Abstract
Fluorescence spectroscopy has emerged in recent years as an effective way to detect cervical cancer. Investigation of the data preprocessing stage uncovered a need for a robust smoothing to extract the signal from the noise. Various robust smoothing methods for estimating fluorescence emission spectra are compared and data driven methods for the selection of smoothing parameter are suggested. The methods currently implemented in R for smoothing parameter selection proved to be unsatisfactory, and a computationally efficient procedure that approximates robust leave-one-out cross validation is presented.
Keywords: Robust smoothing, Smoothing parameter selection, Robust cross validation, Leave out schemes, Fluorescence spectroscopy
1 Introduction
In recent years, fluorescence spectroscopy has shown promise for early detection of cancer (Grossman et al., 2002; Chang et al., 2002). Such fluorescence measurements are obtained by illuminating the tissue at one or more excitation wavelengths and measuring the corresponding intensity at a number of emission wavelengths. Thus, for a single measurement we obtain discrete noisy data from several spectroscopic curves which are processed to produce estimated emission spectra. One important step in the processing is smoothing and registering the emission spectra to a common set of emission wavelengths.
However, the raw data typically contain gross outliers, so it is necessary to use a robust smoothing method. For real time applications we need a fast and fully automatic algorithm.
For this application, we consider several existing robust smoothing methods which are available in statistical packages, and each of these smoothers also has a default method for smoothing parameter selection. We found that the default methods of do not work well. To illustrate this point, we present Figure 1 where some robust fits, using the default smoothing parameter selection methods are shown. We have also included the plots of the “best” fit (as developed in this paper) for comparison purposes.
Fig. 1.
Parts (a) and (b) show raw data with the smoothed estimates superimposed. Part (c) is the fit corresponding to the best smoothing parameter value, and part (d) show the fit based on default methods for various robust smoothers. The default method from robust smoothing splines undersmooths, whereas the default methods of robust LOESS and COBS oversmooths.
The best methods we found for smoothing parameter selection required some form of robust cross validation - leaving out a subset of the data, smoothing, then predicting the left out data, and evaluating the prediction error with a robust loss function. Full leave-one-out robust cross validation performed well for smoothing parameter selection but is computationally very time consuming. We present a new method, systematic K-fold robust cross validation, which reduces the computation time and still gives satisfactory results.
There has been previous work on the robust smoothing of spectroscopy data. In particular, Bussian and Härdle (1984) consider the robust kernel method for the estimation from Raman spectroscopy data with huge outliers. However, whereas they only consider one specific robust smoothing method, we make some comparisons of available methods and make some recommendations. Furthermore, Bussian and Härdle (1984) do not treat the problem of automatic bandwidth selection.
The method presented here is not radically new. Rather, we present a combination of simple techniques that, when combined in a sensible manner, gives us a practical solution to a computationally burdensome yet often overlooked problem of robust smoothing parameter selection. Therefore, we show by extensive simulation studies that our method can indeed solve our application problem and possibly many others that require robust smoothing.
The paper is organized as follows. We first review some robust smoothers in Section 2. Then in Section 3, we introduce the robust procedures for robust smoothing parameter selection along with some default methods from the functions in R. We also introduce a method which approximates the usual leave-one-out cross validation scheme to speed up the computation. In Section 4, we perform a preliminary study, determine which smoothers and smoothing parameter methods to include in a large scale simulation study, and then give results on this, along with the results on how the method works with real data. Finally, we conclude the paper with some recommendations and directions of possible future research in Section 5.
2 Robust Smoothing Methodologies
In this section, we define the robust smoothing methods that will be considered in our study. Consider the observation model
where the εi are i.i.d. random errors, and we wish to obtain an estimate m̂ of m. In order for this to be well defined, we need to assume something about the “center” of the error distribution. The traditional assumption that the errors have zero mean is not useful here as we will consider possibly heavy tailed distributions for the errors. In fact, each different robust smoother in effect defines a “center” for the error distribution, and the corresponding regression functions m(x) could differ by a constant. If the error distribution is symmetric about 0, then all of the methods we consider will be estimating the conditional median of Y given X = x.
2.1 Robust Smoothing Splines
The robust smoothing spline is defined through an optimization problem: find m̂ that minimizes
for an appropriately chosen ρ(·) function and subject to m̂ and m̂' absolutely continuous with ∫m̂′′2 < ∞. Of course, the choice of ρ is up to the investigator. We generally require that ρ be even symmetric, convex, and grows slower than O(x2) as |x| gets large. Here, λ > 0 is the smoothing parameter, with larger values of λ corresponding to smoother estimates.
The implementation of the robust smoothing splines we have used is the R function qsreg from the package fields, version 4.3 (Oh et al., 2004), which uses
| (1) |
It is easily verified that the ρ(x) function in (1) satisfies the requirements above. The break point C in (1) is a scale factor usually determined from the data. See Oh et al. (2004) for details. For the qsreg function, the default is .
2.2 Robust LOESS
We used the R function loess from the package stats (version 2.7.2) to perform robust locally weighted polynomial regression (Cleveland, 1979; Chambers and Hastie, 1992). At each x, we find β̂0,β̂1,…,β̂p to minimize
where the resulting estimate is . We consider only the degree p = 2, which is the maximum allowed. With the family = "symmetric" option, the robust estimate is computed via the Iteratively Reweighted Least Squares algorithm using Tukey’s biweight function for the reweighting. K(·) is a compactly supported kernel (local weight) function that downweights xi that are far away from x. The kernel function used is the “tri-cubic”
The parameter h in the kernel is the bandwidth, which is specified by the fraction of the data λ (0 < λ ≤ 1) within the support of K(·/h), with larger values of λ giving smoother estimates.
2.3 COBS
Constrained B-spline smoothing (COBS) is implemented in R as the function cobs in the cobs package, version 1.1–5 (He and Ng, 1999). The estimator has the form
where N is the number of internal knots, q is the order (polynomial degree plus 1), Bj(x) are B-spline basis functions (de Boor, 1978), and âj are the estimated coefficients. There are two versions of COBS depending on the roughness penalty: an L1 version and an L∞ version. For the L1 version the coefficients â1, … , âN+q are chosen to minimize
where t1, …, tN+2q are the knots for the B-splines. The L∞ version is obtained by minimizing
The λ > 0 is a smoothing parameter similarly to the λ in robust smoothing splines. Here not only λ has to be determined as with other robust smoothers, but we also need to determine the number of internal knots, N, which acts as a smoothing parameter as well. In the cobs program, the degree = option determines the penalty: degree=1 gives an L1 constrained B-spline fit, and degree=2 gives an L∞ fit.
3 Smoothing Parameter Selection
In all robust smoothers, a critical problem is the selection of the smoothing parameter λ. One way we can do this is through subjective judgment or inspection. But for the application to automated real time diagnostic devices, this is not practical. Thus, we need to develop an accurate, rapid method to determine the smoothing parameter automatically.
For many nonparametric function estimation problems, the method of leave-one-out cross validation (CV) is often used for smoothing parameter selection (Simonoff, 1996). The Least Squares CV function is defined as
where is the estimator with smoothing parameter λ and the ith observation (xi,yi) deleted. One would choose λ by minimizing LSCV (λ).
However, this method does not work well for the robust smoothers because the LSCV function itself will be strongly influenced by outliers (Wang and Scott, 1994).
3.1 Default Methods
All of the packaged programs described above have a default method for smoothing parameter selection which is alleged to be robust.
The robust smoothing splines from qsreg function provides a default smoothing parameter selection method using pseudo-data, based on results in Cox (1983). The implementation is done via generalized cross validation (GCV) with empirical pseudo-data, and the reader is referred to Oh et al. (2004, 2007) for details.
In COBS, the default method for selecting λ is a robust version of the Bayes information criterion (BIC) due to Schwarz (1978). This is defined by He and Ng (1999) as
where pλ is the number of interpolated data points.
The robust LOESS default method for smoothing parameter selection is to use the fixed value λ = 3/4. This is clearly arbitrary and one cannot expect it to work well in all cases.
Moreover, the pseudo-data method is known to produce an undersmoothed curve, and BIC method usually gives an oversmoothed curve. These claims can be verified by inspecting the plots in the corresponding papers referenced above (Oh et al., 2004, 2007; He and Ng, 1999) and can also be seen in Figure 1 of present paper. Such results led to an investigation of improved robust smoothing parameter selection methods.
3.2 Robust Cross Validation
To solve the problem of smoothing parameter selection in the presence of gross outliers, we propose a Robust CV (RCV ) function
| (2) |
where ρ(·) again is an appropriately chosen criterion function. Similarly to LSCV, the is the robust estimate with the ith data point left out. We consider various ρ functions in RCV. For each of the methods considered, there is an interpolation (predict) function to compute at the left out xi’s.
We first considered the absolute cross validation (ACV) method proposed by Wang and Scott (1994). With this method, we find a λ value that minimizes
So this is a version of RCV with ρ(·) = |·|. Intuitively, the absolute value criteria is resistant to outliers, because the absolute error is much smaller than the squared error for large values. Wang and Scott (1994) found that the ACV criteria worked well for local L1 smoothing.
Because the ACV function (as a function of λ) can be wiggly and can have multiple minima (Boente et al., 1997), we also considered Huber’s ρ function to possibly alleviate this problem. Plugging in Huber’s ρ into (2), we have a Huberized RCV (HRCV )
To determine the quantity C in ρ(x), we used C = 1.28 * MAD/.6745, where MAD is the median absolute deviation of the residuals from an initial estimate (Hogg 1979) . Our initial estimate is obtained from the smooth using the ACV estimate of λ. The constant 1.28/.6745 gives a conservative estimate since it corresponds to about 20% outliers from a normal distribution.
We have also tried the robust version of the Rice-type scale estimate (Boente et al., 1997; Ghement et al., 2008). This is obtained by considering
where Q(0.5) is the median of the absolute difference |Yi+1–Yi|, i = 1, … , n–1. This is the robust version of scale estimate suggested by Rice (1994) and was extensively studied by Boente et al. (1997) and Ghement et al. (2008). This has the advantage over Hogg’s method that the Rice-type does not require an initial estimate and has shown to possess nice asymptotic properties.
However, the preliminary simulation has shown that there is virtually no difference in performance of the Rice-type estimate versus the Hogg’s method for our setting. Moreover, either methods for the HRCV had a very little improvement over the ACV in preliminary studies. Thus, since the extra steps needed for HRCV do not result in significant increase in performance, we decided not to include the HRCV in our investigation.
3.3 Computationally Efficient Leave Out Schemes
We anticipate that the ordinary leave-one-out scheme for cross validation as described above will take a long time, especially for a dataset with large n. This is further exacerbated by the fact that the computation of the estimate for a fixed λ uses an iterative algorithm to solve the nonlinear optimization problem. Thus, we devise a scheme that leaves out many points at once and still gives a satisfactory result.
Our approach is motivated by idea of K-fold cross validation (Hastie et al., 2001) in which the data are divided into K randomly chosen blocks of (approximately) equal size. The data in each of the blocks is predicted by computing an estimate with that block left out. Our approach is to choose the blocks systematically so as to maximize the distance between xi’s within the blocks (which is easy to do with equispaced one dimensional xi’s as in our application). We call this method systematic K-fold cross validation.
Define the sequence (i : d : n) = {i,i + d , … ,i + kd} where k is the largest integer such that i + kd ≤ n. Let denote the estimate with (xi,yi), (xi+d,yi+d), …, (xi+kd,yi+kd) left out. Define the robust systematic K-fold cross-validation with phase r (where K = d/r) by
If r = 1 so that K = d,we simply call this a systematic K-fold CV (without any reference to phase r). There are two parameters for this scheme: the d which determines how far apart are the left out xi’s, and the phase r which determines the sequence of starting values i. Note that we compute d/r curve estimates for each λ, which substantially reduces the time for computing the RCV function. We will have versions RCV(d,r) for each of the criterion functions discussed above.
The choice of d and r must be determined from data. Preliminary results show that r > 1 (which will not leave out all data points) does not give satisfactory results, and hence we will only consider the case r = 1.
3.4 Justifications, Heuristics and Asymptotics
We briefly discuss the asymptotic properties of the robustified cross validation functions. From the results in Leung (2005), we expect that the λ chosen by minimizing HRCV will be asymptotically optimal for minimizing the Mean Average Squared Error:
We conjecture that the λ chosen by minimizing ACV will also be asymptotically optimal for minimizing MASE. This is perhaps unexpected because the absolute error criterion function seems to have no resemblance to squared error loss. We will give some heuristics to justify this conjecture. First we recall the heuristics behind the asymptotic optimality of the λ chosen by minimizing LSCV (when the errors are not heavy tailed). The LSCV may be written in the form
Now T1 doesn’t depend on λ. For λ in an asymptotic neighborhood of the optimal value, T2 is asymptotically negligible assuming the εi are i.i.d. with mean 0 with other moment assumptions, and T3 is asymptotically equivalent to the MASE. The ACV function may be written as
where IAi,n,λ is the indicator of the event
Now S1 doesn’t depend on λ, and we conjecture that S2 is asymptotically negligible, provided the εi are i.i.d. with a distribution symmetric about 0 (similarly to the assumptions made in Leung, 2005). The justification for this should follow the same type of argument as showing T2 is negligible for the LSCV. The term S3 is a little more complicated. Write it as
Note that for each i, εi is independent of , and assuming the εi have a density fε which is positive and continuous at 0, then
Of course, here we are assuming that is small. Thus, it is reasonable to conjecture that
Turning now to S32 note that the conditional distribution of |εi| given is approximately uniform on the interval and thus |εi| is approximately conditionally independent of Ai,n,λ given and
Combining this with our previous result on the conditional probability of Ai,n,λ, we see that it is reasonable to conjecture that
Combining these results, we have the conjecture
| (3) |
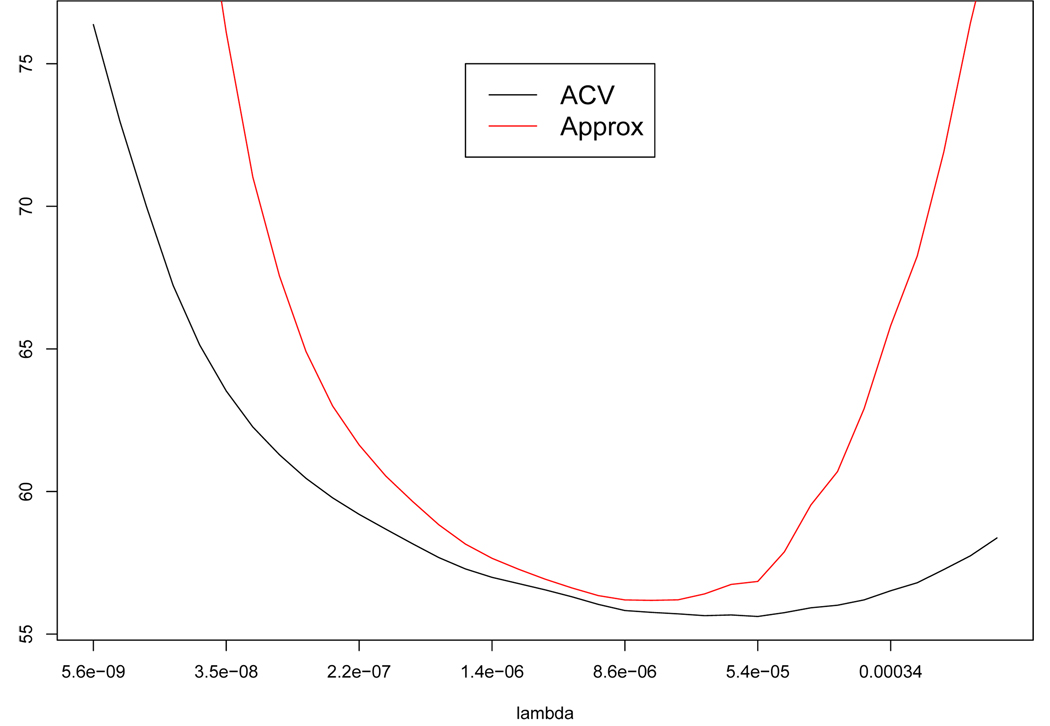
Of course, the last term should approximate MASE. In Figure 2 we show a plot of this last expression along with the ACV function for one simulated data set. The simulation was done with a beta distribution as the “true” curve m(x) and we have added Cauchy error with location 0 and scale 10 at each xi, i = 1, … ,1550. To obtain the ACV curve and its approximation, the right hand side of the equation (3), we used the robust smoothing splines with full leave-one-out scheme. From looking at Figure 2, we see that this conjectured approximation looks reasonable near where the ACV is minimized.
Fig. 2.
Comparison of ACV versus its approximation, the right hand side of the equation (3).
Now, here is the justification for the K-fold systematic CV. We expect the predictions at left out data points to be uniformly close to the original leave-one-out scheme. With an appropriate choice of K, for small bandwidths the influence of other left out points on prediction of a given value will be small because the effective span of the local smoother is small. For large bandwidths, the influence in prediction of left out points will be small because the effective span of the local smoother is large so that no point has much influence.
We will investigate rigorous justifications of these heuristically motivated conjectures in future research.
3.5 Methods to Evaluate the Smoothing Parameter Selection Schemes
For the simulation study, we use the following criteria to assess the performance of the smoothing parameter selection methods.
If we know the values of the true curve, we may compute the integrated squared error (ISE)
for each estimate and for various values of λ. (Note: many authors would refer to ISE in our formula as ASE or “Average Squared Error.” However, the difference is so small that we will ignore it.) Similarly, we can compute integrated absolute error (IAE)
We can determine which value of the λ will give us the “best” fit, based on L1 (i.e., IAE) or L2 (i.e., ISE) criteria. For comparison, we will take the squared root of ISE so that we show √ISE in the results.
Thus, we may find , and as our optimal λ values that gives us the best fit. We then make a comparison with the default λ values obtained by the methods in Section 3.1. In addition, we compute the λ̂’s from the ACV (call this λ̂ACV) and compare them with the optimal values. To further evaluate the performances of ACV, we may compute the “loss functions” ISE(λ̂) (ISE(λ) with λ̂ as its argument) and IAE(λ̂) where λ̂ is obtained from each of default or ACV. We then compare ISE(λ̂) against (and similarly for IAE).
It may be easier to employ what we call an “inefficiency measure,”
| (4) |
where in our case λ̂ = λ̂ACV, but keep in mind that λ̂ may be obtained by any method. Note that this compares the loss for λ̂ with the best possible loss for the given sample. We can easily construct Ineff IAE(λ̂), an inefficiency measure for IAE, as well. The IneffISE(λ̂) (or IneffIAE(λ̂)) has distributions on [0, ∞), and if this distribution is concentrated near 1, then we have a confidence in the methodology behind λ̂. Again, we will present √IneffISE(λ̂) in all the results, so that it is in the same unit as the IneffIAE(λ̂).
4 Simulations and Application
4.1 Pilot Study
Before performing a large scale simulation study, we first did a pilot study for several reasons. First, we want to make sure that all the robust smoothers work well on our data. Second, we want to look at the performance of default smoothing parameter selection procedures from the R routines to see if they should be included in the larger scale simulation study. Lastly, in examining the robust cross validation, we would like to see the effect of various leave out schemes in terms of speed and accuracy. We use the simulated data for this pilot study.
4.1.1 Simulation Models
There are two types of simulation models we use.
First, we construct a simulated model which resemble the data in our problem, because we are primarily interested in our application. To this end, we first pick a real data set (noisy data from one spectroscopic curve) that was shown in Figure 1 and fit a robust smoother, with the smoothing parameter chosen by inspection. We use the resulting smooth curve, m(x), as the “true curve” in this simulation, and then we carefully study the residuals from this fit and get an approximate error distribution to generate simulated data points. We determined that most of the errors were well modeled by a normal distribution with mean 0 and variance 2025. We then selected 40 x values at random between x = 420 and x = 1670 and added in an independent gamma random variate with mean 315 and variance 28,350. Additionally, we put in two gross outliers similar to the ones encountered in raw data of Figure 1.
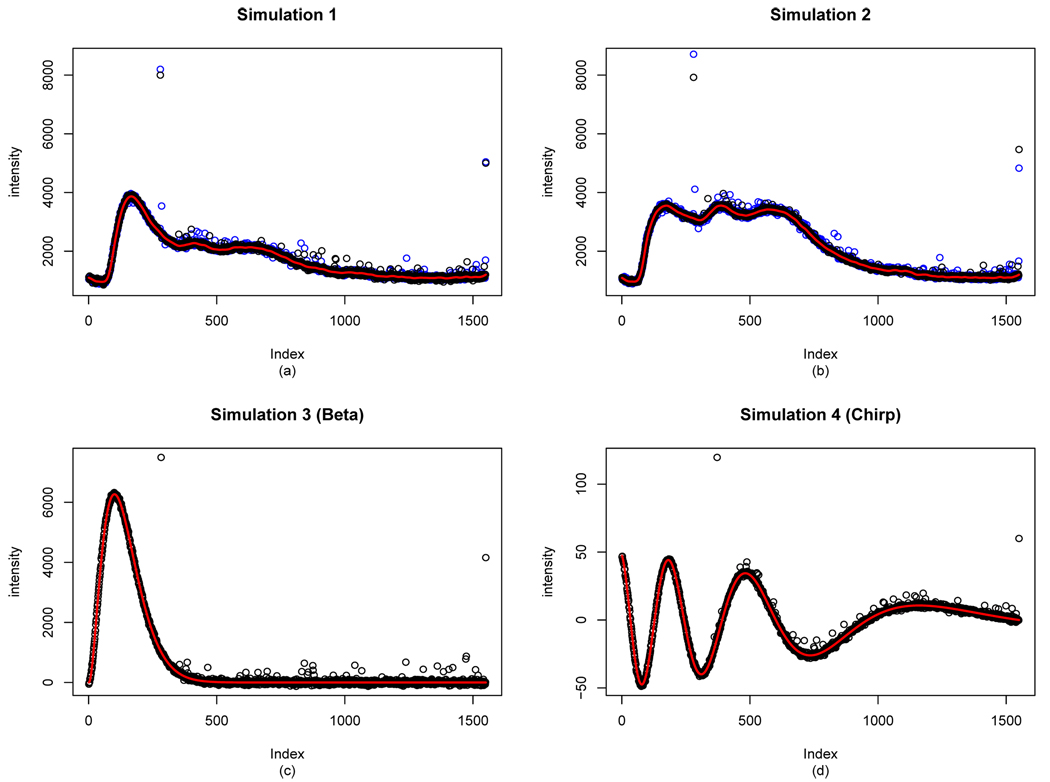
We call this the Simulation 1. See Figure 3 (a), and note how similar our simulated data is to the real data. We have also chosen a fitted curve based on another sample data (data from a different spectroscopic curve), with an error distribution very similar to Simulation 1. See Figure 3 (b). Call this Simulation 2.
Fig. 3.
All simulation models considered here. The blue points in (a) and (b) are the real data points, whereas the black points are simulated data points.
The second type of simulation models is based purely on mathematical models. We also have two curves for this type. First, we choose a beta curve, with the true curve as m(x) = 700beta(3, 30)(x),0 ≤ x ≤ 1 (where beta(3,30) is the pdf of Beta(3,30) distribution), which looks very much like some of the fitted curves from real data. Second, we have included so-called a chirp function (m(x) = 50(1–x) sin(exp(3(1–x))),0 ≤ x ≤ 1), just to see how our methodology works in very general setting. Note the scale difference in the chirp function, so we adjusted the error distribution accordingly by changing the scale. We then create the error distributions closely resembling the errors in previous simulation models. We call the simulation based on beta function the Simulation 3, and we call the simulation with chirp function the Simulation 4. See Figure 3 (c) and (d).
In all simulation examples in this paper, we have n = 1, 550 equispaced points.
4.1.2 Comparing Robust Smoothers and Smoothing Parameter Selection Methods
We compare the robust smoothers from Section 2 by applying them to the simulated data and comparing their computation time and performance.
Let us first step through some of the details regarding the smoothers. First, the robust smoothing splines, using the R function qsreg, has a default method as described in Section 3.1. It uses a range of 80 λ values as candidates for the default λ̂ . Since the range is wide enough to cover all the methods we try, we use these same λ values for all investigation of qsreg.
Next, we consider the robust LOESS using loess function in R. In this case, the program does not supply us with the λ values, so we used a range from 0.01 to 0.1, with length of 80. This range is sufficiently wide to cover all the reasonable λ values and more.
When we use COBS with the R function cobs, we do need to make few adjustments. First, we tried both the L1 (degree=1) and L∞ (degree=2) versions of COBS, and the L1 version was quite rough with discontinuous derivatives, so was dropped from further consideration. In addition, recall that for COBS, not only we have to determine λ but also the number of knots N as well. The default for the number of knots is 20 (N = 20), and we use this N for the default (BIC) method for the smoothing parameter selection of λ (using both the defaults by fixing N = 20 and λ selected by BIC). However, if we fix the number of knots at the default level, it gives unsatisfactory results, and some preliminary studies has shown that the increasing the number of knot to 50 (N = 50) will make the COBS fit better. But either way, the computing time of this smoother suffers dramatically. The computing times for obtaining the estimates (for one curve) were 0.03 seconds for qsreg, 0.06 seconds for loess, but 0.16 seconds for cobs (for both N = 20 and N = 50). For these reasons, we decided to increase the number of knots to 50 for the COBS experiments. Here, the candidate λ values range from 0.1 to 1,000,000.
Now, we present the L1 and L2 loss functions (IAE(λ) and √ISE(λ) for each of the λ̂ values obtained by default and ACV, and compare them with optimal λ values. The results are shown in Table 1. This reveals that all default methods perform very poorly, as in Figure 1. This is especially apparent in default methods for robust smoothing splines and robust LOESS with fixed λ = 3/4. The default method for COBS (with N = 20) also does not work well. If we increase to number of knots to 50 (N = 50), then the default is not as bad as the default methods of the competing robust smoothers. However, the default method still performs more poorly than the ACV method. Furthermore, COBS’s overall performance based on CV is poorer than those of robust smoothing splines or robust LOESS.
Table 1.
A table of L1 and L2 loss function values for default, ACV, and optimum (IAE or ISE) criteria. For the COBS default, two sets of values are shown; Default with N = 20 or Default.50 where N = 50.
| Simulation 1 | Simulation 2 | Simulation 3 | Simulation 4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IAE | √ISE | IAE | √ISE | IAE | √ISE | IAE | √ISE | ||
| qsreg | Default | 24.45 | 31.80 | 16.30 | 21.05 | 23.46 | 29.99 | 0.53 | 0.69 |
| ACV | 9.90 | 12.64 | 7.69 | 9.47 | 9.74 | 13.05 | 0.19 | 0.24 | |
| Optimum | 9.89 | 12.16 | 6.80 | 8.48 | 8.39 | 12.73 | 0.18 | 0.24 | |
| loess | Default | 229.05 | 478.19 | 194.44 | 324.32 | 427.32 | 914.51 | 13.15 | 19.57 |
| ACV | 8.61 | 10.76 | 6.19 | 7.50 | 8.79 | 11.30 | 0.19 | 0.24 | |
| Optimum | 8.12 | 10.33 | 5.73 | 6.86 | 7.92 | 11.27 | 0.18 | 0.24 | |
| cobs | Default | 68.36 | 162.19 | 53.22 | 118.64 | 48.09 | 162.83 | 2.29 | 6.32 |
| Default.50 | 14.26 | 23.65 | 10.65 | 18.76 | 7.88 | 13.79 | 0.27 | 0.47 | |
| ACV | 10.11 | 12.72 | 9.52 | 14.72 | 6.98 | 9.83 | 0.19 | 0.25 | |
| Optimum | 10.11 | 12.72 | 9.45 | 14.71 | 6.98 | 9.80 | 0.19 | 0.25 | |
We remarked that COBS suffers from lack of speed compared to its competitors. This deficiency is not overcome by improvements in performance. Given all the complication with COBS, we dropped it from the rest of the pilot study and the simulations.
All in all, we conclude that the ACV method gives very satisfactory results. We see clearly that any method that does not incorporate the cross validation scheme does poorly. The only disadvantage of the leave-one-out cross validation methods is its computation time. Nevertheless, we demonstrate in the next section that there are ways to overcome this deficiency of ACV method.
4.1.3 Systematic K-fold Method
We investigate ACV(d,r) presented in Section 3.3 for faster computation and comparable performance.
Recall that we will only consider r = 1, the systematic K-fold scheme. (see Section 3.3). For our problem, with d = 5 and r = 1 (the systematic 5-fold CV), λ̂ is almost optimal, but the computation is 365 times faster than the full leave-one-out (LOO) scheme! And the computation time is proportional (to a high accuracy) to how many times we compute a robust smoothing spline. Table 2 confirms that the performance of K-fold CV schemes (d = K, r =1) are superior to default methods and comparable to the full LOO method.
Table 2.
A table of loss functions in Simulation 1, using robust smoothing splines (qsreg).
| Scheme | IAE | √ISE | Time (seconds) |
|---|---|---|---|
| Default | 24.45 | 31.80 | 2.96 |
| Full LOO | 9.90 | 12.64 | 4,477.47 |
| d = 50, r = 1 | 9.90 | 12.64 | 145.50 |
| d = 25, r = 1 | 9.89 | 12.16 | 70.53 |
| d= 5, r = 1 | 9.89 | 12.16 | 12.26 |
| Optimum | 9.89 | 12.16 |
Hence, based on all the results presented, we use the systematic K-fold CV as a main alternative to the full LOO CV.
4.1.4 Comparing Systematic K-fold CV with Random K-fold CV
We would like to see how the random K-fold schemes compare to our systematic schemes. In particular, since the systematic 5-fold CV worked very well in our example, we compare systematic 5-fold CV against the random 5-fold CV.
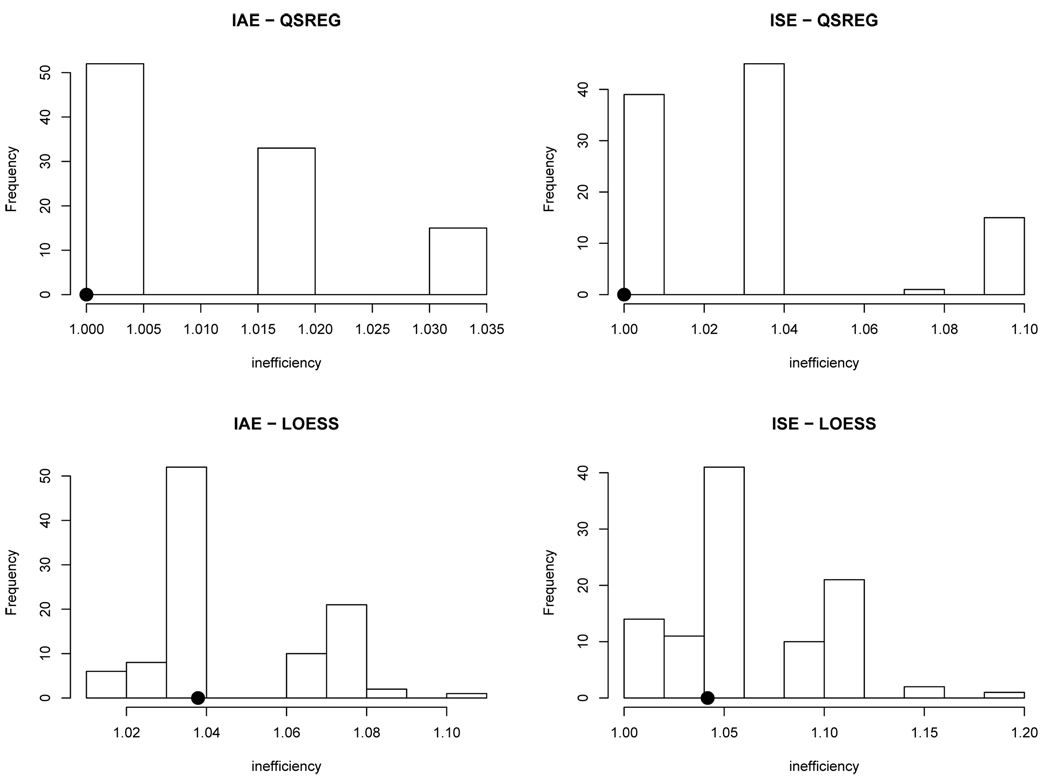
We take a λ̂ from each of 100 draws (i.e., 100 different partitions) of random 5-fold ACVs. Then, we compute the inefficiency measure (4) introduced in the previous section, for both IAE and ISE. We do this for each of the 100 draws, and we compare them with the inefficiency measure of the systematic 5-fold CV. We present histograms of the inefficiency values in Figure 4. The results in the figures suggest that the systematic 5-fold does well relative to random 5-fold and optimal value. Experience with other values of K and other simulations yielded similar results.
Fig. 4.
A histogram of inefficiencies obtained from the random 5-fold ACV. The systematic 5-fold inefficiency value is shown as a dot. The results are based on 100 draws of random 5-fold CVs, in Simulation 1.
One problem with the random K-fold CV is that it introduces another source of variation that can lead to an undesirable consequence. For example, when the random K-fold CV randomly leaves out many points that are in the neighborhood of each other, it will do a poor job of predicting those left out points and hence produces a suboptimal result. It is known that a random K-fold CV result can be hugely biased for the true prediction error (Hastie et al. 2001). Therefore, we decided not to consider the random K-fold CV any further.
4.2 Large Simulation Study
We now report on the results of a large scale simulation study to further assess the performance of the robust estimators. Specifically, we compare the robust smoothing spline and robust LOESS.
The data is obtained just as in Section 4.1.1, where we take a vectorized “true” curve m(x) and add a vector of random errors to it (with error distribution as described in that section), and repeat this M times with the same m(x).
If we obtain ISE(λ) and IAE(λ) functions for each simulation, we can easily estimate mean integrated squared error (E[ISE(λ)]) by averaging over the number of replications, M. The E[IAE(λ)] may likewise be obtained.
Our results are based on M = 100.
4.2.1 Detailed Evaluation with Full Leave-One-Out CV
We begin by reporting our results based on the full LOO validation with ACV for the Simulation 1 model (we defer all other results until next subsection).
We want to assess the performance of the two robust smoothers of interest by comparing E[ISE(λ̂)] values, with λ̂ = λ̂ACV, and similarly for E[IAE(λ̂)]. Also, we present the results of the inefficiency measures. Again, we will take squared roots of those quantities involving ISE so that we present √E[ISE(λ̂)] and √IneffISE(λ̂)] in the tables. The results are presented in Table 3. Clearly, all the integrated error measures of robust LOESS are lower than those of the robust smoothing splines, although these results by themselves do not indicate which smoothing method is more accurate.
Table 3.
A table comparing robust smoothers and loss functions and the mean and median inefficiency measure values.
| √E[ISE(λ̂)] | E[IAE(λ̂)] | √IneffISE(λ̂) | IneffISE(λ̂) | |||
|---|---|---|---|---|---|---|
| mean | median | mean | median | |||
| qsreg | 12.61 | 9.89 | 1.11 | 1.05 | 1.05 | 1.03 |
| loess | 10.45 | 8.21 | 1.07 | 1.03 | 1.03 | 1.01 |
4.2.2 Results on All Simulations
We now report on all simulation results, including those of the systematic K-fold CV as described in section 3.3. See Table 4 for the results, where we only report 5-fold systematic CV since 25- and 50-fold results are very similar. The result is that all of √IneffISE(λ̂) and IneffIAE(λ̂) values are near 1 for all λ̂ values obtained from full LOO schemes, for ACV. This is true for all four simulation models we considered. The numbers are very similar for all the K-fold CV schemes (where we considered K = 5, 25, and 50).
Table 4.
A table of median values of inefficiencies in all simulations. Each entry consists of either √IneffISE(λ̂) or IneffIAE(λ̂) with λ̂ value corresponding to different rows.
| Simulation 1 | Simulation 2 | ||||
|---|---|---|---|---|---|
| IAE | √ ISE | IAE | √ ISE | ||
| qsreg | Default | 2.59 | 2.68 | 2.34 | 2.41 |
| Full LOO | 1.03 | 1.05 | 1.02 | 1.03 | |
| d = 5,r = 1 | 1.02 | 1.02 | 1.00 | 1.00 | |
| loess | Default | 28.67 | 47.29 | 32.95 | 43.48 |
| Full LOO | 1.01 | 1.03 | 1.02 | 1.01 | |
| d = 5, r = 1 | 1.01 | 1.03 | 1.00 | 1.00 | |
| Simulation 3 | Simulation 4 | ||||
|---|---|---|---|---|---|
| IAE | √ ISE | IAE | √ ISE | ||
| qsreg | Default | 2.36 | 2.17 | 2.67 | 2.54 |
| Full LOO | 1.09 | 1.06 | 1.05 | 1.04 | |
| d = 5, r = 1 | 1.05 | 1.06 | 1.02 | 1.02 | |
| loess | Default | 46.88 | 73.53 | 70.73 | 77.85 |
| Full LOO | 1.04 | 1.02 | 1.05 | 1.01 | |
| d = 5, r = 1 | 1.02 | 1.04 | 1.03 | 1.03 | |
In contrast, the √IneffISE(λ̂) and IneffIAE(λ̂) values for λ̂default are at least 2 and can be as much as 77!
The results again demonstrate and reconfirm what we already claimed: our cross validation method is far superior to default methods and is basically as good as - if not better than - the full LOO method.
In conclusion, we have seen that robust LOESS gives the best accuracy for our application, although the computational efficiency of robust smoothing splines is better. At this point, we are not sure if the increase in accuracy is worth the increase in computational effort, since both procedures work very well. Either one will serve well in our application and in many situations to be encountered in practice. For our application, we recommend the systematic 5-fold CV scheme with the robust LOESS, based on our simulation results.
4.3 Application
Now, we discuss the results of applying our methods to real data. We have done most of the work in previous sections, and all we need for the application is to apply the smoother with appropriate smoothing parameter selection procedure to other curves.
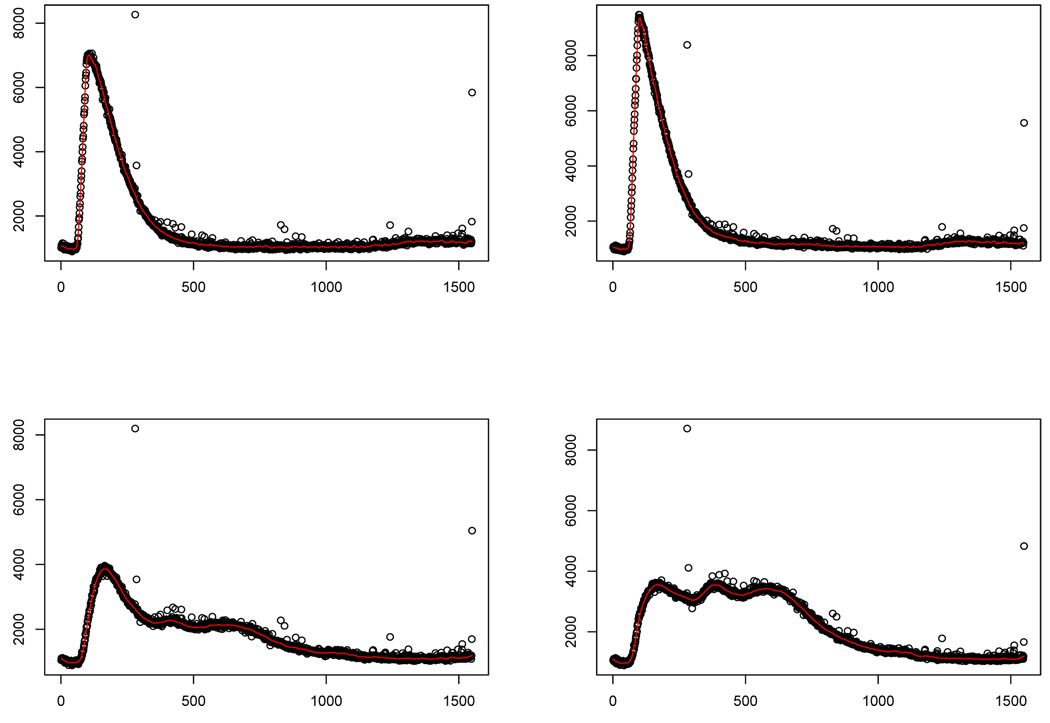
Following the recommendations from the previous section, we used the robust LOESS with ACV based on systematic 5-fold CV (d = 5, r = 1) for smoothing parameter selection. We found that this worked well. See Figure 5 for a sample of results.
Fig. 5.
Plots of raw data with robust smoother superimposed in four of the emission spectra.
We have also performed some diagnostics from fitting to the real data, such as plotting residuals versus fitted values and Quantile-Quantile plot (Q-Q plot), as well as looking at the autocorrelation of the residuals to determine whether there is much correlation between adjacent grid points. The diagnostics did not reveal any problems, and therefore we are confident of the usefulness of our method.
5 Conclusion and Discussion
We have seen how well our robust cross validation schemes perform in our application. Specifically, we are able to implement a smoothing parameter selection procedure in a fast, accurate way. We can confidently say that with either robust smoothing splines or robust LOESS using the ACV based on systematic K-fold cross validation works well in practice.
There may exists some questions raised regarding our method. One such issue may be the fact that we have only considered absolute value and Huber’s rho functions in the RCV function. But we have shown that ACV with absolute value works very well, and most other criterion functions will undoubtedly bring more complications, so we do not foresee any marked improvements over ACV . In addition, there may be some concern about the choice of d and r in our leave out schemes. We obtained a ranges of candidate d and r values by trial and error, and this must be done in every problem. Nevertheless, this small preprocessing step will be beneficial in the long run. As demonstrated in the paper, we can save an enormous amount of time once we figure out the appropriate values for d and r to be used in our scheme. Furthermore, if we need to smooth many different curves as was done here, then we need only to do this for some test cases. Some other issues such as use of default smoothing parameter selection methods and the use of random K-fold has been resolved in the course of the paper.
Now, we would like to discuss some possible future research directions. There exist other methods of estimating smoothing parameters in robust smoothers, such as Robust Cp (Cantoni and Ronchetti, 2001) and plug-in estimators (Boente et al., 1997). We have not explored these in the present work, but they could make for an interesting future study.
Another possible problem to consider is the theoretical aspect of robust cross validation schemes. Leung et al. (1993) gives various conjectures, and Leung (2005) shows some partial results. As indicated in Section 3.4, we have some heuristics for the asymptotics for the robust cross validation and we will investigate this rigorously in the future research.
Also, extending the methods proposed here to the function estimation on a multivariate domain presents some challenges. In particular, the implementation of systematic K-fold cross validation was easy for us since our independent variable values were 1-dimensional and equally spaced.
In conclusion, we believe that our methods are flexible and easily implementable in a variety of situations. We have been concerned with applying robust smoothers only to the spectroscopic data from our application so far, but they should be applicable to other areas which involve similar data to ours.
Supplementary Material
Acknowledgments
We would like to thank the editor and the referees for their comments which led to a significant improvement of the paper. We would also like to thank Dan Serachitopol, Michele Follen, David Scott, and Rebecca Richards-Kortum for their assistance, suggestions, and advice. The research is supported by the National Cancer Institute and by the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental material (E-component) available in electronic version.
References
- 1.Boente G, Fraiman R, Meloche J. Robust plug-in bandwidth estimators in nonparametric regression. Journal of Statistical Planning and Inference. 1997;57:109–142. [Google Scholar]
- 2.Bussian B, Härdle W. Robust smoothing applied to white noise and single outlier contaminated Raman spectra. Applied Spectroscopy. 1984;38:309–313. [Google Scholar]
- 3.Cantoni E, Ronchetti E. Resistant selection of the smoothing parameter for smoothing splines. Statistics and Computing. 2001;11:141–146. [Google Scholar]
- 4.Chambers J, Hastie T. Statistical Models. Pacific Grove, CA: S. Wadsworth & Brooks/Cole; 1992. [Google Scholar]
- 5.Chang SK, Follen M, Utzinger U, Staerkel G, Cox DD, Atkinson EN, MacAulay C, Richards-Kortum RR. Optimal excitation wavelengths for discrimination of cervical neoplasia. IEEE Transaction on Biomedical Engineering. 2002;49:1102–1111. doi: 10.1109/TBME.2002.803597. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. Journal of the American Statistical Association. 1979;74:829–836. [Google Scholar]
- 7.Cox DD. Asymptotics for M-type smoothing splines. Annals of Statistics. 1983;11:530–551. [Google Scholar]
- 8.de Boor C. A Practical Guide to Splines. New York: Springer; 1978. [Google Scholar]
- 9.Ghement IR, Ruiz M, Zamar R. Robust estimation of error scale in nonparametric regression models. Journal of Statistical Planning and Inference. 2008;138:3200–3216. [Google Scholar]
- 10.Grossman N, Ilovitz E, Chaims O, Salman A, Jagannathan R, Mark S, Cohen B, Gopas J, Mordechai S. Fluorescence spectroscopy for detection of malignancy: H-ras overexpressing fibroblasts as a model. Journal of Biochemical and Biophysical Methods. 2001;50:53–63. doi: 10.1016/s0165-022x(01)00175-0. [DOI] [PubMed] [Google Scholar]
- 11.Hastie T, Tiibshirani R, Friedman J. The Elements of Statistical Learning. New York: Springer; 2001. [Google Scholar]
- 12.He X, Ng P. COBS: Qualitatively constrained smoothing via linear programming. Computational Statistics. 1999;14:315–337. [Google Scholar]
- 13.Hogg RV. Statistical robustness. American Statistician. 1979;33:108–115. [Google Scholar]
- 14.Leung D, Marriot F, Wu E. Bandwidth selection in robust smoothing. Journal of Nonparametric Statistics. 1993;2:333–339. [Google Scholar]
- 15.Leung D. Cross-validation in nonparametric regression with out-liers. Annals of Statistics. 2005;33:2291–2310. [Google Scholar]
- 16.Oh H, Nychka D, Brown T, Charbonneau P. Period analysis of variable stars by robust smoothing. Journal of the Royal Statistical Society, Series C. 2004;53:15–30. [Google Scholar]
- 17.Oh H, Nychka D, Lee T. The Role of pseudo data for robust smoothing with application to wavelet regression. Biometrika. 2007;94:893–904. [Google Scholar]
- 18.Rice J. Bandwidth choice for nonparametric regression. Annals of Statistics. 1984;12:1215–1230. [Google Scholar]
- 19.Schwarz G. Estimating the dimension of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 20.Simonoff J. Smoothing Methods in Statistics. New York: Springer; 1996. [Google Scholar]
- 21.Wang FT, Scott DW. The L1 method for robust nonparametric regression. Journal of the American Statistical Association. 1994;89:65–76. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.