
Gene expression profiling of liver-enriched transcription factors in different segments of the human intestine
The gene expression of transcription factor was determined relative to mitochondrial ATPase, i.e. a housekeeping gene that was found to be stably expressed. As described in the Material and Methods section, tissue preparations were obtained from surgically removed but healthy gut segments, as part of the Whipple surgery for the removal of pancreatic cancers or alternatively from patients undergoing surgery for colonic cancer. The tissue material was derived from the mucosa and submucosa and processed further as described in the Material and Methods section.
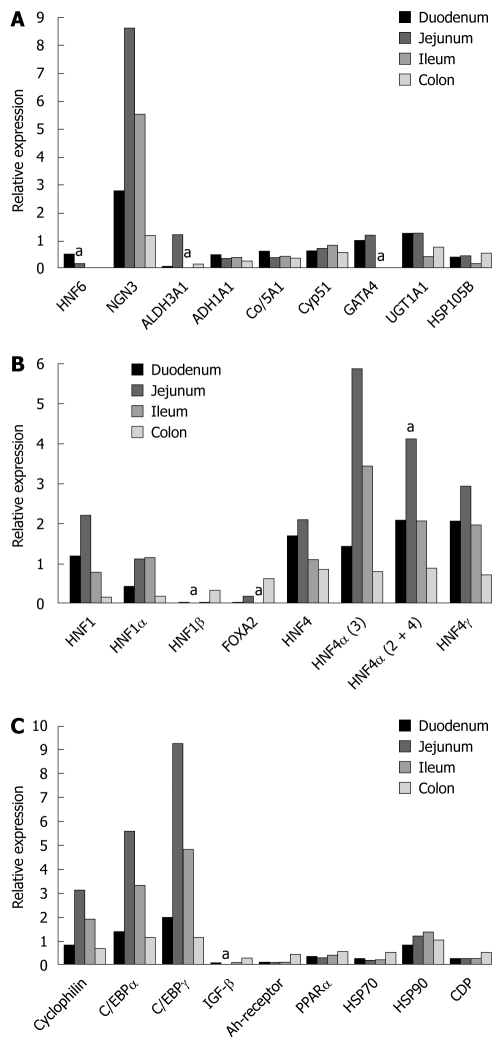
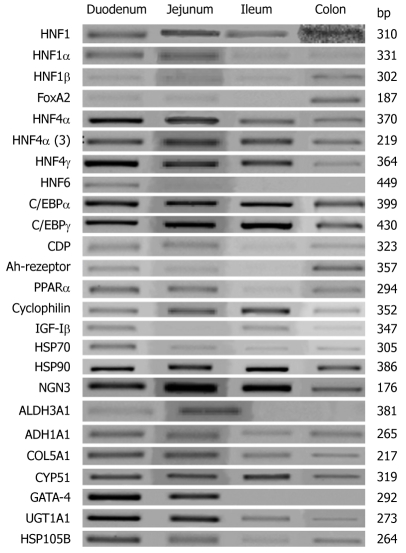
Table 2 gives an overview of the segmental expression pattern of various liver-enriched transcription factors while Figure 1 depicts the expression of HNF6 and of genes targeted by this transcription factor. Note, the data is presented relative to the expression of the mitochondrial ATPase, which serves as a housekeeping gene (Figure 1A). The patterning of individual liver-enriched transcription factors in the duodenum, jejunum, ileum and colon is shown in Figure 1B. Additionally, in Figure 1C expression of some genes of interest in epithelial cell biology are given. In Figure 2 and as described in the Material and Methods section, representative RT-PCR gels are shown. Note, all PCR reactions were done within the linear range of amplification. Essentially, the abundance of transcript expression of HNF1, HNF4 and for some of its splice variants and for C/EBP was significantly more than that of mitochondrial ATPase. Specifically, HNF6 gene expression was most abundant in the duodenum (P < 0.05) when compared with other segments of the intestine, whereas expression of the zinc finger protein GATA4 and of the HNF6 target gene ALDH3A1 was most abundant in the jejunum (Figure 1B, P < 0.05). Likewise, expression of FOXA2 and the splice variants 2 and 4 of HNF4α were most abundantly expressed in the jejunum (P < 0.05). For most of the transcription factors and of genes targeted by these factors, expression of transcripts varied amongst patients rendering statistical significance impossible. With the exception of the colon we were unable to amplify transcripts for FOXA1 and FOXA2 in any of the human intestine tissues examined. Essentially, in the case of HNF6, gene expression declined from the duodenum towards colon, whereas FOXA2 expression was undetectable in the ileum. Likewise, HNF4 expression was most abundant in the jejunum as was expression of its splice variant HNF4α3 and HNF4α2+4 and of HNF4γ. Unlike HNF6 we observed expression of HNF4 and of C/EBPα and γ in colon tissue as well. The expression of HNF6 and of genes targeted by this factor, i.e. NGN3 was most abundant in jejunum followed by ileum and colon but for most of the other genes targeted by HNF6 such as ALDH1A1, ALDH3A1, COL5A1, CYP51, UGT1A1 and HSP105B, expression did not differ amongst the different human intestinal segments. With the exception of heat shock protein 90, the expression of the Ah-receptor and of the nuclear receptor PPARα, HSP70 and the CCAAT enhancer displacement protein CDP, did not differ amongst the different human intestinal segments studied. In contrast, the expression of the insulin growth factor binding protein was more abundant in the colon (P < 0.05).
Table 2.
Expression pattern of liver-enriched transcription factors in the human intestine
| Duodenum | Jejunum | Ileum | Colon | |
| HNF1 | ++ | +++ | ++ | + |
| HNF1α | ++ | +++ | +++ | + |
| HNF1β | + | - | + | ++ |
| FOXA2 | + | ++ | - | ++ |
| HNF4 | ++ | +++ | ++ | + |
| HNF4α | + | +++ | ++ | + |
| HNF4γ | ++ | ++ | ++ | + |
| HNF6 | ++ | + | - | - |
| C/EBPα | + | +++ | ++ | + |
| C/EBPγ | + | +++ | ++ | + |
| MitATPase | ++ | ++ | ++ | ++ |
| PPARα | ++ | + | ++ | ++ |
| IGFβ | + | - | + | ++ |
| AHR | + | + | + | ++ |
| NGN3 | + | +++ | ++ | + |
| ALDH3A1 | + | +++ | - | + |
| ADH1A1 | ++ | + | ++ | + |
| COL5A1 | ++ | + | + | + |
| CYP51 | ++ | ++ | ++ | + |
| UGT1A1 | +++ | +++ | + | ++ |
| HSP105B | ++ | ++ | + | ++ |
| CDP | + | + | + | ++ |
| GATA4 | ++ | +++ | - | - |
+++: Very strong expression; ++: Strong expression; +: Detectable; -: Not detectable. HNF: Hepatocyte nuclear factor; EBP: Enhancer binding protein.
Figure 1.
Gene expression of hepatocyte nuclear factor 6 and of other liver enriched transcription factors in the human intestine. A: Hepatocyte nuclear factor 6 (HNF6) and some of its target genes. Bars hallmarked by "a" refer to statistically significant changed expression (P < 0.05) when compared to other segments of the gut; B: Hepatic nuclear transcription factors. Bars hallmarked by "a" refer to statistically significant changed expression (P < 0.05) when compared to other segments of the gut; C: CAAT enhancer binding proteins and the transcription factors Ah-receptor, PPAR alpha as well as the heat shock proteins HSP70 and HSP90 and IGF-β. Bars hallmarked by "a" refer to statistically significant changed expression (P < 0.05) when compared to other segments of the gut.
Figure 2.
Representative reverse transcription polymerase chain reaction gels of gene expression of hepatocyte nuclear factor 6 and of other liver-enriched transcription factors in the human intestine. Note, all polymerase chain reaction reactions were done within the linear range of amplification.
Studies using the human colon epithelial cancer cell line Caco-2
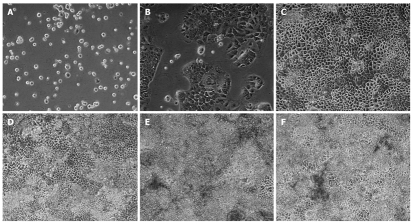
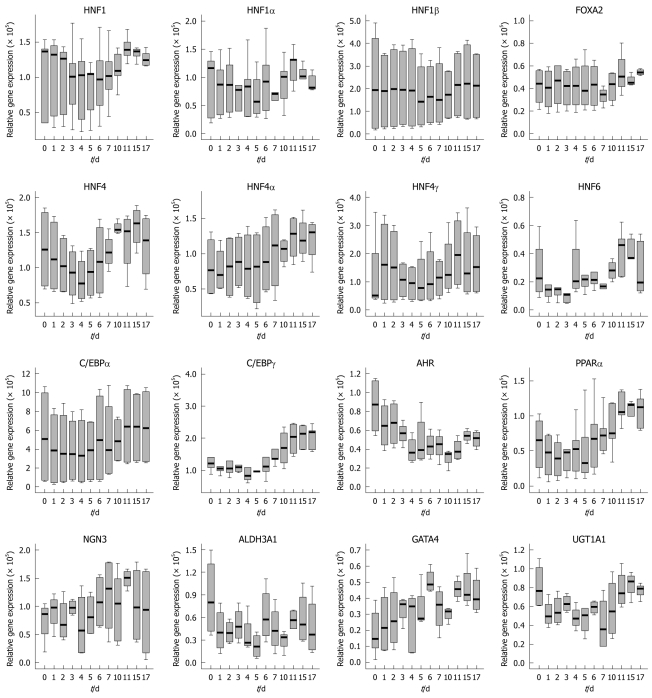
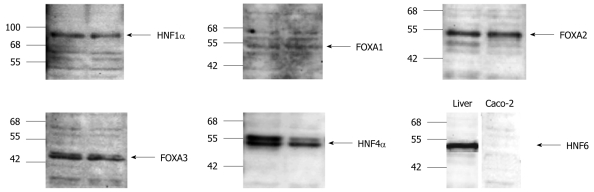
The expression of the aforementioned transcription factors was investigated in cultures of Caco-2 cells. Essentially, with time and cell culture confluency HNF6 gene expression increased up to day 11 but declined thereafter (see Figure 3 for microscopic images and Figure 4 for gene expression data). A similar rise in expression of transcription factor gene expression was seen for HNF1, some splice variants of HNF4 (2, 3 and 4) and of C/EBPγ (data not shown). None of these changes were, however, of statistical significance. Importantly, expression of HNF6 transcripts was unexpected, as neither healthy colon nor primary colonic cancers expressed HNF6. Likely, its expression in Caco-2 cells is the result of the cell culture environment with the supply of optimized culture media. We additionally determined the protein expression of liver-enriched transcription factor and observed expression of HNF1α, FOXA2, FOXA3 and HNF4α (Figure 5). In the case of FOXA3 and HNF4α, 2 immunoreactive bands were observed. Obviously the antibodies recognized these liver-enriched transcription factors posttranslational modifications. No HNF6 protein expression could be determined.
Figure 3.
Microscopic images of time depended sequence of Caco-2 cells. Note, HNF6 gene expression increased with time and cell culture confluency up to day 11 but declined thereafter (see Figure 2 for gene expression data). A: 0 d; B: 3 d; C: 5 d; D: 7 d; E: 11 d; F: 17 d.
Figure 4.
Time-dependent gene expression of liver-enriched transcription factors and some of its target genes in cultures of Caco-2 cells. HNF: Hepatocyte nuclear factor.
Figure 5.
Western blotting of hepatocyte nuclear factor 1α, FOXA1, FOXA2, FOXA3, hepatocyte nuclear factor 4α and hepatocyte nuclear factor 6 in different human colon carcinoma Caco-2 cell line cultures. Note, in case of hepatocyte nuclear factor (HNF) 6 healthy human liver serves as control.
DNA binding studies with HNF6
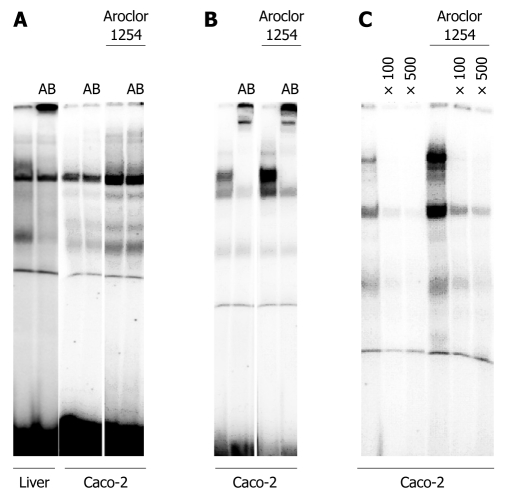
We further investigated the DNA binding of HNF6 and of its downstream target NGN3 as well as HNF4α. As shown in Figure 6A, we were unable to detect HNF6 DNA binding with nuclear extracts of Caco-2 cells. We previously reported abundant HNF6 protein expression in the liver[5], and therefore used nuclear extracts of the liver as a positive control for HNF6 DNA binding activity. In the past we demonstrated that expression of liver-enriched transcription factors can be modulated by treatment of Caco-2 cell cultures with Acrolor 1254[12]. This treatment did not induce DNA binding of HNF6 but increased expression of an unknown nuclear protein to an optimized HNF6 probe (Figure 6A). To further study HNF6 activity we investigated the DNA binding of the transcription factor NGN3. Essentially, NGN3 was reported to be a downstream target of HNF6 and is therefore regulated by this factor[13]. As shown in Figure 6C, NGN3 DNA binding is observed with nuclear extracts of Caco-2 cell cultures. Furthermore, treatment of Caco-2 cell cultures with Aroclor 1254, an inducer of transcription factors[12], resulted in increased DNA binding of NGN3. We also investigated DNA binding of HNF4α and observed abundant DNA binding activity as illustrated in Figure 6B. In the case of HNF4α a suitable antibody was available. The shifted band in the EMSA assay further documents specificity. As observed with NGN3 treatment of Caco-2 cell cultures with Aroclor 1254 resulted in increased HNF4α DNA binding. In the past we reported induction of HNF4α gene expression in cultures of Caco-2 cells[12]. We now extend our initial observation to DNA binding activity of this protein.
Figure 6.
Electromobility shift assay with nuclear extracts of the human colon carcinoma Caco-2 cell line. A: Demonstrates the DNA binding of HNF6 to an optimized oligonucleotide probe using either nuclear protein extract of human liver tissue or Caco-2 cells. Note, the lane labeled as AB refers to the addition of a hepatocyte nuclear factor 6 antibody to demonstrate specificity of the DNA binding assay; B: Demonstrates the DNA binding of HNF4α to an optimized oligonucleotide probe using either nuclear protein extract of human liver tissue or Caco-2 cells. Note, the lane labeled as AB refers to the addition of a HNF4α antibody to demonstrate specificity of the DNA binding assay; C: Demonstrates the DNA binding of NGN3 (a HNF6 target gene) to an optimized oligonucleotide probe using either nuclear protein extract of human liver tissue or Caco-2 cells. Note, the lane labeled as × 100 or × 500 refers to competition assays with excess of unlabeled probe.
DISCUSSION
This study aimed for an improved understanding of the gene expression pattern of HNF6, FOXA2 and other nuclear transcription factors in the descending human intestine. Essentially, expression of liver-enriched transcription factors differed when the duodenum, jejunum, ileum and colon were compared. Mapping of liver-enriched transcription factors to different segments of the human intestine provided valuable insight into gene regulation that may have significant implications for physiology and disease. Indeed, numerous studies have established an important role of liver-enriched transcription factors in organ development and cellular function and there is conclusive evidence of nuclear transcription factors to act in concert in the orchestration of gene expression[1].
Here we report FOXA2 expression to be significantly upregulated in the colon but the coded protein of this transcription factor has been shown to inhibit HNF6 activity. As originally proposed by Rausa et al[14] an interplay of CBP coactivator protein with HNF6 and FOXA2 may regulate steady levels of these transcription factors. In human colorectal liver metastases FOXA2 expression was significantly induced while electromobility shift assays demonstrated HNF6 DNA binding activity to be prevented as previously reported[5].
Notably, no HNF6 DNA binding was observed with nuclear extracts of Caco-2 cells (Figure 6A) whereas DNA binding of HNF4α and of NGN3 was evident. Additionally, we observed increased DNA binding activity for HNF4α and NGN3 upon treatment of Caco-2 cell cultures with Aroclor 1254 (Figure 6B and C). Expression of HNF6 protein was below the limit of detection in untreated cultures of the Caco-2 cell line.
Of all transcription factors investigated, expression of HNF6 was most abundant in duodenum and jejunum whereas expression of GATA4 and of HNF4 including some of its splice variants in addition to C/EBPα and γ was most abundant in the jejunum.
The difference in expression pattern of individual transcription factors amongst the various segments of the human intestine is notable with HNF4 and C/EBP being abundantly expressed in the jejunum and the duodenum. Indeed, HNF4α has been shown to protect the gut against inflammatory bowel disease and there is clear evidence for a role of HNF4α in promoting differentiation of intestinal epithelial cells[15,16]. Furthermore, in the study of Stegmann et al[17] the metabolome, transcriptome and bioformatic analysis identified HNF4 as a central regulator of gene expression during enterocyte differentiation and crypt function, but recent evidence identified forkhead box transcription factors FOXA1 and FOXA2 to be important regulators of mucin expression in intestinal epithelial cells as well[18]. Additionally, the human C/EBPα gene was found to expressed at the highest level in the placenta followed by the liver, lungs, skeletal muscle, pancreas, small intestine, colon and in peripheral blood leucocytes[2]. As was reviewed elsewhere C/EBPα plays an important role in cell cycle control, cellular differentiation, many metabolic processes and the detoxification. The difference in C/EBPα expression may in part be the result of control of epithelial replacement through control of the p21 protein level thereby determining epithelial replacement[2].
In agreement with our previous study on colorectal liver metastases we were unable to detect HNF6 transcript expression in healthy and cancerous colonic tissue while the expression of splice variants of HNF4α in healthy colonic tissue was a significant finding and is likely to impact HNF4α activity, as we have shown recently in the case of human hepatocellular carcinoma[19]. Indeed, mice lacking HNF4α exhibited decreased levels of polysaccharides and acetic mucopolysaccharides with some altered expression of mucins and aquaporins[15].
The use of the human colon carcinoma cell line Caco-2 allowed us to compare expression of transcription factors in healthy colon and colonic epithelium of adenocarcinoma. Initially, we investigated expression of individual liver-enriched transcription factors as a function of time and confluency of the Caco-2 cell culture. With the exception of HNF6 there was no statistically significant difference in the expression of transcription factors as a function of cell culture time (up to 17 d) or confluency while cellular differentiation of Caco-2 cells was dependent upon the activity of the transcription factor NF-Y and E2F[20,21]. We also compared expression of transcription factors with those in the healthy colon but did not identify a significant difference.
Overall, our study identified local and segmental differences in the expression patterning of liver-enriched transcription factors in the human intestine. Tissue specific transcription factor expression provides a regulatory circuitry for the control of gene expression and cellular differentiation. The present study identifies significant differences in the expression of liver-enriched transcription factors amongst different segments of the human intestine that impacts on the epithelial cell biology of the gut.
COMMENTS
Background
Liver-enriched transcription factors are versatile proteins pertinent for cellular growth and differentiation of the liver. Growing evidence suggests these factors play a wider role in epithelial biology and cancerous diseases of the digestive tract. Indeed, recent studies provided evidence for hepatocyte nuclear factor 6 (HNF6) and FOXA2 as key regulators in colorectal liver metastases. As HNF6 was only expressed in colorectal liver metastases but not in primary colonic cancer or healthy colonic tissue, the authors investigated regulation of HNF6 and of other liver-enriched transcription factors in different segments of the human intestine thereby providing information on their expression pattern. Regional differences in the expression and regulation of transcription factors may be related to growth of intestinal tumors and may influence metastatic spread.
Research frontiers
The authors examined the gene expression pattern of liver-enriched transcription factors in different segments of the human intestine and compared the findings with results obtained from a human colon carcinoma cell line for a better understanding of their regulation in gut biology and disease.
Innovations and breakthroughs
This is the first study to investigate the expression of liver-enriched transcription factors in the human intestine and compare the data with the human colon carcinoma cell line Caco-2. Knowledge of the expression pattern of liver-enriched transcription factors in different segments of the human intestine help to better understand the importance of HNF6 and FOXA2 in colorectal liver metastases.
Applications
Tissue specific transcription factor expression provides a regulatory circuitry for the control of gene expression and cellular differentiation of the gut epithelium. Restoring attenuated transcription factor DNA binding activity represents a novel strategy for the treatment of secondary malignancies of the liver.
Terminology
Liver-enriched transcription factors play a pivotal role in disease. Essentially, transcription factors are master regulatory proteins and interact with many different molecules including coactivators, repressors, enzymes, DNA and RNA to control gene expression. Such interactions will inevitably repress or activate gene expression and therefore determine cellular phenotype.
Peer review
This paper examines the distribution of liver-enriched transcription factors and related genes in human intestinal tissues. It represents a useful and novel contribution to the field.
Footnotes
Supported by (in part) Novartis Pharma GmbH, Germany, BU Transplantation and Immunology (to Lehner F) and by the Lower Saxony Ministry of Culture and Sciences and the Volkswagen foundation, Germany, Grant No. 25A.5-7251-99-3/00
Peer reviewers: Lodewijk AA Brosens, MD, PhD, Department of Pathology, University Medical Center Utrecht, Postbus 85500, 3508 GA, Utrecht, The Netherlands; Ross McManus, PhD, Senior Lecturer in Molecular Medicine, Institute of Molecular Medicine and Department of Clinical Medicine, Trinity Centre for Health Science, St. James’s Hospital, Dublin 8, Ireland
S- Editor Wang YR L- Editor Cant MR E- Editor Zheng XM
References
- 1.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev. 2004;56:291–330. doi: 10.1124/pr.56.2.5. [DOI] [PubMed] [Google Scholar]
- 3.Rausa F, Samadani U, Ye H, Lim L, Fletcher CF, Jenkins NA, Copeland NG, Costa RH. The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Dev Biol. 1997;192:228–246. doi: 10.1006/dbio.1997.8744. [DOI] [PubMed] [Google Scholar]
- 4.Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology. 2004;39:1038–1047. doi: 10.1002/hep.20155. [DOI] [PubMed] [Google Scholar]
- 5.Lehner F, Kulik U, Klempnauer J, Borlak J. The hepatocyte nuclear factor 6 (HNF6) and FOXA2 are key regulators in colorectal liver metastases. FASEB J. 2007;21:1445–1462. doi: 10.1096/fj.06-6575com. [DOI] [PubMed] [Google Scholar]
- 6.Pantaleo MA, Astolfi A, Nannini M, Paterini P, Piazzi G, Ercolani G, Brandi G, Martinelli G, Pession A, Pinna AD, et al. Gene expression profiling of liver metastases from colorectal cancer as potential basis for treatment choice. Br J Cancer. 2008;99:1729–1734. doi: 10.1038/sj.bjc.6604681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh KH, Rhee H, Kang HJ, Yang E, You KT, Lee H, Min BS, Kim NK, Nam SW, Kim H. Differential gene expression profiles of metastases in paired primary and metastatic colorectal carcinomas. Oncology. 2008;75:92–101. doi: 10.1159/000155211. [DOI] [PubMed] [Google Scholar]
- 8.Engle MJ, Goetz GS, Alpers DH. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J Cell Physiol. 1998;174:362–369. doi: 10.1002/(SICI)1097-4652(199803)174:3<362::AID-JCP10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Ferraretto A, Gravaghi C, Donetti E, Cosentino S, Donida BM, Bedoni M, Lombardi G, Fiorilli A, Tettamanti G. New methodological approach to induce a differentiation phenotype in Caco-2 cells prior to post-confluence stage. Anticancer Res. 2007;27:3919–3925. [PubMed] [Google Scholar]
- 10.Lampen A, Bader A, Bestmann T, Winkler M, Witte L, Borlak JT. Catalytic activities, protein- and mRNA-expression of cytochrome P450 isoenzymes in intestinal cell lines. Xenobiotica. 1998;28:429–441. doi: 10.1080/004982598239362. [DOI] [PubMed] [Google Scholar]
- 11.Niehof M, Streetz K, Rakemann T, Bischoff SC, Manns MP, Horn F, Trautwein C. Interleukin-6-induced tethering of STAT3 to the LAP/C/EBPbeta promoter suggests a new mechanism of transcriptional regulation by STAT3. J Biol Chem. 2001;276:9016–9027. doi: 10.1074/jbc.M009284200. [DOI] [PubMed] [Google Scholar]
- 12.Borlak J, Zwadlo C. Expression of drug-metabolizing enzymes, nuclear transcription factors and ABC transporters in Caco-2 cells. Xenobiotica. 2003;33:927–943. doi: 10.1080/00498250310001614286. [DOI] [PubMed] [Google Scholar]
- 13.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rausa FM 3rd, Hughes DE, Costa RH. Stability of the hepatocyte nuclear factor 6 transcription factor requires acetylation by the CREB-binding protein coactivator. J Biol Chem. 2004;279:43070–43076. doi: 10.1074/jbc.M407472200. [DOI] [PubMed] [Google Scholar]
- 15.Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, et al. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908–920. doi: 10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lussier CR, Babeu JP, Auclair BA, Perreault N, Boudreau F. Hepatocyte nuclear factor-4alpha promotes differentiation of intestinal epithelial cells in a coculture system. Am J Physiol Gastrointest Liver Physiol. 2008;294:G418–G428. doi: 10.1152/ajpgi.00418.2007. [DOI] [PubMed] [Google Scholar]
- 17.Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritié L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT, et al. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141–155. doi: 10.1152/physiolgenomics.00314.2005. [DOI] [PubMed] [Google Scholar]
- 18.van der Sluis M, Vincent A, Bouma J, Korteland-Van Male A, van Goudoever JB, Renes IB, Van Seuningen I. Forkhead box transcription factors Foxa1 and Foxa2 are important regulators of Muc2 mucin expression in intestinal epithelial cells. Biochem Biophys Res Commun. 2008;369:1108–1113. doi: 10.1016/j.bbrc.2008.02.158. [DOI] [PubMed] [Google Scholar]
- 19.Niehof M, Borlak J. EPS15R, TASP1, and PRPF3 are novel disease candidate genes targeted by HNF4alpha splice variants in hepatocellular carcinomas. Gastroenterology. 2008;134:1191–1202. doi: 10.1053/j.gastro.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Bevilacqua MA, Faniello MC, Iovine B, Russo T, Cimino F, Costanzo F. Transcription factor NF-Y regulates differentiation of CaCo-2 cells. Arch Biochem Biophys. 2002;407:39–44. doi: 10.1016/s0003-9861(02)00436-8. [DOI] [PubMed] [Google Scholar]
- 21.Ding Q, Wang Q, Dong Z, Evers BM. Characterization and regulation of E2F activity during Caco-2 cell differentiation. Am J Physiol Cell Physiol. 2000;278:C110–C117. doi: 10.1152/ajpcell.2000.278.1.C110. [DOI] [PubMed] [Google Scholar]