Summary
The abundant nuclear enzyme PARP-1, a multifunctional regulator of chromatin structure, transcription, and genomic integrity, plays key roles in a wide variety of processes in the nucleus. Recent studies have begun to connect the molecular functions of PARP-1 to specific physiological and pathological outcomes, many of which can be altered by an expanding array of chemical inhibitors of PARP enzymatic activity.
Keywords: cancer, chromatin, DNA repair, inflammation, inhibitor, macrodomain, PARP, PARP-binding zinc finger, poly(ADP-ribose), poly(ADP-ribose) polymerase, transcription
Introduction
Nuclear processes involving access to or modification of the genome, such as transcription and DNA repair, require a host of structural and regulatory proteins. Poly(ADP-ribose) polymerase-1 (PARP-1), a ubiquitous and abundant nuclear protein and the founding member of the PARP family, has a number of distinct biochemical activities that make it well suited for both structural and regulatory roles across the genome (Hassa and Hottiger, 2008; Kim et al., 2005; Schreiber et al., 2006). As discussed below, PARP-1 can bind to various DNA structures and nucleosomes, and it possesses an NAD+-dependent catalytic activity that synthesizes a negatively charged polymer on target proteins called poly(ADP-ribose) or PAR. Although historically studied in the context of DNA damage detection and repair, PARP-1 has more recently been linked to the regulation of chromatin structure and transcription, DNA methylation and imprinting, insulator activity, and chromosome organization. In this review, we provide an overview of PARP-1's structure and activities, as well as an in depth review of papers published in the past few years that have provided new insights into the molecular functions of PARP-1 in the nucleus. In addition, we highlight emerging information about the roles of PARP-1 in physiological and pathological outcomes, its interplay with nuclear NAD+ metabolic enzymes, and the chemical biology of PAR.
PARP-1 and the PARP family
Poly(ADP-ribosyl)ation reactions and PARP-like genes have been identified in a wide variety of single and multicellular eukaryotes, from fungi to mammals (but surprisingly not the yeasts S. cerevisae and S. pombe), as well as eubacteria, archaebacteria, and double stranded DNA viruses (Hassa et al., 2006; Otto et al., 2005). In mammalian cells, the bulk of the PAR production is catalyzed by PARP-1, although recent studies have begun to characterize the structure and function of related PARP proteins.
PARP-1 structure and biochemical activities
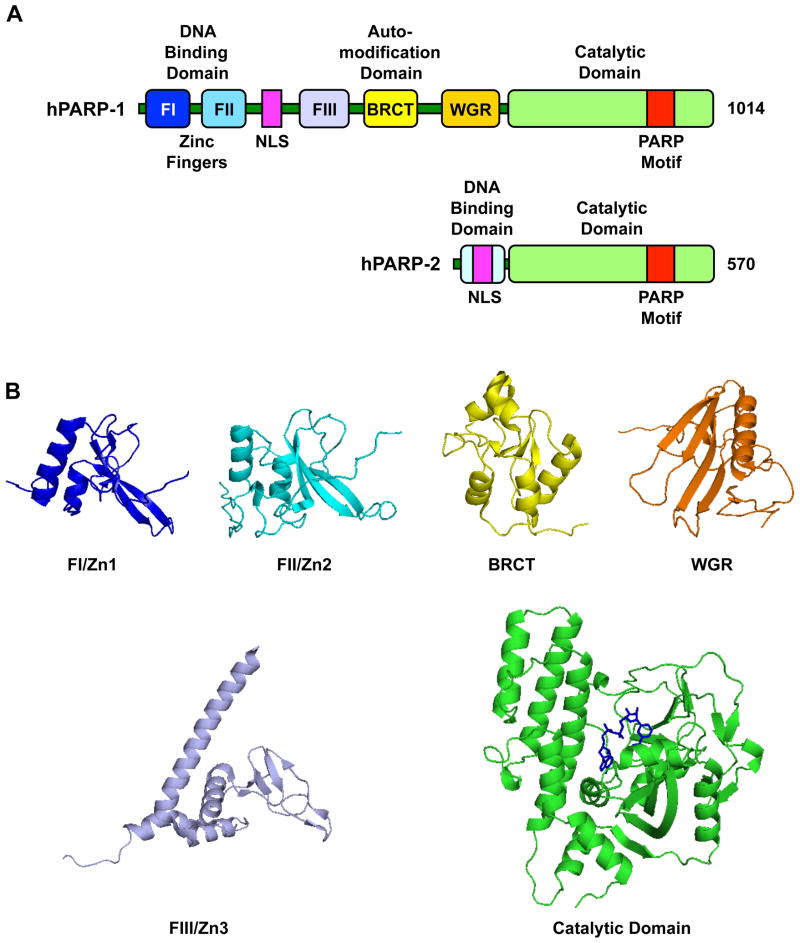
PARP-1 is a highly conserved protein of ~116 kDal (D'Amours et al., 1999). Like many other chromatin- and transcription- related proteins, it has a modular structure comprising multiple independently folded domains. The major functional units of PARP-1 are an amino terminal DNA binding domain (DBD), a central automodification domain (AMD), and a carboxyl-terminal catalytic domain (CD) (Hakme et al., 2008; Schreiber et al., 2006) (Fig. 1A, top). The DBD contains two Cys-Cys-His-Cys zinc fingers (FI/Zn1 and FII/Zn2) that mediate binding to DNA, a newly discovered third zinc binding domain (FIII/Zn3) that mediates inter-domain contacts important for DNA-dependent enzyme activation (Langelier et al., 2010; Langelier et al., 2008), a nuclear localization signal (NLS), and a caspase-3 cleavage site (Hakme et al., 2008; Schreiber et al., 2006). The AMD contains a BRCT (BRCA1 C-terminus) fold, which mediates protein-protein interactions (e.g., with DNA repair enzymes). The CD, which is the most conserved domain across the PARP family, contains a PARP signature motif, which binds NAD+, as well as a "WGR" motif, which is named after the most conserved amino acid sequence in the motif (Trp, Gly, Arg) and has an unknown function. The structures of these domains and motifs are shown in Fig. 1B. Together, the structural and functional domains of PARP-1 confer the activities required for the broad range of functions of PARP-1 in the nucleus.
Figure 1. Structural and functional organization of PARP-1 and PARP-2.
(A) Schematic representation of human PARP-1 and PARP-2 with the functional domains noted in the text.
(B) Structures of the six structural and functional domains in human PARP-1. FI (PDB 2DMJ), FII (PDB 2CS2), FIII (PDB 2RIQ), BRCT (PDB 2COK), WGR (PDB: 2CR9), catalytic domain (PDB 1A26; NAD+ has been modeled in based on a structure of diptheria toxin, PDB 1TOX).
Other PARP family members
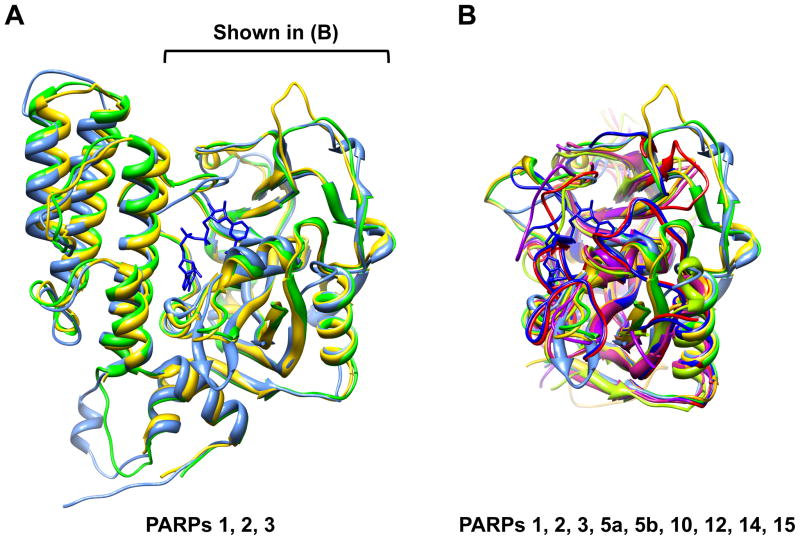
Although much of the focus has been on PARP-1, studies over the past decade have identified a family of as many as 17 proteins that share homology to the catalytic domain of PARP-1 (Ame et al., 2004; Hakme et al., 2008; Hassa and Hottiger, 2008; Schreiber et al., 2006). In addition to the PARP-like domain, the PARP family members are “functionalized” with a wide variety of other structural and functional domains (e.g., DNA-binding domains, RNA-binding domains, subcellular localization signals, macrodomains, BRCT motifs, ankyrin repeats, zinc fingers) that determine their overall biological activities. Recently, a unified nomenclature referring to this family of proteins as ADP-ribosyl transferases (ARTs) has been proposed to recognize that fact that: (1) PARPs catalyze a transferase reaction, not a template-dependent polymerization reaction and (2) not all family members have PARP activity; some are likely to function as mono(ADP-ribosyl) transferases (mARTs) (Hottiger et al., 2010). This new nomenclature is reflected in a recent structure-based classification of PARP family members into three groups based on their catalytic domains: (1) PARPs 1-5, which are bona fide PARPs containing a conserved glutamate (Glu 988 in PARP-1) that defines the PARP catalytic activity, (2) PARPs 6-8, 10-12, and 14-16, which are confirmed or putative mARTs, and (3) PARPs 9 and 13, which lack key NAD+-binding residues and the catalytic glutamate, and are likely inactive (Kleine et al., 2008) (Fig. 2). The standardization of assays for PARP activity, such as the identification of the PRAMP product by HPLC (Alvarez-Gonzalez and Jacobson, 1987), will help to provide direct evidence for the production of PAR by PARP family members.
Figure 2. The PARP catalytic domain is highly conserved across the PARP family.
(A) Alignment of the catalytic domain structures from mammalian PARP-1 (PDB 1A26; green), PARP-2 (PDB 1GSO; yellow), and PARP-3 (PDB 3FHB; blue).
(B) Alignment of the catalytic domain structures from mammalian PARPs 1, 2, 3, 5a, 5b, 10, 12, 14, 15 (PDB 1A26, 1GSO, 3FHB, 2RF5, 3KR7, 3HKV, 2PQF, 3GOY, 3GEY, respectively). In (A) and (B), NAD+ has been modeled in based on a structure of diptheria toxin (PDB 1TOX).
PARP family members localize to various cellular compartments, including the nucleus, cytoplasm, mitochondria, and vault particles, although the subcellular localization and function of many of the PARPs are unknown (Ame et al., 2004; Hassa and Hottiger, 2008). The primary nuclear PARPs are PARP-1, PARP-2 (the closest paralog to PARP-1), PARP-3, and tankyrases 1 and 2 (PARP-5a and -5b) (Ame et al., 2004; Hakme et al., 2008; Hassa and Hottiger, 2008; Schreiber et al., 2006). Also found in the nucleus, although not exclusively, are v-PARP (PARP-4), PARP-6, PARP-8, PARP-9, the Bal proteins Bal 1-3 (PARP-13, -14, -15), and PARP-10. The known functions of the PARP family members span a wide range of cellular processes, including DNA repair, transcription, cellular signaling, cell cycle regulation, and mitosis (Ame et al., 2004; Chang et al., 2004; Hakme et al., 2008; Hassa and Hottiger, 2008; Schreiber et al., 2006). This diverse array of processes plays key roles in a wide variety of biological outcomes, including differentiation, development, stress responses, inflammation, and cancer. Although the focus of this review is on PARP-1, we draw parallels to other PARP family members when applicable.
Molecular Biology and Biochemistry of PARP-1
DNA binding, chromatin binding, and genomic localization
Studies over the past few decades have shown that PARP-1 associates with chromatin in specific patterns that relate to its function (Kraus, 2008; Kraus and Lis, 2003; Tulin et al., 2003). This association is driven by interactions with DNA, nucleosomes, or other chromatin-associated proteins, which are not mutually exclusive. PARP-1 binds to a variety of DNA structures, including single- and double-strand breaks, crossovers, cruciforms, and supercoils, as well as some specific double-stranded DNA sequences (Kraus, 2008; Kraus and Lis, 2003). PARP-1 also binds to nucleosomes in a specific manner, interacting with both DNA and histones at or near the dyad axis where the DNA enters and exits the nucleosome (Kim et al., 2004). Finally, PARP-1 can interact with a wide variety of chromatin-associated proteins, including components of the transcription machinery, sequence-specific DNA-binding transcription factors, chromatin modifying enzymes, and histone variants (e.g., macroH2A; see below) (Kim et al., 2005; Kraus, 2008; Kraus and Lis, 2003; Tulin et al., 2003). Interactions with these proteins allows for indirect association of PARP-1 with chromatin. By binding to chromatin, PARP-1 can alter the structure of nucleosomes, as well as the composition or compaction state of chromatin (Kim et al., 2004; Kraus, 2008; Kraus and Lis, 2003; Langelier et al., 2010; Tulin et al., 2003; Wacker et al., 2007). This may occur through target protein modification by PARP-1’s enzymatic activity, as well as competition for binding sites on nucleosomes. For example, PARP-1 may displace the linker histone H1 from nucleosomes by PARylating it or by competing for overlapping binding sites on the nucleosomes (Ju et al., 2006; Kim et al., 2004; Krishnakumar et al., 2008).
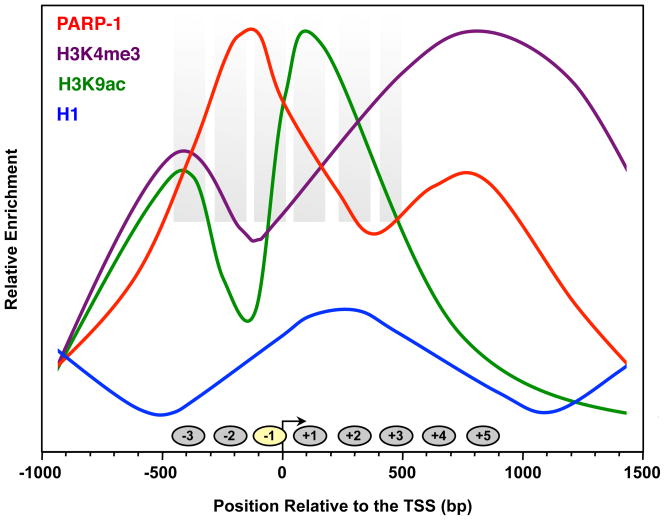
A recent genomic localization study has shown that PARP-1 binds at the promoters of most actively transcribed genes (Krishnakumar et al., 2008). The binding of PARP-1 at promoters correlates with the binding of Pol II, gene expression, and the presence of histone H3 lysine 4 trimethylation (H3K4me3), a histone modification that marks active promoters (Fig. 3). PARP-1 also binds to chromatin outside of promoter regions, including enhancers (Krishnakumar et al., 2008). In response to genotoxic stress, PARP-1 relocalizes to sites of DNA damage (i.e., nicks, breaks) (Haince et al., 2008; Mortusewicz et al., 2007). Whether this DNA damage-induced relocalization results in a global redistribution of PARP-1 away from promoters, as was shown recently for the NAD+-dependent chromatin regulator SIRT1 (Oberdoerffer et al., 2008), remains to be determined. This is an attractive model that fits well with the global reduction in transcription observed in response to DNA damage.
Figure 3. The chromatin landscape at the promoters of highly expressed genes.
Schematic of average genomic ChIP and nucleosome mapping data across the promoters of the most highly expressed genes (top quartile) in cells. The graphs are based on data from the literature: PARP- 1 and H1 (Krishnakumar et al., 2008), H3K4me3 (Barski et al., 2007), H3K9ac (Wang et al., 2008b), and nucleosome positioning (Schones et al., 2008).
Catalytic activity, binding partners, and targets
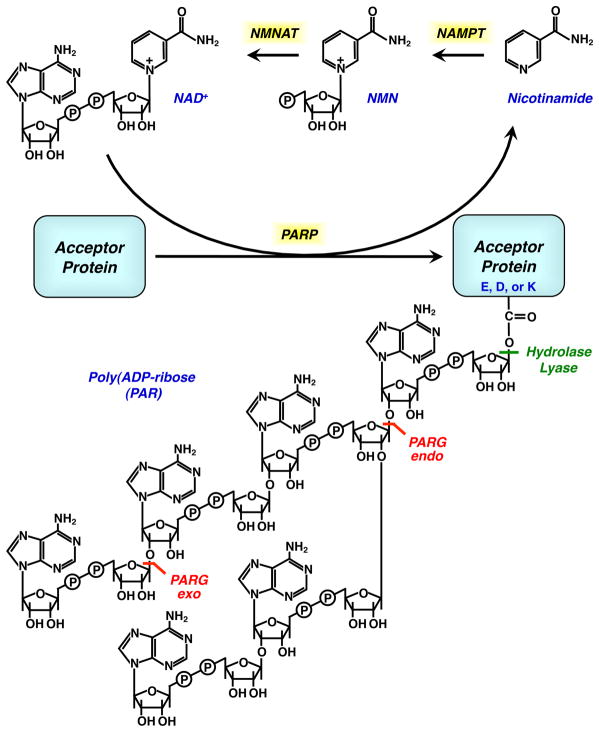
PAR is a large, negatively charged polymer that functions as a post-translational modification, as well as a free polymer. Most of the PAR in the cell is produced by the catalytic activity of PARP-1, which catalyzes the polymerization of ADP-ribose units from donor NAD+ molecules on target proteins (D'Amours et al., 1999) (Fig. 4). The ADP-ribose units are linked to each other via glycosidic ribose-ribose bonds, and the resulting PAR polymers may be linear or branched (D'Amours et al., 1999). The modification most likely occurs on glutamate, aspartate, or lysine residues, although historically the evidence for covalent modification of specific residues has been weak (D'Amours et al., 1999; Hassa and Hottiger, 2008). In fact, some have even argued for strong non-covalent binding of free PAR polymers, rather than covalent modification (Hassa and Hottiger, 2008). Recent studies, however, have begun to make progress on defining specific sites of PAR attachment on target proteins (Altmeyer et al., 2009; Haenni et al., 2008; Kanai et al., 2007) (see below).
Figure 4. Biosynthesis of NAD+ and PAR.
Chemical structures of NAD+, PAR, and metabolites. The enzymes that catalyze the synthesis of NAD+ in the mammalian salvage pathway are shown. The enzymatic actions of PARP, PARG, (ADP-ribosyl) protein hydrolase, and (ADP-ribosyl) protein lyase are also indicated.
PARP-1 catalytic activity is regulated through allosteric mechanisms involving a range of binding partners, including damaged DNA, histones, nucleosomes, and an assortment of nuclear proteins (D'Amours et al., 1999; Kraus and Lis, 2003; Tulin et al., 2003). PARP-1 catalytic activity is also regulated by post-translational modifications; autoPARylation of PARP-1 inhibits its catalytic activity, while phosphorylation by Erk1/2 enhances its catalytic activity (Kauppinen et al., 2006). PARP-1 catalytic activity may also be regulated by nicotinamide mononucleotide adenylyltransferase-1 (NMNAT-1), a nuclear NAD+ synthase that interacts with PARP-1 and can produce NAD+ locally for use by nuclear enzymes that require NAD+, such as PARP-1 and SIRT1 (Kim et al., 2004; Zhang et al., 2009; Zhang and Kraus, 2009). Regulated catalysis, such as that exhibited by PARP-1, may be a more common mode of action for chromatin-modifying enzymes than has generally been considered, and there are likely to be some general principles that can be learned from the study of PARP-1’s catalytic activity.
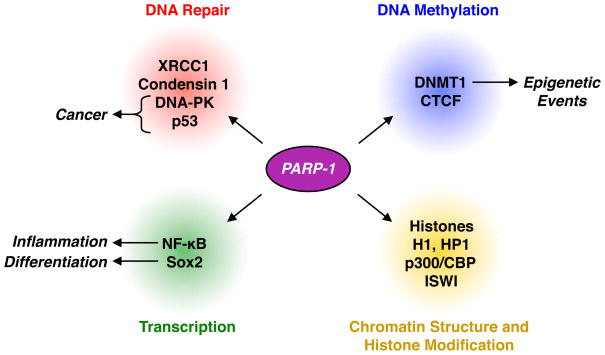
PARP-1, which has many protein binding partners in the nucleus, has been identified as a component of a wide variety of protein complexes, including those that (1) repair DNA damage (e.g., condensin I/XRCC1), (2) regulate transcription (e.g., Mediator; TLE corepressor), (3) function as insulators (e.g., CTCF), and (4) methylate DNA (e.g., DNMT-1) (Fig. 5) (Caiafa et al., 2009; Caiafa and Zlatanova, 2009; El-Khamisy et al., 2003; Farrar et al., 2010; Guastafierro et al., 2008; Hassa et al., 2005; Heale et al., 2006; Ju et al., 2004; Malanga and Althaus, 2005; Pavri et al., 2005; Pleschke et al., 2000; Zampieri et al., 2009). Many of these binding partners have been reported to be PARylated as targets of PARP-1 catalytic activity (Kim et al., 2005; Kraus, 2008; Kraus and Lis, 2003). Covalent attachment of PAR is thought to alter the activity of target proteins through both steric and charge effects, ultimately preventing protein-protein interactions, protein-nucleic acid interactions, enzymatic activity, or subcellular localization (Hassa and Hottiger, 2008; Schreiber et al., 2006).
Figure 5. Interactions and functions of PARP-1 in the nucleus.
PARP-1 interacts with and PARylates proteins involved in DNA repair, transcription, DNA methylation, and the regulation of chromatin structure and histone modification to control physiological and pathological outcomes.
Known or suspected targets of PARP-1 catalytic activity include histones, transcription factors, nuclear enzymes, and nuclear structural proteins. For example, PARP-1 can PARylate histones, especially H1, H2A and H2B, which may play a role in the regulation of chromatin structure, although the extent of histone modification and its relevance to nuclear processes remains to be clarified (D'Amours et al., 1999; Kim et al., 2005; Kraus, 2008; Kraus and Lis, 2003). PARP-1 also PARylates a number of DNA repair proteins, including p53 (Kanai et al., 2007; Mendoza-Alvarez and Alvarez-Gonzalez, 2001), which is not surprising given PARP-1’s well characterized role in DNA repair. Although the functional significance of p53 PARylation has been elusive, a recent study suggests that PARylation of p53 on specific sites (likely Glu 255, Asp 256 and Glu 268) can prevent p53 export from the nucleus by blocking its interaction with the nuclear export receptor Crm1 (Kanai et al., 2007). PARP-1 has also been reported to PARylate and alter the function of numerous other transcription factors, including CTCF, AP-1, YY1 and NF-κB (Kraus, 2008), as well as nuclear enzymes, such as aurora B kinase (Monaco et al., 2005), thereby inhibiting their function. As these examples suggest, the PARylation of target proteins by PARP-1 plays a central role in determining the cellular functions of PARP-1.
Post-translational modifications of PARP-1
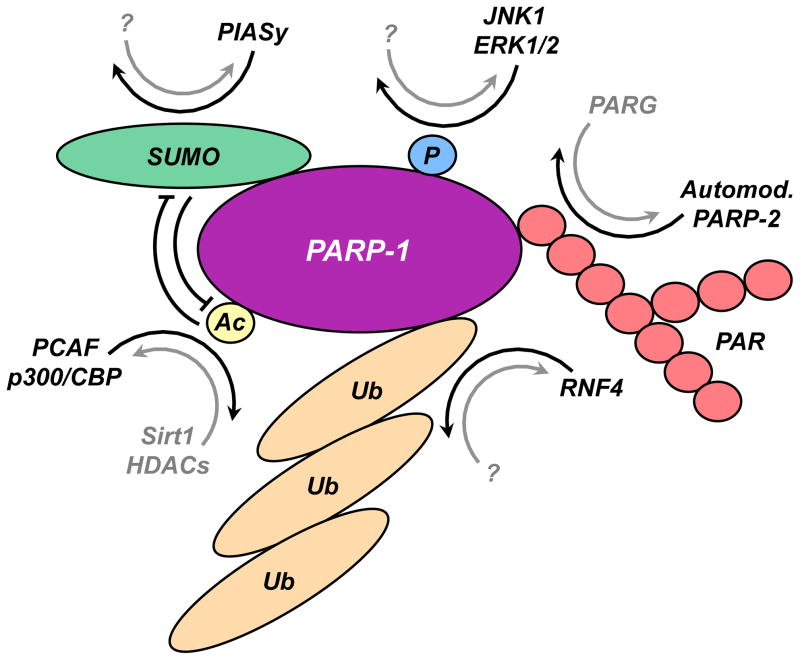
Like other nuclear proteins that play key roles in regulatory processes, PARP-1 is subject to a variety of covalent post-translational modifications as endpoints of cellular signaling pathways. These include PARylation, acetylation, phosphorylation, ubiquitylation, and SUMOylation (Fig. 6); the latter two were more recently discovered and are less well characterized (Cohen-Armon et al., 2007; Hassa et al., 2005; Kauppinen et al., 2006; Martin et al., 2009; Messner et al., 2009; Wang et al., 2008a).
Figure 6. Covalent post-translational modifications of PARP-1.
Schematic representation of PARP-1 modifications: PARylation, phosphorylation, acetylation, SUMOylation, ubiquitylation, as described in the text.
PARylation
PARP-1 is PARylated by itself, PARP-2, and possibly other PARPs. Automodification of PARP-1 (i.e., autoPARylation) may occur as an extensive addition of ADP-ribose in chains >200 units in length or as a more modest addition of a single unit or chains up to 20 units in length (i.e., mono- or oligoPARylation, respectively) (D'Amours et al., 1999; Mendoza-Alvarez and Alvarez-Gonzalez, 1999). Whether this occurs primarily in cis or in trans (i.e., intra- or inter-molecularly, respectively) has been debated in the literature, but is typically considered intermolecular (Altmeyer et al., 2009; Alvarez-Gonzalez and Mendoza-Alvarez, 1995; Mendoza-Alvarez and Alvarez-Gonzalez, 1993, 1999). Extensive autoPARylation of PARP-1 (e.g., in response to DNA damage) inhibits its DNA binding and catalytic activities (D'Amours et al., 1999). Biochemical and cell-based assays have shown that activation and autoPARylation of PARP-1 results in its release from chromatin (Kim et al., 2004; Petesch and Lis, 2008; Tulin and Spradling, 2003; Wacker et al., 2007). The effect of less extensive autoPARylation of PARP-1 is not clear; modestly modified PARP-1 may have altered activities, but retain its association with chromatin.
Initial reports suggested that PARylation of PARP-1 occurred on as many as 28 glutamate residues, primarily in the AMD and DBD (D'Amours et al., 1999; Schreiber et al., 2006). In contrast, a recent study has shown that the glutamate residues in the AMD are not required for PARylation of PARP-1 (Altmeyer et al., 2009). Rather, based on amino acid substitutions (i.e., Lys to Arg), the authors conclude that at least three lysines residues in the AMD (Lys 498, 521, and 524) are sites of automodification on PARP-1 (Altmeyer et al., 2009). A similar approach was used to identify lysines 36 and 37 of PARP-2 as sites of auto-mono(ADP-ribosyl)ation (Haenni et al., 2008). Although these results could significantly change the expectations of the field both in terms of PARP-1 autoregulation, as well as sites of modification on other PARP target proteins, they should be interpreted with caution. Mutation of specific residues in PARP-1 or PARP-2 could reduce automodification without necessarily being sites for covalent attachment of PAR. Furthermore, PARylation seems to be promiscuous; deletion or mutation of one site may allow for modification of another site. The identification of ADP-ribose adducts on targets residues by mass spectrometry will be required to conclusively address this issue.
Phosphorylation and acetylation
PARP-1 is phosphorylated by ERK1/2 at Ser 372 and Thr 373, and JNK1 at undetermined sites (Kauppinen et al., 2006; Zhang et al., 2007). The former is required for maximal PARP-1 activation after DNA damage (Kauppinen et al., 2006), whereas the latter promotes sustained PARP-1 activation during H2O2-induced non-apoptotic cell death (Zhang et al., 2007). A recent proteomic analysis has identified additional phosphorylation sites in PARP-1, as well as sites in PARG, that will be good candidates for further functional analyses (Gagne et al., 2009).
PARP-1 is acetylated by the acetyltransferases p300/CBP and PCAF (Hassa et al., 2003; Hassa et al., 2005; Rajamohan et al., 2009). The acetylation of PARP-1 is reversed by a number of deacetylases, including Sirt1 (Hassa et al., 2005; Rajamohan et al., 2009). Acetylation of PARP-1 was first identified in the context of NF-κB-dependent transcription, where it plays a critical role in regulating NF-κB target genes in immune cells (Hassa et al., 2003; Hassa et al., 2005). In cardiomyocytes, PARP-1 is acetylated as an endpoint of stress responses, resulting in the DNA damage-independent activation of PARP-1 (Rajamohan et al., 2009). PARP-2 is also acetylated at Lys 36 and 37 in the NLS, which are the same sites that are auto-mono(ADP-ribosyl)ated (Haenni et al., 2008). Acetylation of PARP-2 reduces its DNA binding and enzymatic activities, and presumably the extent of auto-mono(ADP-ribosyl)ation (Haenni et al., 2008).
SUMOylation and ubiquitylation
Recent studies have shown that PARP-1 is SUMOylated and ubiquitylated, modulating its role as a regulator of chromatin structure and transcription (Martin et al., 2009; Messner et al., 2009). PARP-1 interacts with and is SUMOylated by PIASy, a SUMO E3 ligase (Martin et al., 2009; Stilmann et al., 2009). In Drosophila, dPARP (a homolog of mammalian PARP-1) is SUMOylated in response to heat shock, which is required for the full activation of the Hsp70 gene (Martin et al., 2009). PIASy is recruited and released at the Hsp70 locus during the heat shock response with kinetics that mirror those of both PARP-1 and the SUMO-conjugating enzyme Ubc9 (Martin et al., 2009). Interestingly, the SUMO-targeted ubiquitin ligase RNF4 polyubiquitylates dPARP and presumably causes its clearance from the Hsp70 promoter via degradation (Martin et al., 2009). These results fit well with the fact that dPARP regulates the chromatin structure at the Drosophila Hsp70 locus upon heat shock (Petesch and Lis, 2008; Tulin and Spradling, 2003). In mammalian cells, SUMOylation and p300/CBP-dependent acetylation at Lys 486 of PARP-1 are mutually exclusive (Messner et al., 2009). Since acetylation of PARP-1 is required for activated transcription at some target promoters (Hassa et al., 2003; Hassa et al., 2005), SUMOylation of PARP-1 might modulate the transcriptional outcome in this PARP-1-dependent pathway. Similar to what is observed in Drosophila, polyubiquitylation of PARP-1, likely in the DBD, promotes the degradation of PARP-1, thereby regulating its overall activity (Wang et al., 2008a).
Nuclear Actions of PARP-1
PARP-1 contributes in many unique ways to the molecular biology of nuclear processes, playing key roles in the maintenance of genomic integrity, the regulation of chromatin structure and transcription, and the establishment of DNA methylation patterns, as well as a host of other processes (e.g., mitotic apparatus function, cell death pathways) (Fig. 5) (Hassa and Hottiger, 2008; Kim et al., 2005). Below, we highlight the newest results related to some of the key aspects of PARP-1 function.
DNA repair and maintenance of genomic integrity
The earliest functions ascribed to PARP-1 were related to DNA repair and the maintenance of genomic integrity, and much of the PARP-1 literature has been devoted to this aspect of PARP-1 biology (D'Amours et al., 1999). PARP-1 has been implicated in at least three distinct DNA repair pathways: base excision repair (BER), single-strand break (SSB) repair, and double-strand break (DSB) repair (Bouchard et al., 2003; Woodhouse and Dianov, 2008). PARP-2 has also been implicated in DNA repair pathways, including BER (Schreiber et al., 2002; Yelamos et al., 2008). Although neither PARP-1 nor PARP-2 is individually required for viability in mice, Parp-1−/ − or Parp-2−/ − mice or embryonic fibroblasts exhibit a variety of DNA repair defects and chromosomal abnormalities (de Murcia et al., 1997; Menissier de Murcia et al., 2003; Wang et al., 1997). Parp-1−/−/Parp-2−/− mice show embryonic lethality with considerable genomic instability (Menissier de Murcia et al., 2003). PARP-1 and PARP-2 act in concert to detect disrupted replication forks, recruit the repair protein Mre11, stimulate recombination repair, and restart replication (Bryant et al., 2009). PARP-1 and a number of other PARP family members (i.e., PARP-1, PARP-3, v-PARP, and tankyrases 1 and 2) are associated with and control the function of the mitotic apparatus, including centromeres, centrosomes, and the mitotic spindle (Kim et al., 2005). In this regard, tankyrase-1 (PARP-5a) is required for spindle structure and function in a PAR-dependent manner, and tankyrase-1 deficiency results in abnormal chromosome distributions and spindle morphology (Chang et al., 2005). Collectively, the available data indicate critical overlapping, as well as non-redundant functions, for PARP-1, PARP-2, and other PARPs in the maintenance of chromosome stability and genomic integrity.
As with other cellular stresses, DNA damage (e.g., SSBs, DSBs, oxidation, alkylation) elicits an immediate and dramatic PARP-1-dependent PARylation response targeting a variety of nuclear proteins. This response may be transient or sustained depending on the extent of damage and the pathway activated (Bouchard et al., 2003; D'Amours et al., 1999; Woodhouse and Dianov, 2008). In response to low levels of DNA damage, PARP-1 promotes cell survival and DNA repair. With severe DNA damage, PARP-1 promotes cell death through at least two distinct pathways: (1) energy failure-induced necrosis, which results from depletion of NAD+ (and ultimately ATP) and (2) apoptosis-inducing factor-dependent apoptosis (Bouchard et al., 2003; Kim et al., 2005). Thus, PARP-1 has a vital role in determining cellular outcomes in response to DNA damage.
As might be expected, PARP-1 interacts physically and functionally with other key DNA damage detection and response proteins, including the ATM kinase and p53 (Bouchard et al., 2003). For example, PARP-1 deficient cells exhibit impaired ATM kinase activity and reduced formation H2AXγ foci (Aguilar-Quesada et al., 2007; Menisser-de Murcia et al., 2001). PARP-1-dependent PARylation of the Spt6 component of the histone chaperone FACT inhibits the exchange of variant H2AX with conventional H2A in the context of the nucleosome (Heo et al., 2008). PARP-1 also interacts with proteins involved in DNA repair pathways and may play a role in recruiting these proteins to sites of DNA damage (Woodhouse and Dianov, 2008). These include XRCC-1 in the BER pathway, which requires PARP-1 for its recruitment to sites of DNA damage (El-Khamisy et al., 2003; Heale et al., 2006; Masson et al., 1998; Masson and Caldwell, 1998; Okano et al., 2003). Some BER proteins (e.g., XRCC1, DNA ligase III) may also bind PAR (Pleschke et al., 2000), although the functional consequences of this binding are not clear. PARP-2 has also been shown to interact with XRCC1, as well as DNA polymerase β and DNA ligase III (Schreiber et al., 2002), which suggests contributions of PARP-2 to the BER process. Although an initial set of PARP-1 and PARP-2 interactions with genome maintenance factors has been determined, this list is unlikely to be complete. In addition, although these interactions are suggestive of possible mechanisms, the detailed mechanisms that might underlie the contributions of PARP-1 and PARP-2 to DNA damage detection and repair have not yet been revealed.
Chromatin structure and transcription
Although historically the focus has been on PARP-1’s role in DNA damage detection and repair, studies over the past decade have revealed important roles for PARP-1 in transcriptional regulation (Kim et al., 2005; Kraus, 2008; Kraus and Lis, 2003; Tulin et al., 2003). The ability of PARP-1 to modulate chromatin structure and function underlies its contributions to this process. In fact, the ability to disrupt chromatin structure by PARylating histones (e.g., H1 and H2B) and destabilizing nucleosomes was one of the earliest functional effects of PARP-1 to be characterized (Huletsky et al., 1989; Mathis and Althaus, 1987; Poirier et al., 1982). More recent biochemical studies have shown that, in the absence of NAD+ or significant autoPARylation, PARP-1 binds to nucleosomes and promotes the compaction of chromatin by bringing together neighboring nucleosomes (Kim et al., 2004; Wacker et al., 2007). With saturating levels of NAD+, which lead to considerable autoPARylation of PARP-1 in the presence of nucleosomes, the compaction is nearly completely reversed (Kim et al., 2004; Wacker et al., 2007).
PARP-1 localizes to the promoters of almost all actively transcribed genes (Krishnakumar et al., 2008), which suggests that it plays a role in promoting the formation of chromatin structures that are permissive to transcription. In this regard, PARP-1 has been shown to block the binding of the linker histone H1, a repressive chromatin architectural protein, to promoter chromatin (Ju et al., 2006; Kim et al., 2004; Krishnakumar et al., 2008). PARP-1 also PARylates DEK, another repressive chromatin-associated protein, and promotes its release from chromatin (Gamble and Fisher, 2007; Kappes et al., 2008). Yet, PARP-1 only regulates a subset of the genes to which it binds and it has both positive and negative effects of transcription (Frizzell et al., 2009; Krishnakumar et al., 2008). Thus, gene regulation by PARP-1 is a complex process that is likely to involve multiple mechanisms and be modulated by additional inputs.
PARP-1 regulates transcription in multiple ways, including (1) regulating chromatin structure and composition (as discussed in the preceding paragraph), (2) functioning as a classical coregulator with a wide variety of signal-regulated, sequence-specific DNA binding transcriptional activators, (3) functioning as a direct enhancer-binding factor, and (4) regulating the actions of insulators and insulator-binding factors, such as CTCF. These aspects of PARP-1 function have been reviewed extensively elsewhere (Kim et al., 2005; Kraus, 2008; Kraus and Lis, 2003; Tulin et al., 2003). In the ‘coregulator’ mode, PARP-1 may be recruited to target promoters as a functional endpoint of signaling pathways to regulate components of the transcription complex assembled at the promoter. In some cases, the enzymatic activity of PARP-1 is required (e.g., with HES1 and Elk1) (Cohen-Armon et al., 2007; Ju et al., 2004), while in others it is not (e.g., NF-κB and RAR) (Hassa and Hottiger, 2002; Kraus and Lis, 2003; Pavri et al., 2005). When acting as a coregulator during signal-regulated transcriptional responses, PARP-1 can function as a promoter-specific ‘exchange factor’ that promotes the release of inhibitory factors and the recruitment of stimulatory factors. In this regard, PARP-1 has been shown to promote the exchange of (1) a TLE1 corepressor complex for a HAT-containing coactivator complex during signal-dependent gene regulation in neuronal cells (Ju et al., 2004) and (2) an inactive cdk8-positive Mediator for an active cdk8-negative Mediator during retinoic acid-regulated activation (Pavri et al., 2005). PARP-1 has also been reported to promote the recruitment of topoisomerase IIβ (TopoIIβ) to hormone-regulated promoters, leading to promoter DNA cleavage, factor exchange, and transcriptional activation (Ju et al., 2006). The DNA cleavage has been proposed to resolve a topological barrier and allow for favorable structural changes at the promoter (Ju et al., 2006), but this model has yet to be proven.
Drosophila has been a useful model organism for studying the role of PARP-1 in the regulation of chromatin structure and transcription because flies only have two genes encoding PARPs: PARP-1-like (dPARP), which is expressed as three isoforms, and tankyrase-like (Hanai et al., 1998; Miwa et al., 1999). In Drosophila larvae, inhibition of PARP activity or disruption of dPARP gene expression blocks PAR accumulation, chromatin decondensation, and transcription at loci containing highly inducible genes, such as those regulated by heat shock or ecdysone) (Tulin et al., 2003). dPARP may also play a role in maintaining the compaction of heterochromatin (Tulin et al., 2002). Interestingly, in the case of the Hsp70 gene in Drosophila S2 cells, dPARP is required for a heat shock-dependent, transcription-independent disruption of nucleosomes across the entire gene, which occurs within 30 seconds of activation - faster than the rate of Pol II transcription (Petesch and Lis, 2008). These results suggest a heat shock-dependent wholesale opening of the entire Hsp70 locus, as supported by results in fly larvae (Tulin et al., 2003). dPARP exhibits ecdysteroid-regulated localization to Cajal bodies and histone locus bodies, where it PARylates resident proteins and maintains organelle integrity (Kotova et al., 2009). dPARP can also PARylate the nucleosome remodeling ATPase, ISWI, leading to its inactivation (Sala et al., 2008). Together, these studies in Drosophila have helped to uncover and clarify the roles of PARP-1 in regulating chromatin structure and transcription.
DNA methylation
Studies over the past decade have begun to link PARP-1-dependent PARylation with DNA methylation, a stable epigenetic mark that can be passed to daughter cells upon cell division and is associated with the repression of gene expression (Attwood et al., 2002; Caiafa and Zampieri, 2005). One of the ways in which PARP-1 affects DNA methylation is by regulating the expression and activity of the DNA methyltransferase Dnmt1 (Caiafa et al., 2009; Caiafa and Zlatanova, 2009). PARP-1 binds to the promoter of the Dnmt1 gene and protects it from DNA methylation-induced silencing in a PAR-dependent manner (Zampieri et al., 2009). In this regard, overexpression of poly(ADP-ribose) glycohydrolase (PARG), an enzyme that degrades PAR (see below), leads to aberrant methylation of a CpG island in the promoter of the Dnmt1 gene in mouse fibroblasts, which in turn inhibits its transcription (Zampieri et al., 2009). The loss of Dnmt1 expression leads to widespread passive hypomethylation of genomic DNA.
In addition, PARP-1 has also been shown to interact with Dnmt1 in a complex that contains PAR (Reale et al., 2005). The non-covalent binding of PAR polymers by Dnmt1 within the complex inhibits Dnmt1 DNA methyltransferase activity, probably through an inhibitory steric mechanism (Reale et al., 2005). Interestingly, the effects of PARP-1 on DNA methylation are modulated by CTCF, which may promote PARP-1 automodification, CTCF PARylation, accumulation of PAR polymers, and ultimately the inhibition of Dnmt1 DNA methyltransferase activity (Guastafierro et al., 2008). Future studies will be required to determine the extent to which PARP-1 plays a role in the dynamic regulation of DNA methylation in different physiological and pathological states.
Functional interplay with Sirt1
Recent studies have begun to elucidate a functional interplay between PARP-1 and the NAD+-dependent protein deacetylase Sirt1. Sirt1 is an important regulator of metabolism, cell differentiation and senescence, stress responses, and cancer through the regulation of chromatin structure and gene expression (Zhang and Kraus, 2009). PARP-1 and Sirt1 have been shown to function antagonistically; chemical activation of SIRT1 leads to reduced PARP-1 activity and knockout of Sirt1 increases PARP-1 activity (Kolthur-Seetharam et al., 2006). PARP-1 and Sirt1 are thought to compete for nuclear NAD+, and a byproduct of the reactions they catalyze, nicotinamide, can inhibit both of their activities (Kim et al., 2005; Zhang and Kraus, 2009). This sets the stage for a tightly regulated interplay between these two proteins.
The interplay between PARP-1 and SIRT1, however, go beyond simple competition for NAD+. As noted above, acetylation of PARP-1 by PCAF is required for stress-induced cell death pathways. Deacetylation of PARP-1 by Sirt1 promotes cell survival (Rajamohan et al., 2009). Knockout of PARP-1 in a Sirt1−/− background increases the late post-natal lethality before weaning that is observed in Sirt1−/− animals, but also rescues abnormal pericentric heterochromatin formation, nucleolar disorganization, and mitotic defects that are observed in Sirt1−/− cells (El Ramy et al., 2009). Unchecked PARP-1 activity in the absence of Sirt1 results in apoptosis inducing factor-mediated cell death. In mammalian cells, Sirt1 inhibits the expression of the PARP-1 gene, adding another layer of complexity to the functional interplay (Rajamohan et al., 2009). In Drosophila, dPARP and dPARG may promote chromatin silencing by regulating the localization and function of dSir2 (the Drosophila homolog of mammalian Sirt1) (Tulin et al., 2006). These results establish a functional link between PARP-1 and Sirt1 that plays key roles in chromatin structure, the maintenance of genomic integrity, and cell viability.
Chemical Biology and Dynamics of PAR
PAR is a negatively charged molecule that resembles, in some respects, single stranded nucleic acid polymers (D'Amours et al., 1999). As described above, it functions as a covalent post-translational modification, as well as a protein-binding matrix. Much of the focus on PAR to date has been on its synthesis and degradation, both of which occur on the time scale of minutes in the cell.
Dynamic synthesis and degradation of PAR
PAR is synthesized rapidly in response to a variety of physiological (e.g., hormone signaling) and stress-related (e.g., heat shock, DNA damage) stimuli (D'Amours et al., 1999; Hakme et al., 2008). As noted above, these stimuli ultimately result in the allosteric activation of PARP-1 catalytic activity, which in turn can lead to the autoPARylation of PARP-1, as well as the transmodification of other protein targets. If extensive, autoPARylation can inhibit PARP-1 enzymatic activity, which can block further PAR synthesis (D'Amours et al., 1999; Hakme et al., 2008). Very rapidly after synthesis (within seconds to minutes), PAR is degraded to ADP-ribose monomers, which may have signaling functions in the nucleus (see below) (Gagne et al., 2006; Min and Wang, 2009). Structurally different types of PAR are degraded at different rates (i.e., short more rapidly than long, linear more rapidly than branched), which may influence their biological functions (Hassa and Hottiger, 2008).
Most PAR in the cell is degraded by the enzyme poly(ADP-ribose) glycohydrolase PARG), an enzyme with both exo and endoglycosidase activities (actually a family of isoforms all encoded by the same gene) (Gagne et al., 2006; Min and Wang, 2009) (Fig. 4). In mice, targeted deletion of the 110 kDal PARG isoforms results in increased lethality in response to genotoxin exposure and septic shock relative to wild-type animals (Cortes et al., 2004). Mice with complete deletion of all PARG isoforms are embryonic lethal. Trophoblast stem cells from these animals are viable only when cultured in the presence of a PARP inhibitor and they exhibit reduced growth, accumulation of PAR, and increased sensitivity genotoxic stress (Koh et al., 2004). In Drosophila, increasing or decreasing dPARG levels phenocopies dPARP mutation, supporting a role for dPARG in removing PAR and, perhaps, facilitating multiple cycles of catalysis by individual PARP molecules (Tulin et al., 2006). The available data highlight the importance of PAR catabolism for embryonic development, the maintenance of normal physiological states, and protection against genotoxic stress (Cortes et al., 2004; Fisher et al., 2007; Koh et al., 2004; St-Laurent et al., 2007).
Recently, the enzyme ADP-ribose-protein-hydrolase-3 (ARH3) was also shown to possesses intrinsic PARG activity (Oka et al., 2006), suggesting that the mammalian genome may encode additional proteins with PARG activities. Other enzymatic activities, such as poly and mono (ADP-ribosyl) protein hydrolase, as well as mono(ADP-ribosyl) protein lyase, may also act to remove PAR polymers and ADP-ribose monomers from target proteins (Hassa and Hottiger, 2008). Although the dynamic nature of PAR synthesis and degradation has been elucidated, the function of the PAR polymer itself and the nature of its biomolecular interactions have remained elusive.
PAR-binding motifs/domains
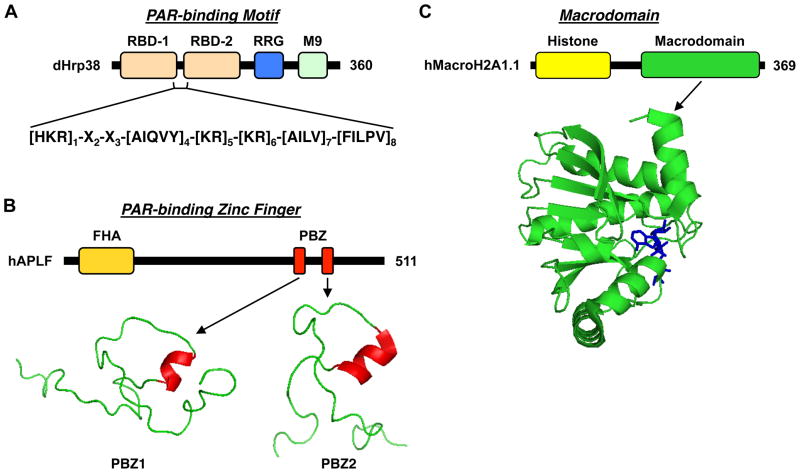
Recent studies have lead to the identification of three different types of motifs or domains that bind PAR, which are found in a variety of proteins involved in DNA repair or chromatin regulation (Fig. 7) (Kleine and Luscher, 2009; Kraus, 2009).
Figure 7. PAR-binding motifs.
(A) PAR binding motifs, as found in dHrp38, a protein that regulates alternative splicing of RNA transcripts.
(B) PAR-binding zinc fingers (PBZs), as found in hAPLF, a protein involved in DNA damage checkpoints. The structures of the two PBZs from hAPLF are shown.
(C) Macrodomains, as found in macroH2A1.1, a histone variant involved in setting the chromatin environment. The structure of the macrodomain of macroH2A1.1 bound to ADP-ribose is shown.
A short conserved motif
A series of studies have lead to the experimental and computational identification of an eight amino acid PAR-binding motif found in PAR-binding proteins: [HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8, (Fig. 7A) (Gagne et al., 2008; Pleschke et al., 2000). Although the function of this motif has not been extensively verified in functional assays, its identification in a large set of proteins suggests a potentially broad role for PAR in regulating the functions of proteins. For example, PAR-binding motifs in the Drosophila hnRNPs Squid/hrp40 and Hrb98DE/hrp38 may play a role in regulating alternative splicing of RNA transcripts (Ji and Tulin, 2009). The prominent role that basic amino acids play as determinants of this consensus sequence, however, raises questions about the specificity of PAR binding or whether the binding reflects the general affinity of the basic amino acids for charged polymers (Kleine and Luscher, 2009).
PAR-binding zinc finger (PBZ)
The (PBZ), a C2H2 zinc finger, represents another motif that can bind PAR (Fig. 7B) (Ahel et al., 2008; Eustermann et al., 2010; Isogai et al., 2010; Rulten et al., 2008). It was originally identified in CHFR (checkpoint protein with FHA and RING domains), APLF (aprataxin PNK-like factor), and other proteins involved in DNA repair and checkpoint control proteins (Ahel et al., 2008; Rulten et al., 2008). Functional analyses have demonstrated that the actions of CHFR in the antephase checkpoint are blocked by mutations in the PBZ motif or by inhibition of poly(ADP-ribose) synthesis (Ahel et al., 2008). PAR binding by the PBZ in APLF is required for targeting the protein to DNA strand breaks and may also serve to suppress further PAR synthesis (Rulten et al., 2008). These results provided the first evidence of functional consequences for PAR binding through a specific motif.
Macrodomains
The macrodomain, an ancient and highly conserved structural domain (Kraus, 2009), represents a third motif that can bind PAR, as well as other metabolites of NAD+ (e.g., ADP-ribose, see above; O-acetyl-ADP-ribose, which is generated as a by product of sirtuin-mediated deacetylation reactions) (Karras et al., 2005; Kustatscher et al., 2005; Neuvonen and Ahola, 2009) (Fig. 7C). Prior reports have suggested a physical and functional link between PARP-1 and the macrodomain-containing histone variant macroH2A (Nusinow et al., 2007; Ouararhni et al., 2006). As highlighted below, three recent papers have gone further to examine the role of PAR binding by macrodomain-containing proteins in the control of nuclear functions (Kraus, 2009).
The macrodomain of macroH2A1.1 is required for the localization of macroH2A1.1 to sites of DNA damage-induced PARP-1 activation and PAR formation in the nucleus (Timinszky et al., 2009). One outcome of macroH2A1.1 localization to PARylated loci is the transient compaction of chromatin, an effect that might play a role in regulating DNA repair responses (Timinszky et al., 2009). The macrodomain of ALC1 (a.k.a. Chd1L), an ATP-dependent nucleosome remodeling enzyme, is required for PAR-dependent interactions with PARP-1 and targeting to sites of PAR formation in the nucleus (Ahel et al., 2009; Gottschalk et al., 2009). Interestingly, the ATPase and nucleosome-remodeling activities of ALC1 are dependent on NAD+-dependent PAR synthesis by PARP-1 (Ahel et al., 2009; Gottschalk et al., 2009). Thus, PAR binding through the macrodomain of ALC1 represents another mechanism by which PARP-1 can alter chromatin structure.
The PAR-binding motifs/domains described herein are likely share at least two common functions: (1) targeting of the proteins that contain them to sites of PAR synthesis and (2) regulating the activity of the proteins that contain them upon PAR-binding. Whether there are additional PAR-binding motifs/domains present in the eukaryotic proteome has yet to be determined, but the future identification of such motifs/domains will give immediate clues as to the function of the proteins that contain them.
PARP-1 and NAD+ Metabolism
As the ADP-ribose donor for PARP-1-catalyzed PARylation reactions, NAD+ plays a central role in determining the function and activity of PARP-1. The synthesis of NAD+ occurs in multiple cellular compartments, including the nucleus, which may be the most relevant source of NAD+ for PARP-1 (Berger et al., 2004; Rongvaux et al., 2003). In mammals, NAD+ is synthesized de novo in a pathway leading from tryptophan, as well as through a salvage pathway leading from nicotinamide and catalyzed by the enzymes nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyltransferase (NMNAT; NMNAT-1 is the nuclear form) (Berger et al., 2004; Rongvaux et al., 2003) (Fig. 4). Interestingly, nicotinamide is a natural endogenous inhibitor of PARP-1 (and Sirt1). Thus, the salvage pathway supports PARP-1 activity by depleting nicotinamide and producing of NAD+.
The enzymatic activities of PARP-1, NAMPT, and NMNAT are functionally linked. For example, stress-induced cell death due to PARP-1-dependent NAD+ depletion in cardiomyocytes can be reversed by overexpression of NAMPT (Pillai et al., 2005), supporting the conclusion that NAMPT catalyzes a rate-limiting step in NAD+ synthesis (Revollo et al., 2004). Furthermore, in addition to producing NAD+ to support PARP-1 catalytic activity, NMNAT-1 also stimulates PARP-1 catalytic activity by binding to activated, automodified PARP-1 (Berger et al., 2007). A recent study has shown that Sirt1 recruits NMNAT-1 to target gene promoters, presumably to supply NAD+ for protein deacetylase reactions at the promoter (Zhang et al., 2009). It is likely that a similar mechanism involving PARP-1 and NMNAT-1 supports PARylation of proteins at the promoters of PARP-1-regulated genes. As noted above, the enzymatic activities of PARP-1 and Sirt1 may also be linked through competition for limiting supplies of nuclear NAD+ (Zhang and Kraus, 2009). Difficulty in accurately determining the concentrations of nuclear NAD+, however, has hampered verification of this conclusion. Although functional interplay between PARP-1 and NAD+-metabolizing enzymes in the nucleus has been established, the molecular mechanisms remain to be clarified.
Cellular Signaling through PARP-1
PARP-1 is a targeted endpoint of a number of distinct cellular signaling pathways, including those regulated by hormones, stress, and DNA damage. As noted above, PARP-1 is subject to a variety of post-translational modifications in response to these pathways, and these modification are likely to play a key role in regulating PARP-1 activity and generating specificity of signaling endpoints.
Cellular signaling pathways and functional outcomes
The ultimate functional outcomes of PARP-1-dependent signaling pathways are varied. For example, PARP-1 can act as an “integrator” in a number of pathways, including stress-dependent gene regulatory pathways, where it facilitates the recruitment of chromatin- and transcription-regulating proteins, and promotes the reorganization of chromatin at PARP-1 target genes (Frizzell et al., 2009; Hassa et al., 2003; Petesch and Lis, 2008; Tulin and Spradling, 2003). PARP-1 can also act as an “exchange factor” at target gene promoters in response to cellular signals, promoting a switch from the binding of repressive complexes to activating complexes at target gene promoters (Ju et al., 2006; Ju et al., 2004; Pavri et al., 2005). The best characterized signaling pathways in which PARP-1 plays a role are NF-κB-dependent pro-inflammatory responses, heat shock, cellular kinase-dependent pathways, and hormone signaling, although the involvement of PARP-1 in a number of other pathways seems likely.
NF-κB-dependent pro-inflammatory pathways
PARP-1 plays a key role in pro-inflammatory gene expression responses. Much of PARP-1’s function in this regard is as a transcriptional coregulator of NF-κB in signaling pathways leading from Toll-like receptors (Hassa and Hottiger, 2002). In this regard, PARP-1 functions as a coactivator of NF-κB to regulate the expression of pro-inflammatory target genes. This involves the acetylation of PARP-1 by p300/CBP, which is required for the interaction of PARP-1 with NF-κB and coactivation by the Mediator complex in response to inflammatory stimuli (Hassa et al., 2003; Hassa et al., 2005). PARP-1 was recently shown to be required for DNA damage-induced activation of IκB kinase (IKK), a key protein in the pathway leading to activation of NF-κB (Stilmann et al., 2009). In this regard, PARP-1 promotes the PAR-dependent assembly of a complex containing PIASy and ATM, both of which contain PAR-binding motifs, as well as IKKγ, which is subsequently SUMOylated.
Heat shock
In Drosophila, PAR rapidly accumulates at heat shock loci in response to heat shock (Tulin and Spradling, 2003). dPARP is required for heat shock-induced “puffing” (i.e., chromatin decondensation) at these loci, as described above (Tulin and Spradling, 2003, Petesch, 2008 #7). Knockdown of dPARP or treatment with a PARP inhibitor prevents heat shock-induced nucleosome loss and enhanced transcription at the Hsp70 gene (Petesch and Lis, 2008). In fact, with dPARP knockdown or in the presence of the PARP inhibitor, the nucleosomes remain in a non-heat shock state even after heat shock (Petesch and Lis, 2008). Currently, the mechanism by which PARP-1 senses the heat shock signal is unknown, but it may involve interactions with heat shock factor, a DNA-binding transcription factor that is phosphorylated in response to heat shock.
Hormone- and kinase-dependent signaling
PARP-1 plays critical roles in signal-dependent gene regulation as an endpoint of neurogenic, steroid, retinoid, and other hormone signaling pathways (Ju et al., 2006; Ju et al., 2004; Kim et al., 2004; Kim et al., 2005; Kraus, 2008; Pavri et al., 2005). PARP-1 alters the chromatin structure and the set of factors bound at the promoters of the target genes whose expression is regulated by these signaling pathways. Some of these pathways involve cellular kinases, such as ERK1/2, JNK1, PKC, and CaMKIIδ (Berger et al., 2007; Cohen-Armon et al., 2007; Ju et al., 2004; Kauppinen et al., 2006; Zhang et al., 2007). Signaling through ERK1/2 enhances PARP-1 activity, although phosphorylation of PARP-1 does not occur in all contexts (Cohen-Armon et al., 2007; Kauppinen et al., 2006). The stress-activated kinase JNK1 phosphorylates PARP-1, which promotes the sustained activation of PARP-1 when cells are stressed with hydrogen peroxide (Zhang et al., 2007). Furthermore, PKC phosphorylates NMNAT-1, reducing its ability to bind PAR, providing yet another level of PARP-1 regulation by the NAD+ metabolic pathway (Berger et al., 2007).
Convergence of signaling pathways: transcription and DNA repair
A number of parallels exist between PARP-1’s roles in transcription and DNA repair. For example, PARP-1 (1) interacts with and PARylates components of both the transcription and DNA repair machineries, (2) directs components of both machineries to specific sites in chromatin, and (3) is covalently modified in response to the signaling pathways that regulate these processes (Kim et al., 2005). The transcription- and repair-related aspects of PARP-1 function may converge in some contexts. For example, a recent study has suggested that upon estrogen treatment, a topoisomerase IIβ- and PARP-1-containing complex is recruited to target promoters, causing the formation of a double strand break in the promoter DNA (Ju et al., 2006). The function of the double strand break is not known, but it may resolve a topological constraint allowing a critical structural change in the promoter. Alternatively, it may serve as a signal to activate PARP-1 and stimulate its factor exchange functions at the promoter. Whether PARP-1 plays a role in the obligate post-transcriptional DNA repair process has not been determined, but it might explain the presence of PARP-1 at nearly all actively transcribed genes (Krishnakumar et al., 2008). Controlled transcription-coupled DNA damage as means of regulating signal-dependent gene expression might seem to be an inefficient and dangerous way for cells to respond to signals, but this is a conceptually interesting and novel view. These results should be evaluated carefully and are in need of additional confirmation and further mechanistic analyses.
Physiology and Pathology of PARP-1
Studies over the past decade have begun to reveal the ways in which the nuclear functions of PARP-1 contribute to specific physiological and pathological outcomes. However, a greater understanding of the specific biological roles of PARP-1 and how they are regulated is still needed. Based on studies in animal models, PARP-1 has been implicated in development, the function of the immune and nervous systems, aging, and cancer, all of which have been reviewed in detail previously (Beneke and Burkle, 2004; Burkle et al., 2004; Kim et al., 2005; Peralta-Leal et al., 2008). Below we highlight some of the key results from animal models, as well as discuss the roles of PARP-1 in inflammation and development.
Animal models of PARP-1 function
PARP-1 knockout mice are viable and show only mild phenotypes (Wang et al., 1995), although some interesting phenotypes have been revealed in response to certain chemical agents, in some genetic backgrounds, and under certain physiological conditions. For example, Parp-1−/− mice are more sensitive to chemically-induced genotoxic stress (de Murcia et al., 1997; Wang et al., 1995; Wang et al., 1997). They also show resistance in various models of inflammation (Ha, 2004; Mabley et al., 2001; Oliver et al., 1999), as well as increased tumor formation in some genetic backgrounds (e.g., p53−/− and SCID) and in chemically-induced models of cancer (Masutani et al., 2005; Morrison et al., 1997; Tong et al., 2001). Furthermore, Parp-1−/− mice are highly susceptible to diet-induced obesity. They accumulate fat tissue and develop hyperleptinemia, insulin resistance, and glucose intolerance when fed a high fat diet (Devalaraja-Narashimha and Padanilam, 2010).
The mild or context-dependent phenotypes observed in the PARP-1 knockout mice may be due to redundancy with other PARP family members. In this regard, genetic ablation of dPARP in Drosophila, which has only one PARP-1-like gene, causes lethality at the larval stage (Miwa et al., 1999; Tulin et al., 2003; Tulin et al., 2002). Furthermore, double knockout of PARP-1 and PARP-2 in mice causes embryonic lethality (Menissier de Murcia et al., 2003). Likewise, individual tankyrase 1/PARP-5a and tankyrase 2/PARP-5b knockout mice (i.e., Tnks1−/− and Tnks2−/−) are largely normal, but double knockout causes early embryonic lethality, indicating redundancy in mouse development (Chiang et al., 2008; Hsiao et al., 2006).
Inflammatory responses
PARP-1 has long been recognized as a key component of immunity and inflammatory responses, and these are the best characterized PARP-1-dependent biological responses (Cuzzocrea, 2005). PARP-1 is heavily automodified in response to bacterial infection (Nossa et al., 2009) and PARP inhibitors inhibit lymphocyte proliferation and lymphokine induction (Weltin et al., 1995). Parp-1−/− mice are resistant to inflammation in various experimental models, including LPS-induced septic shock and streptozotocin-induced diabetes (Ha, 2004; Mabley et al., 2001; Oliver et al., 1999). Moreover, PARP-deficient Drosophila exhibit defects in innate immunity and are more susceptible to bacterial infection than their wild-type counterparts (Tulin and Spradling, 2003). As these results indicate, PARP-1 plays a central role in supporting inflammatory responses. In pathological states, this can have dire consequences, leading to tissue damage. Hence the potential utility of PARP inhibitors in treating inflammatory disorders (Graziani and Szabo, 2005).
Interestingly, PARP-1-dependent pro-inflammatory responses are not limited to cells of the immune system. Recent studies have implicated PARP-1 in pathological pro-inflammatory stress responses in cells of the central nervous and cardiovascular systems (Moroni, 2008; Pacher and Szabo, 2007). In a mouse model of multiple sclerosis, PARP-1 knockout reduces the severity of the disease outcome (Farez et al., 2009; Selvaraj et al., 2009). Furthermore, PARP-1 knockout has been shown to improve various aspects of cardiac function in mice (Pacher and Szabo, 2007). These results suggest a number of exciting potential therapeutic applications for PARP inhibitors.
Development: stem cells and differentiation
Although PARP-1 knockout mice develop normally (Wang et al., 1995), the embryonic lethal phenotype of PARP-1/PARP-2 double knockout mice indicate that PARPs are critical for embryonic development (Menissier de Murcia et al., 2003). The requirement for PARP-1 and PARP-2 in development is due, at least in part, to the roles they play in the maintenance of genomic stability (Menissier de Murcia et al., 2003). The extent to which they control other specific developmental processes is not clear, although new studies have suggested roles for PARP-1 in stem cells and during differentiation.
Stem cell function
In embryonic stem (ES) cells from Parp-1−/− mice, about 10% of genes analyzed showed altered expression compared to about 3% of genes in livers from the same animals (Ogino et al., 2007). The number of genes down-regulated by PARP-1 knockout was about two-fold more than the number of up-regulated genes in both cases, indicating a major role for PARP-1 in keeping genes active in ES and liver cells (Ogino et al., 2007). The large panel of genes whose expression is dependent on PARP-1 in ES cells suggests a role for PARP-1 in the developmental programming of these cells. A recent study has revealed some of the molecular mechanisms whereby PARP-1 might help to promote the differentiation of stem cells (Gao et al., 2009). Specifically, PARP-1 antagonizes the DNA-binding transcription factor Sox2 to stimulate expression of the gene encoding fibroblast growth factor 4 (FGF4), a growth factor that promotes differentiation. In response to appropriate cellular signals, PARP-1 PARylates Sox2 at the FGF4 enhancer, which promotes the dissociation and degradation of Sox2 and leads to enhanced expression of FGF4. These results indicate that PARP-1 can regulate the pluripotent state of ES cells by controlling the activity of key stem cell transcription factors.
Cellular differentiation programs
PARP-1, as well as PARP-2, has been implicated in the differentiation of other cell types as well. For example, in a model of neuronal differentiation, PARP-1 is required for the exchange of corepressors for coactivators at the promoters of genes regulated by the transcription factor HES1 (Ju et al., 2004). PARP-1 is also required for T-cell dependent immunoglobulin class switching in B-cells (Ambrose et al., 2009; Morrison et al., 1997), and PARP-1 deficiency promotes the differentiation of regulatory (CD4+/CD25+/Foxp3+) T-cells (Nasta et al., 2010). In a model of endodermal differentiation, PARP-1 and PARP-2 play distinct roles in a pathway involving physical and functional interactions with the heterochromatin-associated proteins HP1 and TIF1β: PARP-2 is required for differentiation of mouse embryonal carcinoma cells into primitive endoderm-like cells in response to retinoic acid, while PARP-1 is required for subsequent differentiation into parietal endoderm-like cells in response to retinoic acid and dibutyryl cAMP (Quenet et al., 2008).
PARP-2 is required for adipogenesis (Bai et al., 2007) and spermiogenesis (Dantzer et al., 2006), and T-cell survival during thymopoiesis (Yelamos et al., 2006). During adipogenesis, PARP-2 functions as a coactivator of the adipogenic transcription factor PPARγ (Bai et al., 2007). During thymopoiesis, PARP-2 prevents the activation of a DNA damage-dependent apoptotic response through multiple rounds of T-cell receptor gene rearrangements (Yelamos et al., 2006). Whether PARP-1 plays a similar, or perhaps an antagonistic role, in these same differentiation pathways has yet to be determined.
PARP Inhibitors
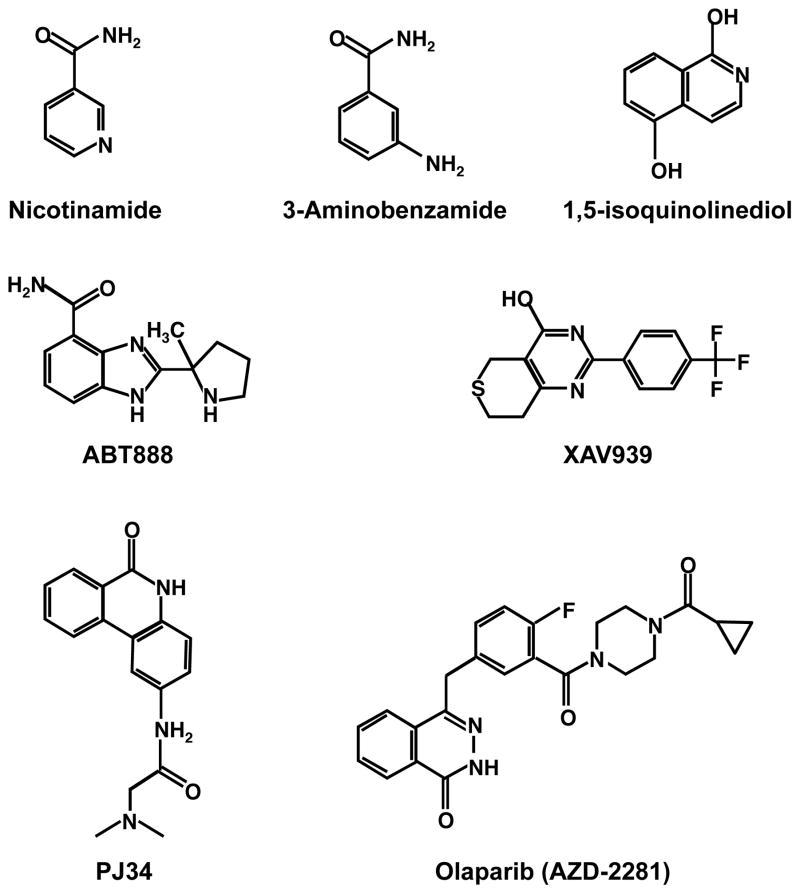
The development of specific, potent, effective, and safe PARP inhibitors has become an area of active research and much recent excitement in the PARP field (Rouleau et al., 2010). The focus has been on competitive inhibitors of PARP catalytic activity (Fig. 8) that may be useful as research tools, as well as clinical therapies.
Figure 8.
Structures of PARP inhibitors.
Pharmacology
3-aminobenzamide (3-AB) (Fig. 8) was the first PARP inhibitor to be extensively characterized, but it lacks the requisite selectivity and potency to be useful as a research tool or in the clinic (Rouleau et al., 2010). Over the past decade, a large number of compounds with the ability to inhibit one or more PARP family members have been synthesized and screened by various laboratories and companies (Pacher and Szabo, 2007; Ratnam and Low, 2007; Rouleau et al., 2010). These include compounds derived from isoquinolines, phenanthridines, and phthalazines, as well as other structural derivatives (Fig. 8), and a number of them are currently being tested in clinical trials as cancer therapies (Ratnam and Low, 2007; Rouleau et al., 2010). Although these inhibitors are highly specific for PARPs and most have nanomolar affinities, developing inhibitors that are specific for a single specific PARP has proven to be considerably more difficult given the high level of conservation of PARP catalytic domains (Hakme et al., 2008; Schreiber et al., 2006) (Fig. 2). Although quinazolinone and quinoxaline derivatives may be more selective for PARP-1 and PARP-2, respectively (Hassa and Hottiger, 2008), increasing specificity is an important area of focus for the future. PARP inhibitors are likely to be useful for treating a wide variety of diseases related to genome integrity (e.g., cancers; (Ratnam and Low, 2007)), as well as stress and inflammatory responses (e.g., cardiovascular disease; (Pacher and Szabo, 2007)).
Clinical trials: focus on cancers
A number of clinical trials are now underway examining the safety and efficacy of PARP inhibitors as treatments for a variety of cancers, including breast, uterine, and ovarian cancers (Rouleau et al., 2010). In many cases, the efficacy of the inhibitors may be due to synthetic lethality between PARP inhibition and a genetic lesion in the cancer cells. For example, p53-deficient breast cancer cells treated with a PARP inhibitor lose resistance to doxorubicin, a clinically active antitumor anthracycline antibiotic that promotes apoptosis (Munoz-Gamez et al., 2005). Similarly, germline mutations in the familial breast cancer genes BRCA1 or BRCA2 sensitize breast cancer cells to PARP inhibitors in a PARP-1-dependent manner (Bryant et al., 2005; Farmer et al., 2005). The goal of this approach is to target cells defective in one DNA repair pathway by inhibiting another. A clinical trial based on this approach has shown selective anti-tumor activity for the PARP inhibitor, olaparib (Fig. 8), in breast and ovarian cancers containing BRCA1 and BRCA2 mutations at safely administrable doses with minimal side-effects (Fong et al., 2009). RNAi-based synthetic lethal screens may be a useful way of identifying other genes that mediate sensitivity to a PARP inhibitors; a recent study has identified a set of kinases whose silencing sensitized cells to a PARP inhibitor (Turner et al., 2008). Another study has suggested that breast cancer cells may be generally sensitive to the PARP inhibitor, PJ34 (Inbar-Rozensal et al., 2009) (Fig. 8), although this may be due synthetic lethality with unknown genetic alterations in the cells examined.
Tankyrase may also be a useful target for the treatment of cancers. In this regard, chemical inhibition of tankyrase shows synthetic lethality with BRCA1 or BRCA2 mutations in breast cancer cells, much like inhibition of PARP-1 (McCabe et al., 2009). Furthermore, XAV939 (Fig. 8), an inhibitor of tankyrases 1 and 2 regulates Wnt signaling in colon cancer cells by prolonging the half-life of axin and promote β-catenin degradation, a target that may be useful for treating Wnt pathway-dependent cancers (Huang et al., 2009). These and other related clinical discoveries have moved PARP-1 and other PARP family members from interesting subjects of molecular analyses to the forefront as clinical targets for cancer treatment (Rouleau et al., 2010).
Future Directions
Based on the literature reviewed herein, it is evident that the functions of PARP-1 are as diverse as they are numerous. In many cases, however, we lack a clear mechanistic understanding of how PARP-1 contributes to the nuclear processes in which it participates. There are many questions and issues that remain to be addressed in future studies. For example, our knowledge of PARP-1 structure is incomplete. A structure of full length PARP-1, alone or in combination with its binding partners (e.g., DNA, nucleosomes, transcription factors), will be required to achieve a full understanding of PARP-1 function. In addition, our understanding of the physiological functions of PARP-1 is limited. More sophisticated and specific animal models, such as tissue-specific knockout mice, will be required to address this issue. Furthermore, our understanding of how the diverse functions of PARP-1 are integrated and controlled is limited. In this regard, the field must reconcile the roles played by PARP-1 in distinct, but inter-related biological processes, such as transcription and DNA repair. Finally, more specific PARP inhibitors will be required both as tools and therapeutics. The next decade promises to be an exciting one for the field.
Acknowledgments
The authors would like to thank members of the Kraus and Lis labs for critical reading of this manuscript and Bryan Gibson for assistance with the structure figures. The research in the Kraus lab is supported by grants from the NIH/NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta A, Valenzuela MT, Matinez-Romero R, Quiles-Perez R, Menissier-de Murcia J, de Murcia G, Ruiz de Almodovar M, Oliver FJ. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Mendoza-Alvarez H. Dissection of ADP-ribose polymer synthesis into individual steps of initiation, elongation, and branching. Biochimie. 1995;77:403–407. doi: 10.1016/0300-9084(96)88153-3. [DOI] [PubMed] [Google Scholar]
- Ambrose HE, Willimott S, Beswick RW, Dantzer F, de Murcia JM, Yelamos J, Wagner SD. Poly(ADP-ribose) polymerase-1 (Parp-1)-deficient mice demonstrate abnormal antibody responses. Immunology. 2009;127:178–186. doi: 10.1111/j.1365-2567.2008.02921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J Biol Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beneke S, Burkle A. Poly(ADP-ribosyl)ation, PARP, and aging. Sci Aging Knowledge Environ. 2004;2004:re9. doi: 10.1126/sageke.2004.49.re9. [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci U S A. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Burkle A, Beneke S, Muiras ML. Poly(ADP-ribosyl)ation and aging. Exp Gerontol. 2004;39:1599–1601. doi: 10.1016/j.exger.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 2009;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J Cell Biochem. 2005;94:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- Caiafa P, Zlatanova J. CCCTC-binding factor meets poly(ADP-ribose) polymerase-1. J Cell Physiol. 2009;219:265–270. doi: 10.1002/jcp.21691. [DOI] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, Hodes RJ. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One. 2008;3:e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzocrea S. Shock, inflammation and PARP. Pharmacol Res. 2005;52:72–82. doi: 10.1016/j.phrs.2005.02.016. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam B. Parp-1 deficiency exacerbates diet-Induced obesity in mice. J Endocrinol. 2010 doi: 10.1677/JOE-09-0402. [DOI] [PubMed] [Google Scholar]
- El Ramy R, Magroun N, Messadecq N, Gauthier LR, Boussin FD, Kolthur-Seetharam U, Schreiber V, McBurney MW, Sassone-Corsi P, Dantzer F. Functional interplay between Parp-1 and SirT1 in genome integrity and chromatin-based processes. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat Struct Mol Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farez MF, Quintana FJ, Gandhi R, Izquierdo G, Lucas M, Weiner HL. Toll-like receptor 2 and poly(ADP-ribose) polymerase 1 promote central nervous system neuroinflammation in progressive EAE. Nat Immunol. 2009;10:958–964. doi: 10.1038/ni.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Farrar D, Rai S, Chernukhin I, Jagodic M, Ito Y, Yammine S, Ohlsson R, Murrell A, Klenova E. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol. 2010;30:1199–1216. doi: 10.1128/MCB.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol. 2006;18:145–151. doi: 10.1016/j.ceb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Moreel X, Gagne P, Labelle Y, Droit A, Chevalier-Pare M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, Poirier GG. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res. 2009;8:1014–1029. doi: 10.1021/pr800810n. [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- Gao F, Kwon SW, Zhao Y, Jin Y. PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation. J Biol Chem. 2009;284:22263–22273. doi: 10.1074/jbc.M109.033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani G, Szabo C. Clinical perspectives of PARP inhibitors. Pharmacol Res. 2005;52:109–118. doi: 10.1016/j.phrs.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–21880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC. Defective transcription factor activation for proinflammatory gene expression in poly(ADP-ribose) polymerase 1-deficient glia. Proc Natl Acad Sci U S A. 2004;101:5087–5092. doi: 10.1073/pnas.0306895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int J Biochem Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl)ation reactions. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 2008;9:1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai S, Uchida M, Kobayashi S, Miwa M, Uchida K. Genomic organization of Drosophila poly(ADP-ribose) polymerase and distribution of its mRNA during development. J Biol Chem. 1998;273:11881–11886. doi: 10.1074/jbc.273.19.11881. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J Biol Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- Inbar-Rozensal D, Castiel A, Visochek L, Castel D, Dantzer F, Izraeli S, Cohen-Armon M. A selective eradication of human nonhereditary breast cancer cells by phenanthridine-derived polyADP-ribose polymerase inhibitors. Breast Cancer Res. 2009;11:R78. doi: 10.1186/bcr2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Kanno S, Ariyoshi M, Tochio H, Ito Y, Yasui A, Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Burkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U S A. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PARlaying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kleine H, Luscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–19. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. New functions for an ancient domain. Nat Struct Mol Biol. 2009;16:904–907. doi: 10.1038/nsmb0909-904. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010 doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- Mabley JG, Suarez-Pinzon WL, Hasko G, Salzman AL, Rabinovitch A, Kun E, Szabo C. Inhibition of poly (ADP-ribose) synthetase by gene disruption or inhibition with 5-iodo-6-amino-1,2-benzopyrone protects mice from multiple-low-dose-streptozotocin-induced diabetes. Br J Pharmacol. 2001;133:909–919. doi: 10.1038/sj.bjp.0704156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 2009 doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson ME, Caldwell JI. Conceptually driven encoding episodes create perceptual misattributions. Acta Psychol (Amst) 1998;98:183–210. doi: 10.1016/s0001-6918(97)00042-5. [DOI] [PubMed] [Google Scholar]
- Masutani M, Gunji A, Tsutsumi M, Ogawa K, Kamada N, Shirai T, Jishage K, Nakagama H, Sugimura T. Role of poly-ADP-ribosylation in cancer development. In: Burkle A, editor. Poly(ADP-Ribosyl)ation. Georgetown, TX: Landes Bioscience; 2005. [Google Scholar]
- Mathis G, Althaus FR. Release of core DNA from nucleosomal core particles following (ADP-ribose)n-modification in vitro. Biochem Biophys Res Commun. 1987;143:1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- McCabe N, Cerone MA, Ohishi T, Seimiya H, Lord CJ, Ashworth A. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene. 2009;28:1465–1470. doi: 10.1038/onc.2008.483. [DOI] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268:22575–22580. [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Biochemical characterization of mono(ADP-ribosyl)ated poly(ADP-ribose) polymerase. Biochemistry. 1999;38:3948–3953. doi: 10.1021/bi982148p. [DOI] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J Biol Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- Menisser-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A, de Murcia G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol. 2001;21:1828–1832. doi: 10.1128/MCB.21.5.1828-1832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Schuermann D, Altmeyer M, Kassner I, Schmidt D, Schar P, Muller S, Hottiger MO. Sumoylation of poly(ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function. FASEB J. 2009 doi: 10.1096/fj.09-137695. [DOI] [PubMed] [Google Scholar]
- Min W, Wang ZQ. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front Biosci. 2009;14:1619–1626. doi: 10.2741/3329. [DOI] [PubMed] [Google Scholar]
- Miwa M, Hanai S, Poltronieri P, Uchida M, Uchida K. Functional analysis of poly(ADP-ribose) polymerase in Drosophila melanogaster. Mol Cell Biochem. 1999;193:103–107. [PubMed] [Google Scholar]
- Monaco L, Kolthur-Seetharam U, Loury R, Murcia JM, de Murcia G, Sassone-Corsi P. Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:14244–14248. doi: 10.1073/pnas.0506252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni F. Poly(ADP-ribose)polymerase 1 (PARP-1) and postischemic brain damage. Curr Opin Pharmacol. 2008;8:96–103. doi: 10.1016/j.coph.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Gamez JA, Martin-Oliva D, Aguilar-Quesada R, Canuelo A, Nunez MI, Valenzuela MT, Ruiz de Almodovar JM, De Murcia G, Oliver FJ. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem J. 2005;386:119–125. doi: 10.1042/BJ20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasta F, Laudisi F, Sambucci M, Rosado MM, Pioli C. Increased Foxp3+ regulatory T cells in poly(ADP-Ribose) polymerase-1 deficiency. J Immunol. 2010;184:3470–3477. doi: 10.4049/jimmunol.0901568. [DOI] [PubMed] [Google Scholar]
- Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossa CW, Jain P, Tamilselvam B, Gupta VR, Chen LF, Schreiber V, Desnoyers S, Blanke SR. Activation of the abundant nuclear factor poly(ADP-ribose) polymerase-1 by Helicobacter pylori. Proc Natl Acad Sci U S A. 2009;106:19998–20003. doi: 10.1073/pnas.0906753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Nozaki T, Gunji A, Maeda M, Suzuki H, Ohta T, Murakami Y, Nakagama H, Sugimura T, Masutani M. Loss of Parp-1 affects gene expression profile in a genome-wide manner in ES cells and liver cells. BMC Genomics. 2007;8:41. doi: 10.1186/1471-2164-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. Embo J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–260. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Peralta-Leal A, Rodriguez MI, Oliver FJ. Poly(ADP-ribose)polymerase-1 (PARP-1) in carcinogenesis: potential role of PARP inhibitors in cancer treatment. Clin Transl Oncol. 2008;10:318–323. doi: 10.1007/s12094-008-0207-8. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F. The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1alpha. FASEB J. 2008;22:3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–690. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010 doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, La Rocca G, Burgio G, Kotova E, Di Gesu D, Collesano M, Ingrassia AM, Tulin AV, Corona DF. The nucleosome-remodeling ATPase ISWI is regulated by poly-ADP-ribosylation. PLoS Biol. 2008;6:e252. doi: 10.1371/journal.pbio.0060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Soundarapandian MM, Chechneva O, Williams AJ, Sidorov MK, Soulika AM, Pleasure DE, Deng W. PARP-1 deficiency increases the severity of disease in a mouse model of multiple sclerosis. J Biol Chem. 2009;284:26070–26084. doi: 10.1074/jbc.M109.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Laurent JF, Gagnon SN, Dequen F, Hardy I, Desnoyers S. Altered DNA damage response in Caenorhabditis elegans with impaired poly(ADP-ribose) glycohydrolases genes expression. DNA Repair (Amst) 2007;6:329–343. doi: 10.1016/j.dnarep.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. 2009. [DOI] [PubMed] [Google Scholar]
- Tong WM, Hande MP, Lansdorp PM, Wang ZQ. DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol Cell Biol. 2001;21:4046–4054. doi: 10.1128/MCB.21.12.4046-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, Ashworth A. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Simbulan-Rosenthal CM, Smulson ME, Chock PB, Yang DC. Polyubiquitylation of PARP-1 through ubiquitin K48 is modulated by activated DNA, NAD+, and dipeptides. J Cell Biochem. 2008a;104:318–328. doi: 10.1002/jcb.21624. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008b;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltin D, Picard V, Aupeix K, Varin M, Oth D, Marchal J, Dufour P, Bischoff P. Immunosuppressive activities of 6(5H)-phenanthridinone, a new poly(ADP-ribose)polymerase inhibitor. Int J Immunopharmacol. 1995;17:265–271. doi: 10.1016/0192-0561(95)00007-o. [DOI] [PubMed] [Google Scholar]
- Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst) 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Yelamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, et al. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, Guastafierro T, Bacalini MG, Reale A, Amicosante G, et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lin Y, Kim YS, Hande MP, Liu ZG, Shen HM. c-Jun N-terminal kinase mediates hydrogen peroxide-induced cell death via sustained poly(ADP-ribose) polymerase-1 activation. Cell Death Differ. 2007;14:1001–1010. doi: 10.1038/sj.cdd.4402088. [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: Linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]