Abstract
Carbohydrate-protein interactions play a critical role in a variety of biological processes, and agonists/antagonists of these interactions are useful as biological probes and therapeutic agents. Most carbohydrate-binding proteins achieve tight binding through formation of a multivalent complex. Therefore, both ligand structure and presentation contribute to recognition. Since there are many potential combinations of structure, spacing, and orientation to consider and the optimal one cannot be predicted, high-throughput approaches for analyzing carbohydrate-protein interactions and designing inhibitors are appealing. In this report, we develop a strategy to vary neoglycoprotein density on a surface of a glycan array. This feature of presentation was combined with variations in glycan structure and glycan density to produce an array with approximately 600 combinations of glycan structure and presentation. The unique array platform allows one to distinguish between different types of multivalent complexes on the array surface. To illustrate the advantages of this format, it was used to rapidly identify multivalent probes for various lectins. The new array was first tested with several plant lectins, including concanavalin A (conA), Vicia villosa isolectin B4 (VVL-B4), and Ricinus communis agglutinin (RCA120). Next, it was used to rapidly identify potent multivalent inhibitors of Pseudomonas aeruginosa lectin I (PA-IL), a key protein involved in opportunistic infections of P. aeruginosa, and mouse macrophage galactose-type lectin (mMGL-2), a protein expressed on antigen presenting cells that may be useful as a vaccine targeting receptor. An advantage of the approach is that structural information about the lectin/receptor is not required to obtain a multivalent inhibitor/probe.
Keywords: Carbohydrate microarray, glycan array, multivalency, lectin inhibitors
Introduction
Carbohydrate-protein interactions play a critical role in many biological processes, such as cell-cell recognition, inflammation, and metastasis.1–3 Due to their importance, there have been significant efforts to identify carbohydrate-binding proteins, determine their natural ligands, and characterize their biological functions. In addition, the development of agonists/antagonists of these interactions has received considerable attention. For example, many bacteria and viruses bind to carbohydrates on the surface of host cells as a key step of infection, and inhibitors of these interactions have been sought after as anti-bacterial or anti-viral agents.4–6 As a second example, migration of leukocytes to sites of infection is mediated by a carbohydrate-binding protein, L-selectin, and inhibitors of this protein have been investigated extensively as anti-inflammatory agents.7–9
Analysis of carbohydrate-protein interactions and development of inhibitors/probes of these interactions is challenging for several reasons. First, there are thousands of glycans found in nature, and it is difficult to predict which ones will bind to a protein. Consequently, one would like to evaluate binding to many glycans to determine if a protein binds carbohydrates and to identify its ligands. Unfortunately, only a small number of structurally-defined glycans are available in homogeneous form, and these are typically only available in small quantities. Therefore, it is difficult to test large numbers of potential ligands for binding. Second, carbohydrate-binding proteins, such as lectins and antibodies, typically bind monovalent carbohydrate ligands with low affinity and poor selectivity.6, 10–12 To compensate for the low intrinsic affinity, carbohydrate-binding proteins normally possess multiple binding sites, which enables them to simultaneously bind two or more carbohydrate ligands on a cell surface or glycoprotein. The resulting multivalent complexes can have much higher overall affinity (referred to as avidity) and selectivity. To produce a multivalent interaction, however, the spacing and orientation of the carbohydrate ligands must be appropriately matched to the binding sites of the receptor. Therefore, proper presentation of the ligands is critical for recognition.
Agonists/antagonists of carbohydrate-binding proteins are very useful for a variety of basic research and clinical applications. While there have been many attempts to design monovalent inhibitors of lectins, high affinity monovalent inhibitors are extremely rare and most have affinities in the micromolar to millimolar range. Therefore, multivalency has been a key design feature for the majority of current inhibitors and probes for lectins. A variety of multivalent scaffolds have been developed, such as dendrimers, proteins, polymers, beads, and liposomes.6, 10–12 Different multivalent platforms can display glycans with varying spacing, orientation, density, flexibility, and overall architecture, but it is difficult to predict the optimal scaffold for a particular glycan-lectin interaction. Optimization of multivalent presentation is especially tough when the spacing and geometries of the binding sites on the target lectin are not known.
Glycan arrays contain many different carbohydrates immobilized on a solid support in a spatially-separated arrangement.13–19 They provide a high-throughput tool to evaluate many potential carbohydrate-protein interactions in parallel while using only tiny amounts of scarce materials. Glycan arrays have great potential for aiding the design and development of lectins inhibitors, but certain challenges exist. For most array platforms, a multivalent display is achieved by presenting multiple copies of a monovalent ligand on the surface. With this approach, the surface acts as the multivalent scaffold and, therefore, the spacing and orientation of ligands are defined by the surface. While this approach is useful for identifying ligands that are recognized by carbohydrate-binding proteins, it is not ideal for the development of multivalent probes since it can be difficult to identify a soluble multivalent scaffold that mimics the presentation of the carbohydrates on the array surface.
We have focused on an alternative approach for the construction of glycan arrays wherein multivalent glycoconjugates are immobilized on the surface.20–29 To produce our arrays, glycans are first covalently attached to a carrier protein, such as albumin, to generate multivalent neoglycoproteins. These conjugates are then immobilized on the surface to produce a neoglycoprotein array. Neoglycoproteins have been used for many years as reagents to study carbohydrate recognition, as multivalent inhibitors of carbohydrate-protein interactions, and as immunogens.4, 30 Since the same multivalent scaffold is used on the array surface and for solution applications, this strategy is well-suited for the identification of multivalent neoglycoprotein probes for lectins. In addition, the approach offers unique opportunities to modulate presentation, including varying the carrier protein, the glycan density, and the neoglycoprotein density.
In this study, we describe the development and evaluation of a strategy to modulate neoglycoprotein density on the array surface. The approach is simple and economical and can readily be combined with other elements of glycan array diversity. To illustrate this, we constructed arrays with variations in neoglycoprotein density, glycan structure, and glycan density. Collectively, the new arrays contained approximately 600 combinations of glycan structure and presentation. One advantage of this unique array format is that it allows one to distinguish between different multivalent binding modes. To illustrate the utility of this new multi-dimensional array format, we demonstrate that it provides rapid access to high affinity multivalent probes for various plant, bacterial, and animal lectins.
Materials and Methods
General methods
Unless otherwise stated, reagents were purchased from commercial suppliers and used without purification. All aqueous solutions were prepared from Milli-Q water with a 0.2 µm filter. Bovine serum albumin (BSA) was purchased from Sigma (St. Louis, MO). Alexa Fluor®488 (AF488) BSA conjugates, Alexa Fluor®594 (AF594) BSA conjugates, AF555 succinimidyl ester, and QSY-7 succinimidyl ester were purchased from Invitrogen Corporation (Carlsbad, CA). Biotinylated BSA, ConA, VVL-B4, and RCA120 were purchased from Vector Laboratories (Burlingame, CA). Cy3-Streptavidin and Alkaline Phosphatase (AP)-Streptavidin were purchased from Zymed Laboratories of Invitrogen Corporation (Carlsbad, CA). His-tagged PA-IL was a gift from Professor Lara Mahal (New York University). Anti-penta-His mouse IgG1 was purchased from Qiagen (Valencia, CA), Cy3-labeled goat anti mouse IgG+IgM(H+L) and AP-goat anti mouse IgG+IgM(H+L) were purchased from Jackson ImmunoResearch (West Grove, PA). AlphaGal-BSA and Pk-HSA were purchased from V-Labs (Covington, LA). Recombinant mMGL-2 and biotinylated goat anti-mouse IgG were purchased from R&D Systems (Minneapolis, MN).
Analysis of lectin binding on the full array
Microarray slides (for array fabrication, see Supporting Information) were assembled with an 8 well slide holder (Grace Bio-Labs, Inc, Bend, OR). All slides were blocked with 3% BSA/PBS at r.t. for 2.0 h before experiments. Dilution series of biotinylated lectins [Concanavalin A (ConA), Vicia villosa lectin (VVL-B4), and Ricinus communis agglutinin (RCA120)] were prepared in 1% BSA/PBST0.05. ConA was prepared in a range from 0.18 nM to 460 nM. VVL-B4 was in a range from 1.2 nM to 620 nM. RCA120 was in a range from 0.41 nM to 105 nM. 200 µL of the lectin solutions was added to each well, covered tightly with seal strips and incubated at r.t for 2.0 h. After washing unbound lectin with 4×400 µL of PBST0.05, streptavidin-Cy3 in 1% BSA/PBS (1:500, 1 µg/mL, 200 µL/well) was added and incubated at r.t. for 2.0 h.
Pseudomonas aeruginosa lectin I (PA-IL) and mouse macrophage galactose-type lectin-2 (mMGL-2) were prepared in 1% BSA/TSMT0.05 (20 mM Tris, 150 mM NaCl, 0.05% tween 20, 2 mM CaCl2, 2 mM MgCl2). PA-IL was diluted in a range from 37 nM to 4700 nM. And mMGL-2 was in a range from 0.38 nM to 24 nM. Unbound lectin was washed off by 4×400 µL TSMT0.05 and tapped dry. Mouse anti-His IgG1 in 1% BSA/TMS (1:200, 1 µg/mL, 200 µL/well) for PA-IL, and biotinylated goat anti-mouse IgG in 1% BSA/TMS (1:200, 2 µg/mL, 200 µL/well) were added and incubated at r.t. for 2.0 h. Slides were washed by 4×400 µL TSMT0.05 and tapped dry. Then goat anti mouse Cy3-IgG+IgM(H+L) 1% BSA/TMS (1:500, 1 µg/mL, 200 µL/well) for PA-IL and Cy3-streptavidin in 1% BSA/PBS (1:500, 1 µg/mL, 200 µL/well) for mMGL-2 were added and incubated at r.t. for 2.0 h. All Slides were washed 4×400 µL of PBST0.05 and tapped dry, removed from the holder, and immersed into PBST0.05 buffer for 10 min. Slides were dried by centrifuging at 1000 rpm for 5 min.
Slides were scanned using a Genepix 4000A microarray scanner at 10 µm resolution (Molecular Devices Corporation, Union City, CA) at a PMT voltage setting of 440 (or 460) at 532 nm and 632 nm. Images were analyzed with Genepix Pro 6.0 analysis software (Molecular Devices Corporation). Spots were defined as circular features of 100 µm. The features were resized manually as needed. The background-corrected mean (F532mean-B532) was used for data analysis. Fluorescence data for each spot for a given neoglycoprotein or glycoprotein was averaged. The apparent Kd values were determined following the method of MacBeath.31 The mean was plotted with the concentration of lectins on a logarithmic scale and a nonlinear curve was fitted using Origin 8.0 (OriginLab, Northampton, MA) according to the equation below:
where Fc is the fluorescent intensity for the lectin binding at any specific concentration, Fmax is the maximum intensity, Kd is the apparent dissociation constant for the lectin and neoglycoprotein on the array, and [L] is the concentration of lectins.
Analysis of solution inhibition and IC50 determination by ELISA-like assays
Wells of a 96-well plate (nontreated Maxisorb NUNC 96-well plate) were incubated with 500 ng/well of neoglycoprotein in 1× PBS buffer, pH 7.4, at 4 °C overnight. The following neoglycoproteins were used for the respective lectins: Man-α–BSA (ConA), Ac-S-Tn(Ser)-S-G-33-BSA (VVL-B4), Ac-S-TF(Thr)-S-G-28-BSA (RCA120), Lac-BSA (PA-IL) and Ac-TF(Thr)-G-24-BSA (mMGL-2). After adsorbing the appropriate neoglycoprotein to the well surfaces, the solutions were removed and the wells were blocked with 3% BSA in PBS buffer at r.t. for 2.0 h. Biotinylated plant lectins (ConA = 1:5000; VVL-B4 = 1:2000; and RCA120 = 1:2500; each in 1% BSA in PBS) were incubated with a series of concentrations of various neoglycoproteins for 30 min. 60 µL of the mixture was then added to each well and incubated at r.t. for 2.0 h. The neoglycoprotein-lectin mixture solutions were removed and the plate was washed four times with PBST0.05 (200 µL/well). AP-streptavidin was diluted 1:500 in 1% BSA in PBS. 65 µL of the solution was added to each well and incubated for 2.0 h. The streptavidin solution was removed, and the plate was washed four times with PBST0.05. Next, 75 µL of 4-methylumbellyferyl phosphate solution (MUP; 100 µM in Tris, pH 9.0) was added to each well and the plate was scanned immediately using a FLX 800 microplate fluorescence reader (BioTek, Winooski, VT; excitation = 360, emission = 440).
Inhibition assays for PA-IL and mMGL-2 were carried out in an analogous manner except that samples were diluted in TSM buffer (20 mM Tris, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2). In addition, binding was detected by mouse anti-His IgG1 (1:200 in 1% BSA/TMS; 65 µL/well) followed by AP-goat anti mouse IgG+IgM (H+L) (1:500 in 1% BSA/TSM; 70 µL/well) for PA-IL, and biotinylated goat anti-mouse IgG (1:200 in 1% BSA/TMS; 65 µL/well) followed by AP-streptavidin (1:500 in 1% BSA/TSM; 70 µL/well) for mMGL-2.
The assays were conducted in triplicate, and the data was normalized by subtracting the negative control (samples without lectin) and then dividing by the maximum [maximum = positive control (lectin without mixing with neoglycoprotein) - negative control]. The mean was plotted and fit using Origin 8.0 software. IC50 values for the neoglycoproteins were obtained from the concentration of neoglycoprotein at 50% of relative intensity.
Results and Discussion
Array design and rationale
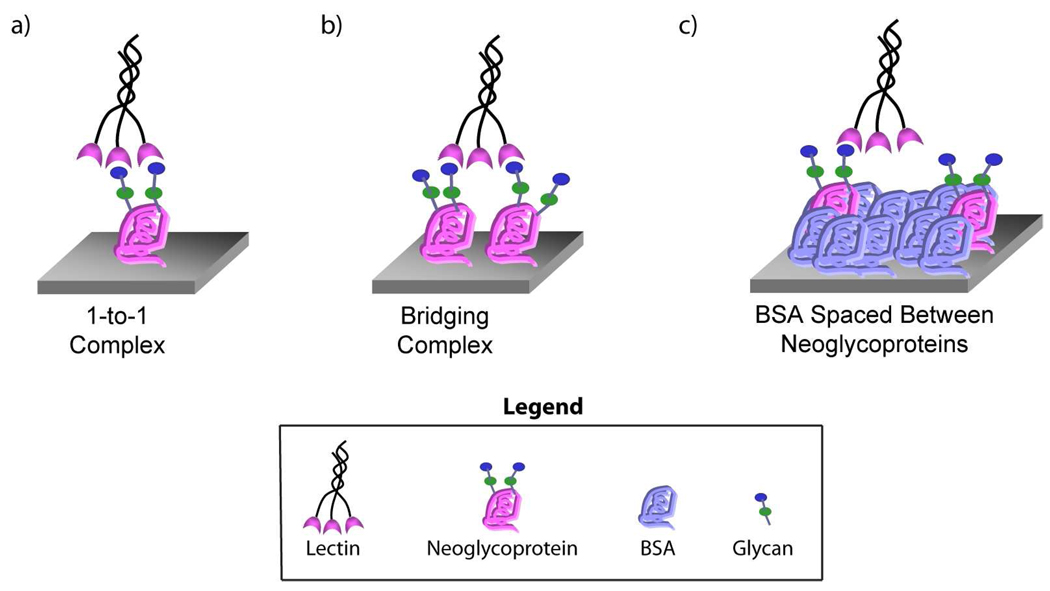
Our objective was to develop an array-based strategy to identify multivalent probes for glycan-binding proteins and to better profile their binding properties. Carbohydrate-binding proteins can adhere to a surface coated with neoglycoproteins in a variety of ways. For example, a carbohydrate-binding protein can form a multivalent complex with a single neoglycoprotein (1-to-1 complex, see Figure 1a) or can recognize carbohydrates on adjacent neoglycoproteins to form a bridging complex (i.e. one lectin binding two or more neoglycoproteins, see Figure 1b). These different types of multivalent complexes are not equivalent, and the ability to distinguish between them could be useful for a number of applications. First, it would be useful for identifying multivalent probes for lectins. Although both types of multivalent complexes produce high avidity interactions on a surface, a bridging complex is difficult to achieve in solution and generally produces aggregates. Therefore, a neoglycoprotein with the ability to form a 1-to-1 complex on a surface is more likely to retain activity as a high affinity, solution-phase probe. Second, methods to distinguish between different types of multivalent complexes could be useful for detecting carbohydrate-binding proteins within complex mixtures. Many carbohydrate-binding proteins have similar specificities but distinct architectures (e.g. different numbers of binding sites, different distances between binding sites). Because the spacing and orientations of the binding sites are different, these proteins could form different types of multivalent complexes on the surface, and methods to discriminate between those complexes could enable one to differentially detect those proteins.
Figure 1.
Different Multivalent Binding Modes and the Array Strategy.
One strategy to distinguish 1-to-1 complexes and bridging complexes is to vary the neoglycoprotein density on the array surface. Formation of a bridging complex requires neoglycoproteins to be positioned in close proximity to each other. If individual neoglycoprotein molecules are spaced farther apart on the surface, bridging complexes should be disfavored (see Figure 1c). One method to achieve the desired spacing involves adding unmodified BSA into the print solutions and then printing these mixtures on the array surface.32 With this strategy, unmodified BSA occupies surface area between neoglycoprotein molecules (see Figure 1c). As the proportion of BSA increases, individual neoglycoproteins will be spaced farther apart, and the neoglycoprotein density decreases. While the relative proportions of neoglycoprotein and BSA would be modulated, the total protein concentration would be maintained at a constant value (125 µg/mL, a concentration that saturates the surface). By comparing binding at high and low neoglycoprotein density, one could distinguish between 1-to-1 complexes and bridging complexes.
We,22 and others,33–39 have previously described methods to vary the glycan density on the surface of an array. In our previous work, glycan density was modulated by varying the average number of glycans per molecule of albumin. The method described above for spacing apart neoglycoproteins involves varying the neoglycoprotein density (the average number of neoglycoprotein molecules per unit surface area). While similar in certain respects, modulation of neoglycoprotein density is functionally distinct and complementary with varying glycan density (for a detailed example illustrating the functional differences between variations in glycan density versus variations in neoglycoprotein density, see Figure S4, Supporting Information). It was our intention to construct arrays with variations in both glycan density and neoglycoprotein density.
Although the design concept was simple, a number of factors could cause problems. First, the neoglycoproteins must have limited movement on the surface. Some degree of flexibility was expected due to the linkers and conformational motion of the carrier protein, but individual molecules of neoglycoprotein should not be able to move or “slide around” on the surface. If this were the case, then neoglycoproteins and molecules of unmodified BSA could rearrange during an assay to form both 1-to-1 and bridging complexes. Second, the immobilization process should result in an even distribution of neoglycoproteins and unmodified BSA on the surface. If the neoglycoproteins cluster together, for example, then the addition of BSA would not generate the expected spacing. Ideally, the spacing on the surface would be predictable, controllable, and consistent for all neoglycoproteins. For example, variations in glycan length, branching, and the number of glycans per molecule of albumin should not significantly affect this relationship. For these reason, our initial studies were aimed at characterizing the surface and validating the design concept.
Surface characterization and model studies
To better characterize the surface, we carried out a set of experiments with model arrays. First, arrays were printed with AF488-BSA, a fluorophore-labeled protein, with or without varying amounts of unmodified BSA and the resulting spots were scanned using a confocal microscope. For each of the neoglycoprotein densities, the fluorescence was found to be consistent over the area of the spot (see Figure S1, Supporting Information). Although the resolution is not sufficient to define positions at a single molecule level, the result supports an even distribution of conjugates. Next, we carried out a photobleaching experiment to determine if the conjugates can move around on the spot. A pattern was photobleached within a section of the spot and the fluorescence was monitored over the next 4 hours (see Figure S2, Supporting Information). If the AF488-BSA conjugates were capable of moving around on the spot, one would expect redistribution of the AF488-BSA molecules and disappearance of the photobleached pattern. In contrast, we found that the photobleached section did not regain fluorescence, at least over the time frame of the experiment, indicating that the conjugates do not move around on the surface.
Next, we estimated the effects of BSA addition on the spacing of neoglycoproteins on the surface. BSA has dimensions of approximately 35 Å×35 Å×70 Å. For simplicity, we treated BSA molecules as rectangular boxes with the above dimensions and assumed that the surface contained a monolayer of closely packed BSA molecules. In addition, we assumed that the BSA molecules formed a regular arrangement with the 70 Å side laying parallel to the surface. Based on this model, we would expect each neoglycoprotein to be, on average, surrounded by a shell of unmodified BSA molecules at a ratio of 1:7 (neoglycoprotein:BSA). This would correspond to an average spacing from the center of one neoglycoprotein to the center of the next neoglycoprotein of about 140 Å (see Figure S3, Supporting Information).
To evaluate this model, we carried out a set of resonance energy transfer experiments. AF555-BSA (donor) and QSY7-BSA (quencher) were mixed in equal amounts and then the mixture was combined with varying amounts of unmodified BSA. The mixtures were then printed on the array surface and the fluorescence was imaged and quantified. In this experiment, QSY7-BSA quenches the fluorescence of AF555-BSA in a distance-dependent manner. Quenching was clearly evident in the absence of added BSA, and the relative amount of quenching decreased as the proportion of BSA increased (see Table S1, Supporting Information), showing that addition of BSA spaced the donors and acceptors farther apart. At a ratio of 1:7, no quenching was observed, indicating that the average distance between acceptor and donor was greater than ~90 Å (based on a Förster critical distance of 45 Å for AF555-QSY7). These results are consistent with our model. As with previous experiments, the fluorescence was uniform over the area of the spot, which is consistent with an even distribution of acceptor and donor conjugates.
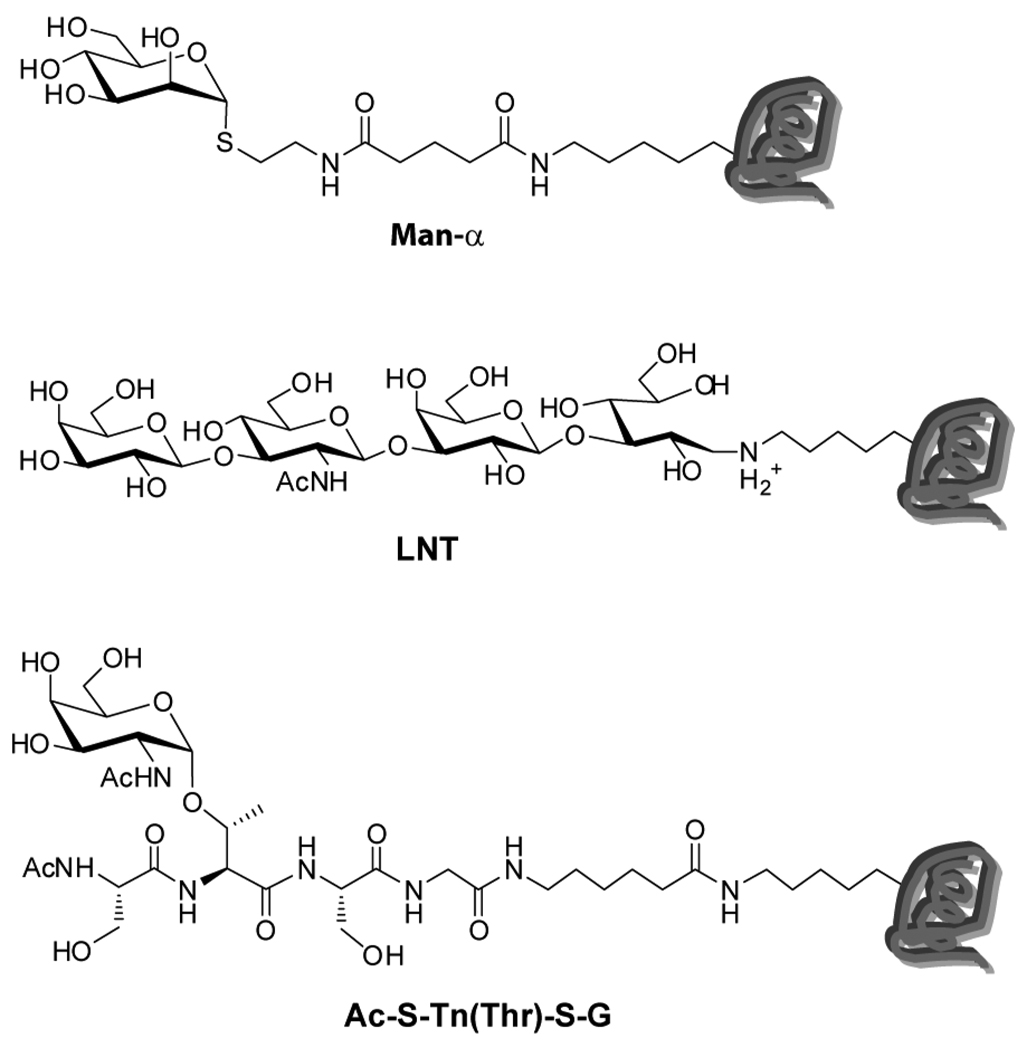
Finally, we were concerned that longer glycans, branched structures, and/or high densities of glycans on BSA might impede or slow immobilization of these neoglycoproteins relative to unmodified BSA. If so, the compositions and spacings of neoglycoproteins on the surface would vary depending on the particular neoglycoprotein, which could complicate analyses and comparisons. For example, long and/or branched glycans on certain neoglycoprotein might slow the rate of immobilization to the surface. If so, unmodified BSA might out-compete those neoglycoproteins, resulting in much lower neoglycoprotein densities than expected on the surface. To test this, we evaluated three glycans at both high and low density on BSA for a total of 6 neoglycoproteins. We included 2 monosaccharide-BSA conjugates (Man-α-4-BSA and Man-α-20-BSA), 2 linear trisaccharides (LNT-4-BSA and LNT-20-BSA; LNT = Galβ1-3GlcNAcβ1-3Galβ), and 2 branched glycopeptides [S-TnThr-S-4-BSA and S-TnThr-S-20-BSA] (see Figure 2). Each was labeled with a fluorophore, AF555, and printed on the microarray slide with varying proportions of BSA (see Figure S6 and Table S2, Supporting Information). The slides were washed and scanned to determine the amount of fluorescence at each spot. The fluorescence intensities of samples were normalized relative to the corresponding spots with 100% neoglycoprotein to get a relative percentage of fluorescence. As shown in Table S3 in the supporting information, each of the samples at a given BSA ratio gave about the same fluorescence relative to the 100% sample, indicating that size, branching, and the density do not significantly affect the immobilization efficiency for these six neoglycoproteins. Although evaluation of all the other array components was not feasible, the results from this subset suggest that the addition of unmodified BSA should have similar effects on the surface composition for most neoglycoproteins in our collection. As with previous experiments, the fluorescence was uniform over the area of the spots, which is consistent with an even distribution of conjugates.
Figure 2.
Neoglycoproteins Used in the Model Studies
Construction of the full array and validation with plant lectins
The next step was to incorporate variations in neoglycoprotein density with our existing glycan array. Arrays were fabricated using our previously published protocols (see also Supporting Information).22, 23 Based on preliminary studies, we chose to construct our first generation array with 4 ratios of added BSA per array component: 1:0 (100%; no BSA), 1:1 (50%), 1:3 (25%), and 1:7 (12.5%). We included 147 different neoglycoproteins and glycoproteins, along with several controls including BSA alone (for a full list of array components, see Table S4 in the Supporting Information), giving a total of 591 combinations of structure and neoglycoprotein/glycoprotein density. Each combination was printed in duplicate within a subarray, and 8 subarrays were printed on each slide, allowing us to carry out 8 independent array experiments in parallel on a slide.
Our next objective was to evaluate binding of several plant lectins to validate the array design strategy. As a first test case, we examined binding of concanavalin A (ConA). ConA is a mannose/glucose binding plant lectin that has been studied extensively using many methods and techniques. ConA was evaluated on the array to determine the effects of neoglycoprotein density on binding. It is important to note that the measured signal intensity for each array component at a given lectin concentration can vary as a function of the amount of ligand and that changes in neoglycoprotein density produce differences in the total amount of ligand on each spot. Therefore, we evaluated binding at a range of concentrations and determined the apparent Kd values for various glycans at each of the 4 BSA ratios (see Table 1 and Table S5 for a full listing of apparent Kd values for ConA), since affinity constants are not dependent on the amount of ligand. Due to the high-throughput design of the array, a full dilution series for a lectin can easily be completed in a single day.
Table 1.
Selected Binding Data for the Plant Lectins
| Lectin | Ligand1 | Without BSA (1:0) | With BSA (1:7) | Inhibition data | |||
|---|---|---|---|---|---|---|---|
| App Kd (nM) |
p-value2 | App Kd (nM) |
p-value2 | IC50 (nM) | p-value2 | ||
| ConA | Man6 | 49±29 | 108±10 | 38±14 | |||
| Man-α | 69±7 | 0.23 | NB | <0.0005 | >758 | <0.0005 | |
| Glc-α | 113±15 | 0.04 | NB | <0.0005 | >758 | <0.0005 | |
| VVL-B4 | Ac-A-TnThr-S-G-23 | 66±9 | 317±115 | 16±4 | |||
| Ac-S-TnSer-S-G-22 | 132±44 | 0.17 | ND | <0.0005 | 650±39 | 0.002 | |
| Ac-V-Tn(Thr)-S-G-19 | 125±58 | 0.29 | ND | <0.0005 | 368±35 | 0.03 | |
| GA2di-37 | 164±27 | 0.04 | ND | <0.0005 | 172±2 | 0.001 | |
| RCA120 | LacNAc (trimeric) | 8.0±0.3 | 83±18 | 11.5±0.8 | |||
| Lac | 8.1±0.5 | 0.83 | 307±42 | 0.02 | 114±3 | 0.0005 | |
For complete array data and apparent Kd values, see the Supporting Information.
Paired t-test. NB denotes no binding. ND (not determined) denotes some binding but apparent Kd too large to measure. Values have not been adjusted for valency. Complete binding data can be found in the supporting information.
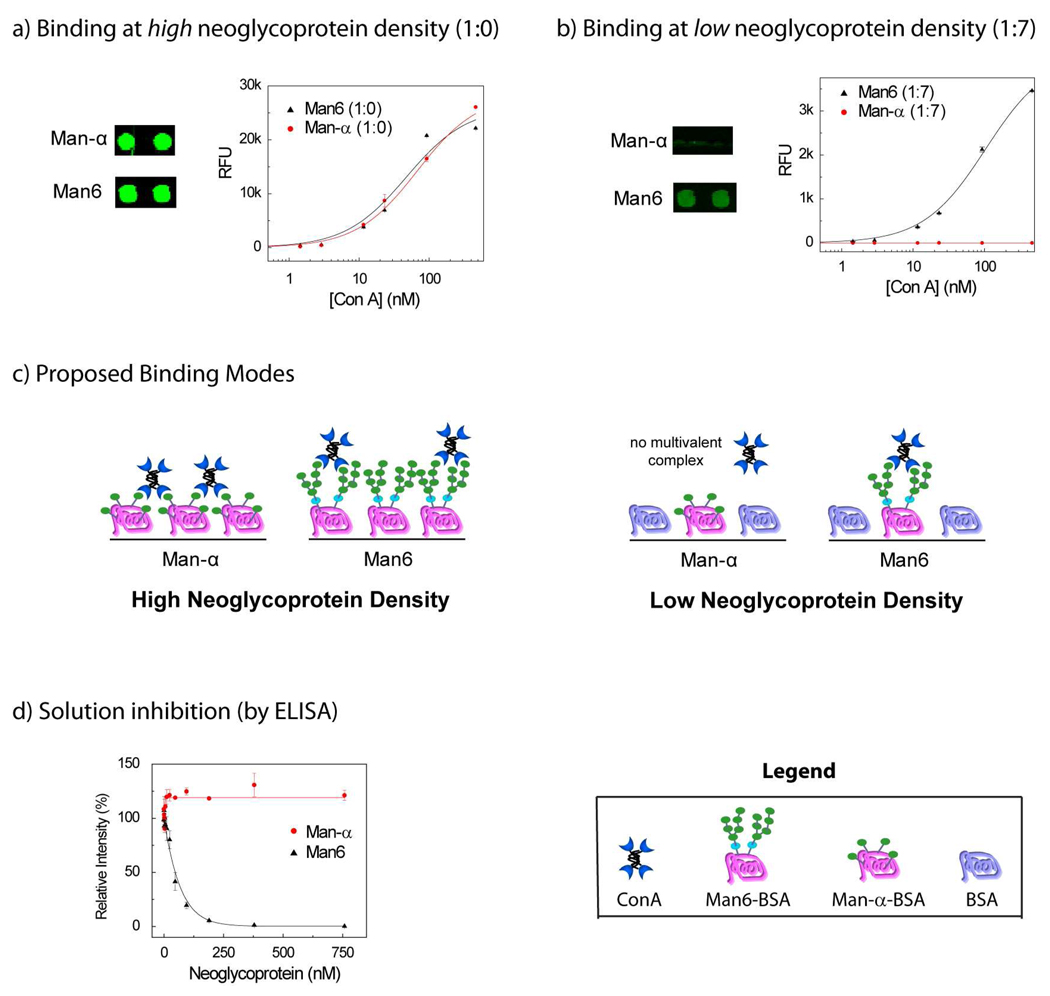
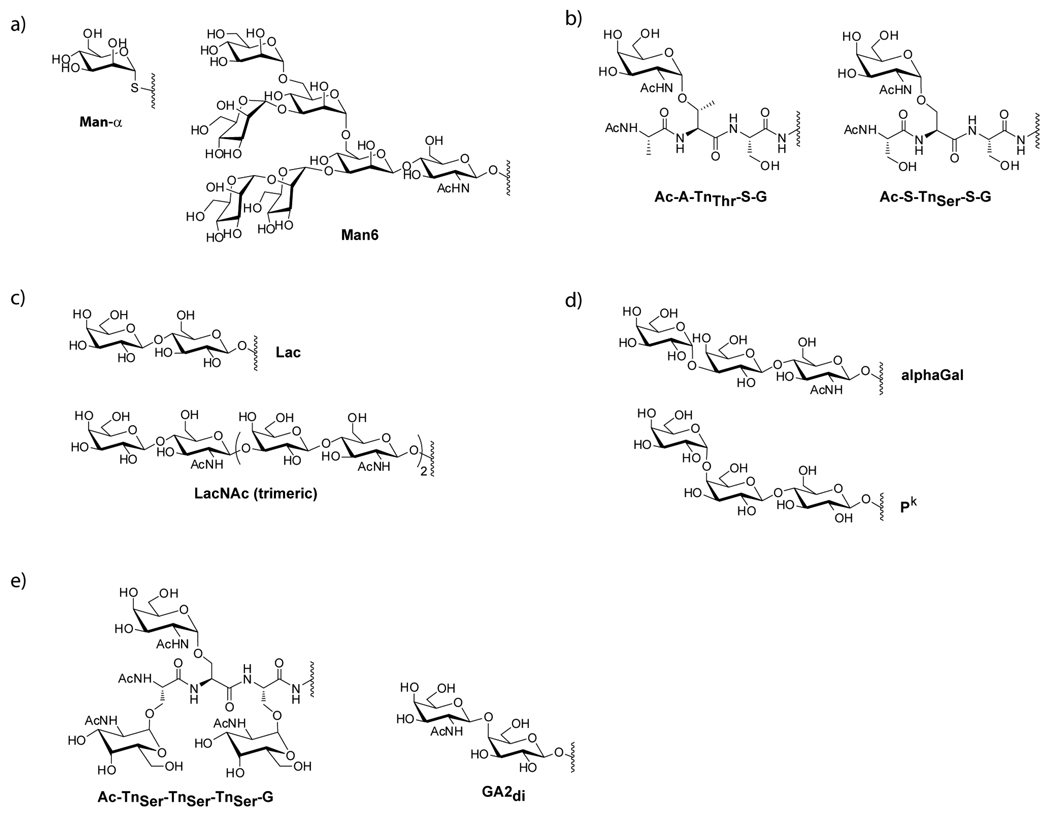
ConA binding was highly dependent on neoglycoprotein density. For the spots with 100% neoglycoprotein, ConA showed similar avidity for a number of mannose-containing neoglycoproteins and for Glc-α-BSA. For example, Man6-BSA and Man-α-BSA had similar apparent Kd values of 49 and 69 nM, respectively (see Figure 3a; see Figure 4a for structures). The apparent Kd for Glc-α-BSA was 113 nM. These values are quite comparable to the apparent Kd values measured on another glycan array37 and affinity constants measured by SPR.33 In contrast, binding to these neoglycoproteins was significantly different at low neoglycoprotein density (ratio of 1:7). In particular, Man6-BSA was bound tightly by ConA even at a ratio of 1:7 (app Kd = 108 nM), whereas Man-α-BSA and Glc-α-BSA were not bound at all by ConA at a ratio of 1:7 (see Figure 3b).
Figure 3.
Comparison of ConA binding to Man6 and Man-α a) ConA binding at high neoglycoprotein density (1:0); shown are pairs of spots for Man-α and Man6 at a single ConA concentration (189 nM) and binding curves over a range of ConA concentrations. b) ConA binding at low neoglycoprotein density (1:7); shown are pairs of spots for Man-α and Man6 at a single ConA concentration (189 nM) and binding curves over a range of ConA concentrations. c) Proposed binding modes at high and low neoglycoprotein density. At high density, ConA binds Man-α via a bridging complex and Man6 via a 1:1 complex. At low neoglycoprotein density, a bridging complex with Man-α cannot be formed. d) Inhibition of ConA binding was evaluated by an ELISA-like assay at a range of neoglycoprotein concentrations and inhibition curves are shown for Man6 and Man-α. Man6 shows good inhibition while Man-α shows no inhibition.
Figure 4.
Chemical Structures of Selected Glycans
Based on these results and the design concept, we inferred that Man6-BSA was forming a 1-to-1 complex with ConA while Man-α-BSA and Glc-α-BSA were forming bridging complexes (see Figure 3c). Therefore, we anticipated that Man6-BSA would be capable of inhibiting ConA in solution while Man-α-BSA and Glc-α-BSA would not. To test this, an ELISA-like assay was used. Neoglycoproteins were incubated with biotinylated ConA at varying concentrations and the mixtures were then added to 96-well plates pre-coated with Man-α-BSA. Binding of ConA was detected by incubating the wells with streptavidin-alkaline phosphatase (SA-AP) followed by a fluorogenic AP substrate (see Figure 3d). The IC50 values, the concentrations of neoglycoprotein required to produce 50% inhibition, were then determined. Man6-BSA was found to have an IC50 of 38±14 nM, while Man-α-BSA and Glc-α-BSA failed to show any significant inhibition at a concentration as high as 758 nM. Therefore, only Man6-BSA acted as a solution-phase inhibitor. The results highlight one of the key advantages of this method. Using the conventional immobilization format (i.e. 100% neoglycoprotein; no added BSA), Man6-BSA, Man-α-BSA, and Glc-α-BSA appeared to have roughly equivalent binding to ConA, both in terms of their apparent Kd values and in terms of the raw fluorescence signals at each lectin concentration (see Figure 3a). By varying neoglycoprotein density, however, major differences could be detected, and these differences enabled us to predict which neoglycoprotein would have activity as a solution-phase inhibitor and which ones would be poor inhibitors.
Next, we evaluated binding to the plant lectin, Vicia villosa lectin B4 (VVL-B4). This lectin is a GalNAc binding protein that has been used for many years to detect expression of the Tn antigen in tumors samples.40–46 Analogous to our studies on ConA, binding was evaluated at a range of concentrations using the array and apparent Kd values were determined. A number of glycans/glycopeptides were bound by VVL-B4, and full binding profiles are provided in the supporting information. Like ConA, neoglycoprotein density had a major impact on binding for VVL-B4. Using the conventional format (i.e. no added BSA), a number of glycopeptides had similar binding signals and apparent Kd values. For example, the apparent Kd values for Ac-A-TnThr-S-G-23-BSA, Ac-S-TnSer-S-G-22-BSA, Ac-V-TnThr-S-G-19-BSA, and GA2di-37-BSA ranged from 66–164 nM (see Table 1). At a ratio of 1:7, however, Ac-A-TnThr-S-G-23-BSA was clearly the best binder (apparent Kd of 317 nM). Ac-S-TnSer-S-G-22-BSA, Ac-V-TnThr-S-G-19-BSA, and GA2di-37-BSA retained some binding at 1:7, but the apparent Kd values were too high to measure on the array (> 3 µM). These results suggested that Ac-S-TnSer-S-G-22-BSA, Ac-V-TnThr-S-G-19-BSA, and GA2di-37-BSA may have some activity as solution inhibitors, but they would be much worse than Ac-A-TnThr-S-G-23-BSA. To test this, an ELISA-like solution inhibition assay was used as before. As expected, Ac-A-TnThr-S-G-23-BSA was a good inhibitor (IC50 value of 16±4 nM; see Table 1) while the other three were found to be 11–40 fold worse. Again, variations in neoglycoprotein density on the array surface revealed differences binding that enabled us to predict which of these neoglycoproteins would be the best inhibitor in solution.
As a third test case, we evaluated binding of Ricinus communis agglutinin (RCA120), a galactose-binding plant lectin.47, 48 RCA120 is one of two toxic lectins isolated from the seeds of the castor bean. The more potent of the two is ricin, a potential biological warfare agent. RCA120 is about 20 times less toxic and is often used as a model of ricin, since the carbohydrate-binding properties are similar. RCA120 was evaluated at a range of concentrations and apparent Kd values were determined. Only a few neoglycoproteins were bound by the lectin, and two clearly stood out as the best binders: Lac-BSA and LacNAc (trimeric)-BSA (see Figure 4c for structures). In the absence of added BSA, these two neoglycoproteins both had apparent Kd values of 8 nM (see Table 1). At a ratio of 1:7, binding was weaker for both conjugates and modest differences in avidities could be detected. In particular, LacNAc(trimeric)-BSA had about a 4 fold better apparent Kd (83 vs 307 nM).
One explanation for the observed binding is that RCA120 can form high avidity, bridging complexes with both neoglycoconjugates at high neoglycoprotein density. At low neoglycoprotein density, RCA120 can still form multivalent complexes with both ligands, but the resulting 1:1 complexes are 10–40 fold weaker than the corresponding bridging complexes formed at high neoglycoprotein density. In addition, RCA120 displays a difference in selectivity when binding in a 1:1 format, with LacNAc(trimeric)-BSA being bound more tightly. Based on this hypothesis, both neoglycoproteins should function as solution inhibitors, but LacNAc(trimeric)-BSA should be better. To test this, an ELISA-like inhibition assay was used. Both neoglycoproteins were found to inhibit binding; however, LacNAc (trimeric)-BSA was about 10 fold better [IC50 for LacNAc (trimeric)-BSA = 11.5±0.8 nM; IC50 for Lac-BSA = 114±3 nM]. The inhibition constants were consistent with our array results, although the absolute difference between the neoglycoproteins at 1:7 was not as large as the difference measured in solution (4 fold vs 10 fold, respectively). By evaluating binding at multiple neoglycoprotein densities, we were able to detect differences in binding that could not have been predicted based on binding at high neoglycoprotein density alone.
Rapid identification of multivalent inhibitors of PA-IL
Based on the success with the plant lectins, we next applied the new array to the identification of inhibitors of a lectin involved in bacterial infections. P. aeruginosa is an opportunistic bacteria causing numerous nosocomial diseases, such as septicaemia, urinary tract infections, pancreatitis, and dermatitis. Pseudomonas aeruginosa lectin I (PA-IL) is a soluble carbohydrate-binding protein from the bacteria P. aeruginosa.49–51 It is a galactose-binding, C-type lectin consisting of 121 amino acids (12.75 kDa), and the crystal structure reveals that it is a tetramer with a rectangular shape.52 PA-IL is a virulence determinant and Pseudomonas aeruginosa lethality is dependent on its expression.51, 53 PA-IL also contributes to biofilm formation, as demonstrated by studies of P. aeruginosa variants lacking or overexpressing the lectin.54 Therefore, PA-IL has become an important molecular target for P. aeruginosa infections and several inhibitors have been reported in the literature.50, 55, 56 PA-IL is known to bind glycans terminating in a Gal-α residue, such as Galα1-4Galβ1-4Glc (Pk) and Galα1-3Galβ1-4GlcNAc (alphaGal).49, 50, 52, 54 Previous studies have also shown that multivalent conjugates with GalNAcβ1-4Galβ (the terminal disaccharide of GA2; GA2di) can inhibit Pseudomonas aeruginosa binding.57 The specificity of PA-IL has also been evaluated previously on several glycan arrays.25, 58, 59
Our objective was to use the array to rapidly identify high avidity, solution-phase inhibitors of PA-IL. His-tagged PA-IL25 was evaluated at a range of concentrations on the array and the apparent Kd values at various neoglycoprotein densities were determined. This lectin bound a number of glycans with terminal alpha-linked Gal or GalNAc residue and the best ligands at low neoglycoprotein density are listed in Table 2 (complete data can be found in the supporting information). Based on the array binding at low neoglycoprotein density, several neoglycoproteins were predicted to be good solution phase inhibitors, including alphaGal, P1, Pk, BG-B, and Bdi. Two of these, alphaGal and Pk, were selected for additional studies to confirm inhibitory activity. Solution inhibition was measured using an ELISA-like assay as before. Both conjugates were excellent inhibitors: alphaGal-BSA had an IC50 of 30±3 nM and Pk-HSA had an IC50 of 64±12 nM. For comparison, galactose showed 32% inhibition at a concentration of 400 mM under the same conditions.
Table 2.
Binding data for Pseudomonas aeruginosa lectin I (PA-IL)
| Ligand | Apparent Kd (nM) | Solution Inhibition | ||
|---|---|---|---|---|
| at 1:7 | p-value1 | IC50 (nM) | p-value2 | |
| Pk | 200±51 | - | 64±12 | - |
| Adi-17 | 336±99 | 0.23 | ||
| alphaGal | 292±87 | 0.33 | 30±3 | 0.06 |
| Bdi | 282±56 | 0.27 | ||
| BG-B (EMD) | 291±91 | 0.34 | ||
| GA2di-37 | ND | <0.0005 | >758 | <0.0005 |
| Galα1-4Galβ | 319±64 | 0.18 | ||
| G2M4 | 138±35 | 0.29 | ||
| LacNAc | 253±24 | 0.32 | ||
| P1 | 128±30 | 0.23 | ||
p values refer to a comparison between the Kd and the IC50 value for Pk and each of the other neoglycoproteins (paired t test). Values have not been adjusted for valency. ND (not determined) denotes some binding but apparent Kd too large to measure. Complete binding data can be found in the supporting information.
PA-IL had poor binding to GalNAcβ1-4Galβ-containing neoglycoproteins on our array, indicating that they should not be good inhibitors of PA-IL. Nevertheless, previous published studies had shown that other multivalent GalNAcβ1-4Galβ conjugates do bind PA-IL. Therefore, we also tested the corresponding neoglycoprotein, GA2di-37-BSA, in the solution inhibition assay. As expected from the array data, it did not show any significant inhibition in solution at a concentration of 758 nM. This result highlights one of the major difficulties in designing multivalent probes. Although a specific carbohydrate may be an excellent ligand when presented on a particular scaffold, it may be a poor ligand when presented on a different multivalent scaffold. Therefore, the ligand and scaffold must be optimized simultaneously. Our array format provides a high-throughput means to identify appropriately matched ligands and scaffolds.
Rapid identification of multivalent inhibitors of mMGL-2
Macrophage galactose-type lectins are C-type lectins expressed as homo-oligomers on myeloid antigen presenting cells (APC), such as macrophages and dendritic cells (DCs).60–62 Mice express two related lectins, mMGL-1 and mMGL-2, while humans express a single MGL. mMGL-1 and mMGL-2 have distinct carbohydrate-binding specificities, and mMGL-2 has been found to be more similar to human MGL.60, 63, 64 The biological roles of mMGL-2 are still being elucidated, but it is thought to be involved in immune recognition of pathogens, internalization of antigens, and regulation of effector T cells.62 Human MGL has also been implicated in viral infection and immune evasion by tumor cells.62 For these reasons, inhibitors with high affinity and selectivity would be useful tools for studying the biological roles of this protein and evaluating its potential as a receptor for antigen targeting for vaccines.
mMGL-2 has been shown previously to bind several tumor-associated carbohydrate antigens, including the Tn antigen (GalNAc-α-Ser/Thr) and the Thomsen-Friedenreich disaccharide (TF, Galβ1-3-GalNAc-α-Ser/Thr).60, 63, 64 In the studies by Irimura and colleagues, mMGL-2 was found to preferentially bind GalNAc-terminal oligosaccharides. The best solution phase inhibitor for mMGL-2 was found to be Gb4, with a Kd value of 19.2 µM.64 No solution phase multivalent inhibitors were reported, but binding to a multivalent surface containing a terminal GalNAc-β provided a Kd value of 2.1 µM. In a separate report, Irimura found that multivalent polyacrylamide polymers containing GalNAc-β provided the best binding to surface immobilized mMGL-2, but solution phase inhibition was not tested.60 In a study by van Kooyk and colleagues, mMGL-2 was found to have broader specificity, with binding to both GalNAc- and Gal-terminal structures. Multivalent polyacrylamide polymers displaying GalNAc-α, GalNAc-β, or core 1 were found to bind to cells expressing mMGL-2, but affinities were not reported63 As a qualitative measure of binding, about 35% of mMGL-2 positive cells were bound by the multivalent polymers in a flow cytometry assay when using a concentration of about 0.5 µM polymer. While these results are promising, it is unlikely that a monosaccharide-based ligand would display sufficient selectivity for most biological applications. Therefore, more potent and selective multivalent inhibitors for mMGL-2 would be useful.
Our objective was to use the array to identify high avidity, solution-phase inhibitors of mMGL-2. As with our previous experiments, mMGL-2 was assessed at a range of concentrations and the apparent Kd values were determined at various neoglycoprotein densities. The earlier reports by van Kooyk and Irimura showed some discrepancies in the binding specificity of mMGL-2. Similar to the van Kooyk study, we observed binding to glycans with both terminal GalNAc and Gal. In general, however, the GalNAc terminal glycans bound better, and the preference for GalNAc over Gal was more prominent at lower neoglycoprotein densities, suggesting that they would be better soluble inhibitors. The best ligands at low neoglycoprotein density are listed in Table 3 (complete data can be found in Table S9, Supporting Information). Solution inhibition was evaluated in an ELISA-based assay for two of the best ligands to confirm the predicted activity. Based on the inhibition curves, GA2di-37-BSA had an IC50 of 15±6 nM and Ac-TnSer-TnSer-TnSer-G-27-BSA had an IC50 of 28±12 nM. Thus, within a single day, we were able to identify potent neoglycoprotein inhibitors for mMGL-2.
Table 3.
Binding data for mouse macrophage galactose-type lectin-2 (mMGL-2)
| Ligand | Apparent Kd (nM) | Solution Inhibition | ||
|---|---|---|---|---|
| at 1:7 | p-value1 | IC50 (nM) | p-value1 | |
| GA2di-37 | 3.6±0.8 | - | 15±6 | - |
| Ac-A-TnThr-S-G - 23 | 4.9±2.0 | 0.48 | ||
| Ac-S-TnThr-A-G - 22 | 4.1±1.4 | 0.70 | ||
| Ac-S-TnThr-G-G - 19 | 4.6±1.9 | 0.56 | ||
| Ac-S-TnThr-S-G - 24 | 3.8±1.5 | 0.88 | ||
| Ac-TnThr-G - 21 | 5.1±1.5 | 0.34 | ||
| Ac-TnSer-TnSer-TnSer-G -27 | 4.4±2.4 | 0.70 | 28±12 | 0.30 |
| Ac-V-TnThr-S-G - 19 | 5.4±1.9 | 0.34 | ||
| GA2di-16 | 5.1±1.7 | 0.38 | ||
| GalNAc-α-22 | 5.0±1.2 | 0.30 | 89±53 | 0.19 |
p values refer to a comparison between the Kd and the IC50 of GA2di-37 and the other neoglycoproteins (paired t test). Values have not been adjusted for valency. Complete binding data can be found in the supporting information.
Conclusion
Multivalent interactions play a critical role in a wide variety of biological processes and are especially important for recognition of carbohydrates. To achieve a multivalent complex, the spacing and orientation of ligands must be properly matched to the binding sites of the receptor. Therefore, both ligand structure and ligand presentation are critical features for binding in natural systems and for the design of inhibitors or probes to modulate natural recognition events. Identification of the optimal combination of ligand structure and multivalent presentation can be challenging, especially in the absence of structural information on the receptor.
The array format described in this study provides a high-throughput tool for rapidly evaluating relationships between ligand structure, presentation, and recognition. In particular, it allows one to discriminate between different types of multivalent complexes on the array surface. To illustrate the utility of this array format, it was used to identify multivalent neoglycoprotein inhibitors/probes of several lectins. The construction of the array is simple and economical, and the screening process can be completed in a single day. In addition, no structural information on the receptor is required to identify a multivalent probe. The array format will be useful for other applications as well. For example, many proteins, especially glycan-binding proteins, have overlapping specificities. The array format described in this study provides one approach to distinguish proteins with similar ligand preferences but distinct architectures. In principle, it could also be used to distinguish different forms or oligomeric states of the same protein. Therefore, variation of neoglycoprotein density provides a more comprehensive view of carbohydrate recognition and is a useful element of diversity for inclusion on glycan arrays. While this study focused on interactions between carbohydrates and proteins, the approach could also be used to discover multivalent inhibitors for interactions between other biomolecules.
Supplementary Material
Acknowledgment
We thank Professor Lara Mahal for the generous gift of PA-IL. We thank the Optical Microscopy and Analysis Laboratory, SAIC-Frederick, Inc., Advanced Technology Program for use of the confocal microscope. This research was supported by the Intramural Research Program of the NIH, NCI.
Footnotes
Supporting Information Available. Confocal microscope images of AF488-BSA on the array surface at 1:0. 1:1, 1:3, and 1:7, characterization of the AF555-BSA conjugates, full array data and fluorescence intensities for ConA, VVL-B4, RCA120, PA-IL, and mMGL-2, and a full list of array components are available free of charge via the internet at http://pubs.acs.org.
References
- 1.Lis H, Sharon N. Chem. Rev. 1998;98:637. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 2.Sharon N, Lis H. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 3.Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 4.Roy R. Curr. Opin. Struct. Biol. 1996;6:692–702. doi: 10.1016/s0959-440x(96)80037-6. [DOI] [PubMed] [Google Scholar]
- 5.Ernst B, Magnani JL. Nat. Rev. Drug Discovery. 2009;8:661–677. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayaraman N. Chem. Soc. Rev. 2009;38:3463–3483. doi: 10.1039/b815961k. [DOI] [PubMed] [Google Scholar]
- 7.Kaila N, Thomas BE. Exp. Opin. Therap. Pat. 2003;13:305–317. [Google Scholar]
- 8.Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Exp. Opin. Therap. Targets. 2007;11:1473–1491. doi: 10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simanek EE, McGarvey GJ, Jablonowski JA, Wong CH. Chem. Rev. 1998;98:833–862. doi: 10.1021/cr940226i. [DOI] [PubMed] [Google Scholar]
- 10.Lee RT, Lee YC. Glycoconj. J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 11.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. Engl. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J. Am. Chem. Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 13.Song E-H, Pohl NLB. Curr. Opin. Chem. Biol. 2009;13:626–632. doi: 10.1016/j.cbpa.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Oyelaran O, Gildersleeve JC. Curr. Opin. Chem. Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Palma AS, Feizi T. Biol. Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- 16.Liang P-H, Wu C-Y, Greenberg WA, Wong C-H. Curr. Opin. Chem. Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horlacher T, Seeberger PH. Chem. Soc. Rev. 2008;37:1414. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- 18.Oyelaran O, Gildersleeve JC. Expert Rev. Vaccines. 2007;6:957–969. doi: 10.1586/14760584.6.6.957. [DOI] [PubMed] [Google Scholar]
- 19.Paulson JC, Blixt O, Collins BE. Nat. Chem. Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 20.Oyelaran O, Gildersleeve JC. Proteomics- Clin. Appl. 2010;4:285–294. doi: 10.1002/prca.200900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Anver MR, Li Z, Butcher DO, Gildersleeve JC. Int. J. Cancer. 2010;126:459–468. doi: 10.1002/ijc.24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyelaran OO, Li Q, Farnsworth DF, Gildersleeve JC. J. Proteome Res. 2009;8:3529–3538. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. J. Proteome Res. 2009;8:4301–4310. doi: 10.1021/pr900515y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Anver MR, Butcher DO, Gildersleeve JC. Mol. Cancer. Ther. 2009;8:971–979. doi: 10.1158/1535-7163.MCT-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu KL, Gildersleeve JC, Mahal LK. Mol. BioSyst. 2008;4:654–662. doi: 10.1039/b800725j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gildersleeve JC, Oyelaran O, Simpson JT, Allred B. Bioconjugate Chem. 2008;19:1485–1490. doi: 10.1021/bc800153t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manimala JC, Roach TA, Li Z, Gildersleeve JC. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 28.Manimala JC, Roach TA, Li ZT, Gildersleeve JC. Angew. Chem. Int. Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 29.Manimala J, Li Z, Jain A, VedBrat S, Gildersleeve JC. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 30.Stowell CP, Lee VC. Adv. Carbohydr. Chem. Biochem. 1980;37:225–281. doi: 10.1016/s0065-2318(08)60022-0. [DOI] [PubMed] [Google Scholar]
- 31.Gordus A, MacBeath G. J. Am. Chem. Soc. 2006;128:13668–13669. doi: 10.1021/ja065381g. [DOI] [PubMed] [Google Scholar]
- 32.An alternative method to space neoglycoproteins apart on the array surface is to print sub-saturating amounts of neoglycoprotein; however, we have found previously that when the array surface is not saturated, uneven distributions of neoglycoproteins can be produced.
- 33.Smith EA, Thomas WD, Kiessling LL, Corn RM. J. Am. Chem. Soc. 2003;125:6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 34.Houseman BT, Mrksich M. Chem. Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 35.Ngundi MM, Taitt CR, McMurry SA, Kahne D, Ligler FS. Biosens. Bioelectron. 2006;21:1195–1201. doi: 10.1016/j.bios.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevolot Y, Bouillon C, Vidal S, Morvan F, Meyer A, Cloarec JP, Jochum A, Praly JP, Vasseur JJ, Souteyrand E. Angew. Chem. Int. Ed. 2007;46:2398–2402. doi: 10.1002/anie.200604955. [DOI] [PubMed] [Google Scholar]
- 37.Liang PH, Wang SK, Wong CH. J. Am. Chem. Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 38.Mercey E, Sadir R, Maillart E, Roget A, Baleux F, Lortat-Jacob H, Livache T. Anal. Chem. 2008;80:3476. doi: 10.1021/ac800226k. [DOI] [PubMed] [Google Scholar]
- 39.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Glycoconj. J. 2008;25:15. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 40.Tollefsen S, Kornfeld R. J. Biol. Chem. 1983;258:5172–5176. [PubMed] [Google Scholar]
- 41.Puri KD, Gopalakrishnan B, Surolia A. FEBS Lett. 1992;312:208–212. doi: 10.1016/0014-5793(92)80937-c. [DOI] [PubMed] [Google Scholar]
- 42.Medeiros A, Bianchi S, Calvete JJ, Balter H, Bay S, Robles A, Cantacuzene D, Nimtz M, Alzari PM, Osinaga E. Eur. J. Biochem. 2000;267:1434–1440. doi: 10.1046/j.1432-1327.2000.01141.x. [DOI] [PubMed] [Google Scholar]
- 43.Osinaga E, Bay S, Tello D, Babino A, Pritsch O, Assemat K, Cantacuzene D, Nakada H, Alzari P. FEBS Lett. 2000;469:24–28. doi: 10.1016/s0014-5793(00)01248-5. [DOI] [PubMed] [Google Scholar]
- 44.Babino A, Tello D, Rojas A, Bay S, Osinaga E, Alzari PM. FEBS Lett. 2003;536:106–110. doi: 10.1016/s0014-5793(03)00037-1. [DOI] [PubMed] [Google Scholar]
- 45.Wu AM. FEBS Lett. 2004;562:51–58. doi: 10.1016/S0014-5793(04)00180-2. [DOI] [PubMed] [Google Scholar]
- 46.Kato K, Takeuchi H, Ohki T, Waki M, Usami K, Hassan H, Clausen H, Irimura T. Biochem. Biophys. Res. Comm. 2008;371:698–701. doi: 10.1016/j.bbrc.2008.04.120. [DOI] [PubMed] [Google Scholar]
- 47.Itakura Y, Nakamura-Tsuruta S, Kominami J, Sharon N, Kasai KI, Hirabayashi J. J. Biochem. (Tokyo) 2007;142:459–469. doi: 10.1093/jb/mvm153. [DOI] [PubMed] [Google Scholar]
- 48.Podder SK, Surolia A, Bachhawat BK. Eur. J. Biochem. 1974;44:151–160. doi: 10.1111/j.1432-1033.1974.tb03468.x. [DOI] [PubMed] [Google Scholar]
- 49.Imberty A, Wimmerová M, Mitchell EP, Gilboa-Garber N. Microbes Infect. 2004;6:221–228. doi: 10.1016/j.micinf.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Kirkeby S, Hansen AK, d'Apice A, Moe D. Microb. Pathogenesis. 2006;40:191–197. doi: 10.1016/j.micpath.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Chemani C, Imberty A, De Bentzmann S, Pierre M, Wimmerova M, Guery BP, Faure K. Infect. Immun. 2009;77:2065–2075. doi: 10.1128/IAI.01204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cioci G, Mitchell EP, Gautier C, Wimmerova M, Sudakevitz D, Perez S, Gilboa-Garber N, Imberty A. FEBS Lett. 2003;555:297–301. doi: 10.1016/s0014-5793(03)01249-3. [DOI] [PubMed] [Google Scholar]
- 53.Laughlin RS, Musch MW, Hollbrook CJ, Rocha FM, Chang EB, Alverdy JC. Ann. Surgery. 2000;232:133–142. doi: 10.1097/00000658-200007000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. Environ. Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- 55.Cioci G, Mitchell EP, Gautier C, Wimmerová M, Sudakevitz D, Pérez S, Gilboa-Garber N, Imberty A. FEBS Lett. 2003;555:297–301. doi: 10.1016/s0014-5793(03)01249-3. [DOI] [PubMed] [Google Scholar]
- 56.Hauber HP, Schulz M, Pforte A, Mack D, Zabel P, Schumacher U. Int. J. Med. Sci. 2008;5:371–376. doi: 10.7150/ijms.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Autar R, Khan AS, Schad M, Hacker J, Liskamp RMJ, Pieters RJ. ChemBioChem. 2003;4:1317–1325. doi: 10.1002/cbic.200300719. [DOI] [PubMed] [Google Scholar]
- 58.Moni L, Pourceau G, Zhang J, Meyer A, Vidal S, Souteyrand E, Dondoni A, Morvan F, Chevolot Y, Vasseur JJ, Marra A. ChemBioChem. 2009;10:1369–1378. doi: 10.1002/cbic.200900024. [DOI] [PubMed] [Google Scholar]
- 59.ConA, VVL-B4, RCA120, PA-IL, and mMGL-2 have been screened by the Consortium for Functional Glycomics and binding data can be found at http://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp.
- 60.Tsuiji M, Fujimori M, Ohashi Y, Higashi N, Onami TM, Hedrick SM, Irimura T. J. Biol. Chem. 2002;277:28892–28901. doi: 10.1074/jbc.M203774200. [DOI] [PubMed] [Google Scholar]
- 61.Singh SK, Streng-Ouwehand I, Litjens M, Weelij DR, GarcÃa-Vallejo JJ, van Vliet SJ, Saeland E, van Kooyk Y. Mol. Immunol. 2009;46:1240–1249. doi: 10.1016/j.molimm.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 62.van Vliet SJ, Saeland E, van Kooyk Y. Trends Immunol. 2008;29:83–90. doi: 10.1016/j.it.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Singh SK, Streng-Ouwehand I, Litjens M, Weelij DR, Garcia-Vallejo JJ, van Vliet SJ, Saeland E, van Kooyk Y. Mol. Immunol. 2009;46:1240–1249. doi: 10.1016/j.molimm.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 64.Oo-puthinan S, Maenuma K, Sakakura M, Denda-Nagai K, Tsuiji M, Shimada I, Nakamura-Tsuruta S, Hirabayashi J, Bovin NV, Irimura T. Biochim Biophys Acta. 2008;1780:89–100. doi: 10.1016/j.bbagen.2007.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.