Abstract
Suberin is a protective hydrophobic barrier consisting of phenolics, glycerol, and a variety of fatty acid derivatives, including C18:0-C22:0 primary fatty alcohols. An eight-member gene family encoding alcohol-forming fatty acyl-coenzyme A reductases (FARs) has been identified in Arabidopsis (Arabidopsis thaliana). Promoter-driven expression of the β-glucuronidase reporter gene indicated that three of these genes, FAR1(At5g22500), FAR4(At3g44540), and FAR5(At3g44550), are expressed in root endodermal cells. The three genes were transcriptionally induced by wounding and salt stress. These patterns of gene expression coincide with known sites of suberin deposition. We then characterized a set of mutants with T-DNA insertions in FAR1, FAR4, or FAR5 and found that the suberin compositions of roots and seed coats were modified in each far mutant. Specifically, C18:0-OH was reduced in far5-1, C20:0-OH was reduced in far4-1, and C22:0-OH was reduced in far1-1. We also analyzed the composition of polymer-bound lipids of leaves before and after wounding and found that the basal levels of C18:0-C22:0 primary alcohols in wild-type leaves were increased by wounding. In contrast, C18:0-OH and C22:0-OH were not increased by wounding in far5-1 and far1-1 mutants, respectively. Heterologous expression of FAR1, FAR4, and FAR5 in yeast confirmed that they are indeed active alcohol-forming FARs with distinct, but overlapping, chain length specificities ranging from C18:0 to C24:0. Altogether, these results indicate that Arabidopsis FAR1, FAR4, and FAR5 generate the fatty alcohols found in root, seed coat, and wound-induced leaf tissue.
Suberin is a lipid- and phenolic-based polymer present at the inner face of primary cell walls of various external and internal tissue layers (Franke and Schreiber, 2007). In roots, suberin deposition occurs in the endodermis surrounding the central vasculature, the exodermis near the soil interface, and the peridermis of underground storage tubers (Ma and Peterson, 2003). In aerial tissues, suberin deposition occurs in bark tissue, the bundle sheath of C4 plants, cotton (Gossypium hirsutum) fibers, the chalazae plug of seeds, seed coat integuments, and in response to wounding (Kolattukudy, 2001). Suberin often consists of alternating dark and light bands, termed suberin lamellae, when viewed by transmission electron microscopy. The phenylpropanoid-derived aromatic portion of suberin mostly consists of ferulate, which is a p-hydroxycinnamic acid (Bernards et al., 1995). The aliphatic portion of suberin is a polyester largely composed of ω-hydroxy fatty acids, α,ω-dicarboxylic acids, mid-chain oxygenated fatty acids, unsubstituted fatty acids, and primary fatty alcohols (Pollard et al., 2008). The relative abundance of suberin monomer constituents varies considerably between species (Pollard et al., 2008). Another major component of the suberin aliphatic domain is glycerol (Moire et al., 1999), which in partial depolymerization experiments is found esterified to ω-hydroxy and α,ω-dicarboxylic fatty acids (Graça and Pereira, 2000; Graça and Santos, 2006). The overall organization of the interunit linkages, including those involving unsubstituted fatty acids and primary fatty alcohols, remains to be determined.
The composition of suberin indicates that the following enzymatic activities, at least, are required for suberin biosynthesis: ω-carbon oxidation, further oxidation of ω-hydroxy fatty acids to α,ω-dicarboxylic acids, fatty acid elongation of long-chain acyl precursors, activation of fatty acids to fatty acyl-CoA thioesters, reduction of acyl chains to primary alcohols, and various acylations, including those involving glycerol, to generate a polyester matrix. The identification of many of these enzymes has been facilitated by the elucidation of the chemical composition of suberin in the model plant Arabidopsis (Arabidopsis thaliana; Franke et al., 2005) and the isolation of Arabidopsis mutants with altered suberin composition (Pollard et al., 2008). Arabidopsis cytochrome P450 monooxygenases, CYP86A1 and CYP86B1, are chain-length-specific fatty acid hydroxylases that generate ω-hydroxy acids and α,ω-dicarboxylic acids, which are subsequently incorporated into the suberin polymer (Li et al., 2007a; Hofer et al., 2008; Compagnon et al., 2009). Similarly, a CYP86A family member in potato (Solanum tuberosum) has been shown to be important for production of ω-functionalized suberin monomers of the tuber periderm (Serra et al., 2009b). DAISY/KCS2 and KCS20 from Arabidopsis are β-ketoacyl-CoA synthases that are important for elongation of C20 acyl chain suberin precursors (Franke et al., 2009; Lee et al., 2009). Mutations in these KCS genes result in reductions of C22 and C24 very-long-chain derivatives in root suberin, and the double mutant kcs20 kcs2/daisy is affected more than either single mutant, indicating that the enzymes function in a partially redundant manner. Silencing of a potato KCS gene, called StKCS6, causes reductions of suberin monomers with chain lengths of C28 and higher in the tuber peridermis (Serra et al., 2009a). Arabidopsis mutants of GPAT5, which encodes for an acyl-CoA:glycerol-3-phosphate acyltransferase, exhibit large reductions of C20-C24 unsubstituted fatty acids, ω-hydroxy acids, and α,ω-dicarboxylic acids in both root and seed coat suberin (Beisson et al., 2007). GPAT5 is thus likely involved in the formation of very-long-chain fatty acid-containing acylglycerol precursors for suberin biosynthesis. A feruloyl-CoA transferase called AFST from Arabidopsis, as well as the ortholog from potato called FHT, are required for incorporation of ferulate into the suberin polyester (Gao et al., 2009; Molina et al., 2009; Serra et al., 2010).
Saturated primary fatty alcohols, typically of chain lengths C18, C20, and C22, are common components of root suberin (Zeier et al., 1999; Franke et al., 2005; Schreiber et al., 2005b; Beisson et al., 2007). Fatty acyl-coenzyme A reductase (FAR) enzymes catalyze the reduction of fatty acyl-CoAs to primary fatty alcohols in a NADPH-dependent reaction. This reduction is a two-step process proceeding via an aldehyde intermediate, which can be carried out by a single alcohol-forming FAR (Kolattukudy, 1970; Pollard et al., 1979; Vioque and Kolattukudy, 1997). Purification of an alcohol-forming FAR from jojoba (Simmondsia chinensis) embryos facilitated the cloning of the corresponding cDNA, and heterologous expression in bacteria confirmed that reduction of fatty acyl-CoAs to primary alcohols is carried out by a single enzyme in plants (Metz et al., 2000). FAR enzymes related to the jojoba FAR have subsequently been identified and characterized in wheat (Triticum aestivum; Wang et al., 2002), silkmoth (Moto et al., 2003), mouse and human (Cheng and Russell, 2004), and Arabidopsis (Rowland et al., 2006; Doan et al., 2009). Expression of these FARs in various heterologous systems indicated that each has distinct substrate specificity in terms of both acyl chain length and degree of saturation.
Eight genes encoding putative FARs have been identified in Arabidopsis, but to date the biological functions of only two of the corresponding proteins have been described. MS2 (FAR2) produces primary alcohols that are incorporated into sporopollenin of the pollen exine layer (Aarts et al., 1997; Dobritsa et al., 2009), while CER4 (FAR3) generates C24:0-C30:0 primary alcohols present in cuticular wax of aerial tissues (Rowland et al., 2006). We demonstrate here that the gene expression patterns of FAR1, FAR4, and FAR5 coincide with known sites of suberin deposition and that the encoded enzymes have activities for acyl chain lengths consistent with those of suberin-associated primary alcohols. Furthermore, mutations in each of these genes differentially affected primary alcohol formation in root, seed coat, and wound-induced leaf tissue.
RESULTS
FAR1, FAR4, and FAR5 Gene Expression Is Associated with Suberin Deposition
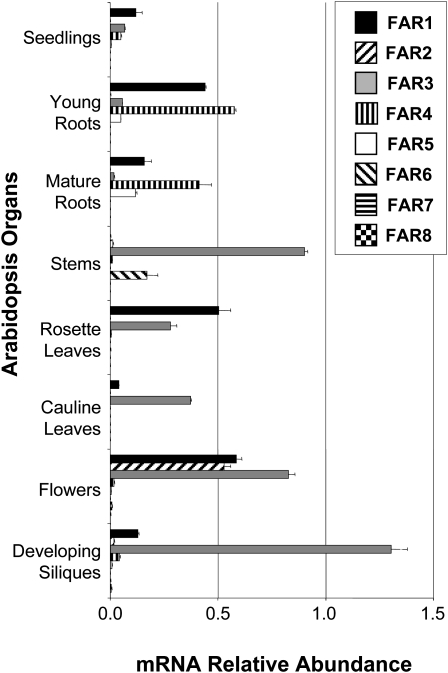
Arabidopsis contains eight FAR genes that, in agreement with The Arabidopsis Information Resource, we have named FAR1to FAR8 (Supplemental Table S1). The tissue-specific gene expression patterns of the FAR genes were determined by quantitative reverse transcription (RT)-PCR using RNA derived from seedlings and young roots from 15-d-old plants grown in tissue culture and using RNA derived from various organs (mature roots, stems, cauline and rosette leaves, flowers, and developing siliques) of 10-week-old plants grown on soil (Fig. 1). This analysis indicated that FAR3/CER4 is highly expressed in aerial organs of the plant while FAR2/MS2 expression is restricted to flowers, in agreement with their respective roles in wax biosynthesis (Rowland et al., 2006) and sporopollenin formation (Aarts et al., 1997). The FAR6 gene was found to be mainly expressed in stems, while the levels of expression for FAR7 and FAR8 were very low under our conditions, suggesting that these latter two genes are expressed at very specific developmental stages. These results are in agreement with DNA microarray data suggesting that FAR6 is expressed in stem epidermis while expression of both FAR7 and FAR8 is restricted to stigmas (Zimmermann et al., 2004; Suh et al., 2005). FAR1 was widely expressed, most notably in young roots, rosette leaves, and flowers. FAR1 transcripts were also detected in cauline leaves and developing siliques, but not in stems. Finally, FAR4 and FAR5 transcripts were largely restricted to young and mature roots, with slight accumulation detected in seedlings, flowers, and developing siliques.
Figure 1.
Tissue-specific gene expression patterns of Arabidopsis FAR genes. Total RNA was isolated from 15-d-old plants grown in tissue culture (seedlings and young roots) or 10-week-old mature plants grown on soil (mature roots, stems, cauline leaves, rosette leaves, flowers, and developing siliques). Gene expression levels were determined by quantitative RT-PCR analysis. Results are presented as relative transcript abundances and normalized through geometric averaging of five constitutively expressed genes (ACT2, EF-1α, eIF-4A-1, UBQ10, and PP2A). The data represent means ± sd of three biological replicates.
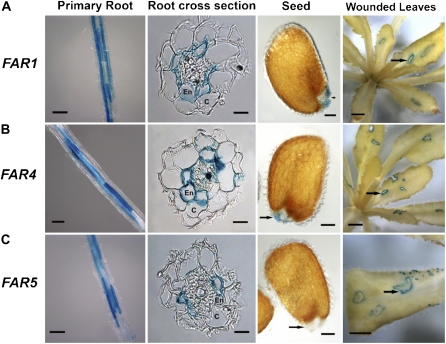
In order to identify FAR enzymes associated with suberin biosynthesis, we focused our subsequent analyses on the root-expressed FAR1, FAR4, and FAR5 genes. Since FAR3/CER4 was previously shown to be important for the production of C24:0-C30:0 chain length fatty alcohols of cuticular wax (Rowland et al., 2006) and its expression in young and mature roots was very low (Fig. 1), it is unlikely to have a significant role in suberin biosynthesis. The gene expression patterns of FAR1, FAR4, and FAR5 were further investigated by fusing about 2,500 bp of their upstream sequences, relative to the start codon, in frame with the coding region of the GUS reporter gene. The T-DNA plant binary constructs containing the promoter:GUS fusions were used to transform Arabidopsis. Histochemical staining for GUS activity was performed on at least 20 independent T1 transformants and then repeated in several T2 or T3 transgenic lines that showed representative expression patterns. For all three FAR promoters, strong GUS activity was found along the length of primary roots, and cross sections indicated that it was restricted to the endodermal cell layer surrounding the central vasculature (Fig. 2). The promoters of both FAR1 and FAR4, but not FAR5, drove GUS expression in the micropyle region (hilum) of the seed (Fig. 2). We also occasionally observed patchy staining in the seed coats of plants expressing the FAR1 and FAR4 promoter:GUS fusions. The root endodermal layer, the hilum region, which is where sealing of tissue after seed detachment takes place, and the seed coat are sites of suberin deposition. Finally, high levels of GUS activity driven by the FAR1 promoter were also observed in the elongation zone of lateral root tips, cotyledons, the shoot apex, young leaves, petals, filaments of stamens, and the receptacle of siliques (Supplemental Fig. S1A). FAR4 promoter-directed GUS activity in aerial tissues was restricted to filaments of stamens and the receptacle of siliques (Supplemental Fig. S1B), while that driven by the FAR5 promoter was observed only in floral organs of very young unopened buds and the receptacle of siliques (Supplemental Fig. S1C).
Figure 2.
Expression patterns of FAR1 (A), FAR4 (B), and FAR5 (C) detected in transgenic promoter:GUS lines. Primary root tissues were collected from 10-d-old seedlings grown in tissue culture. Seeds were collected from recently matured siliques, and arrows point to the micropyle region (hilum). The leaves of 6-week-old plants were punctured with a needle and stained for GUS activity 24 h after wounding. Bars = 100, 25, 10, and 500 μm for primary root, root cross section, seed, and wounded leaves, respectively.
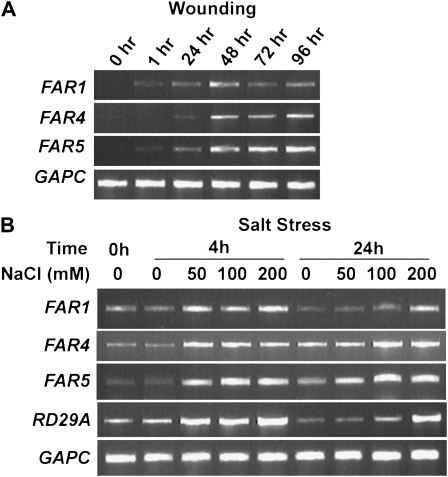
Suberin has also been reported to be produced in response to wounding and abiotic stress (Dean and Kolattukudy, 1976; Kolattukudy, 2001). Six-week-old transgenic plants containing the FAR1, FAR4, or FAR5 promoter:GUS fusion were wounded, and GUS activity was assayed over a 24-h period. Histochemical staining was evident in wounded, but not unwounded, leaf tissue of transgenic plants containing each of the three promoter:GUS fusions after 24 h, all along the perimeter of sections that had been punctured (Fig. 2). The same pattern of staining was also observed when stems of the transgenic plants were wounded (Supplemental Fig. S1). GUS activity was not observed immediately after wounding (data not shown), indicating that apparent wound-induced staining was not simply due to increased access of substrate to the reporter enzyme. To further investigate the kinetics of wound-induced gene expression, a detailed time course of wound induction of the endogenous FAR genes was monitored by RT-PCR in wounded stem tissues (Fig. 3A). At the cycle number used for RT-PCR, the transcripts of all three FARs were not detected in unwounded stems. In contrast, the transcripts of FAR1 and FAR5 were detectable by RT-PCR at 1 h after wounding and that of FAR4 at 24 h after wounding. The transcript levels of all three FAR genes peaked at 48 h and remained high after 96 h. We also exposed seedlings to NaCl-induced salt stress (50, 100, and 200 mm NaCl) and observed the influence on FAR1, FAR4, and FAR5 gene expression by RT-PCR (Fig. 3B). All three genes were expressed at low levels in seedlings grown on plates in tissue culture prior to salt stress, but all showed a rapid induction after 4 h at all three salt concentrations. Induced expression levels remained high after 24 h at all three salt concentrations for FAR4 and FAR5, but FAR1 levels remained induced at 24 h only in the presence of 200 mm NaCl. For comparison, the well-characterized RESPONSIVE TO DEHYDRATION29A (RD29A; Yamaguchi-Shinozaki and Shinozaki, 1993) gene was used as a reference for salt stress induction and was strongly up-regulated at 4 h by all concentrations, but only by 200 mm NaCl at 24 h (Fig. 3B).
Figure 3.
Induction of FAR1, FAR4, and FAR5 gene expression by wounding and salt stress. Transcripts were detected by RT-PCR analysis. A, Wounding. Stems of 6-week-old plants were slit at multiple positions using a needle, and RNA from stems was extracted at the indicated time points. B, Salt stress. Seven-day-old seedlings grown on solid MS medium were transferred to solid MS medium containing different NaCl concentrations as indicated. RNA samples from whole seedlings were taken at the indicated time points. The RD29A gene was used as a positive control for salt-induced expression. The GAPC gene was used as a constitutively expressed control in both A and B.
Isolation of Mutants Disrupted in the FAR1, FAR4, and FAR5 Genes
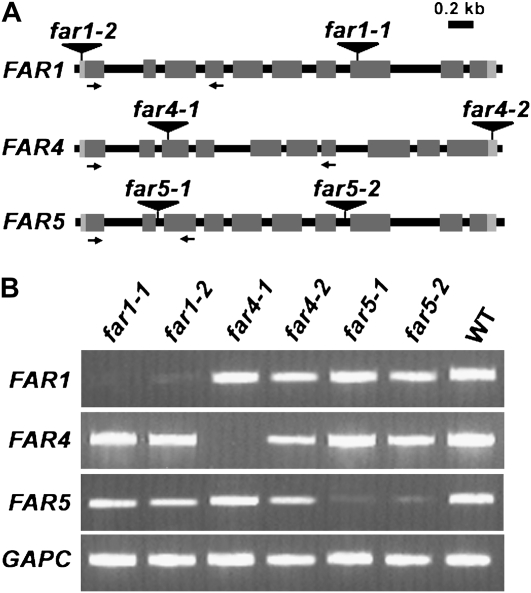
We identified two T-DNA insertion lines in each of FAR1, FAR4, and FAR5 to investigate their respective roles in fatty alcohol production associated with suberin deposition. The sites of T-DNA insertion were determined by sequencing, and in some cases, they did not precisely match the sites of insertion reported on the T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress). The far1-1 (SALK_068605) and far1-2 (SALK_149469) alleles have T-DNA inserts in the eighth exon (nucleotide +2,346 relative to the start codon) and 5′ untranslated region (nucleotide −94), respectively (Fig. 4A). The transcript levels of FAR1 were nearly absent in both of these lines (Fig. 4B). The far4-1 (SALK_000229) and far4-2 (SALK_147493) alleles have T-DNA inserts in the third exon (nucleotide +883) and 3′ untranslated region (87 bp after the stop codon), respectively (Fig. 4A). The far4-1 allele was confirmed to be a transcriptional null of FAR4, whereas the far4-2 allele had very slight, if any, effect on transcript abundance (Fig. 4B). The far5-1 (SALK_152963) and far5-2 (SALK_070363) alleles have T-DNA inserts in the second and seventh introns (nucleotides +562 and +1,966), respectively (Fig. 4A). The transcript levels of both of these FAR5 alleles were substantially reduced, but nonetheless detectable, indicating that they are likely not null alleles (Fig. 4B). No obvious developmental phenotypes were visually observed in the homozygous lines of any of these mutant plants when compared with the wild type.
Figure 4.
T-DNA insertion lines of FAR1, FAR4, and FAR5, and transcript levels in the far mutants. A, Schematic of FAR1, FAR4, and FAR5 gene structures indicating the positions of the T-DNA insertions in each pair of far1, far4, and far5 mutant alleles. Dark gray boxes indicate exons, black lines indicate introns, and light gray boxes indicate 5′ and 3′ untranslated regions. The arrows underneath the gene structures are the positions of convergent primers used for RT-PCR to analyze transcript abundance in the homozygous insertion lines. B, RT-PCR analysis of steady-state FAR mRNAs in roots of far mutants compared with the wild type. The GAPC gene was used as a constitutively expressed control.
Alterations of Fatty Alcohol Composition Associated with Suberin Isolated from Roots and Seed Coats of far Mutants
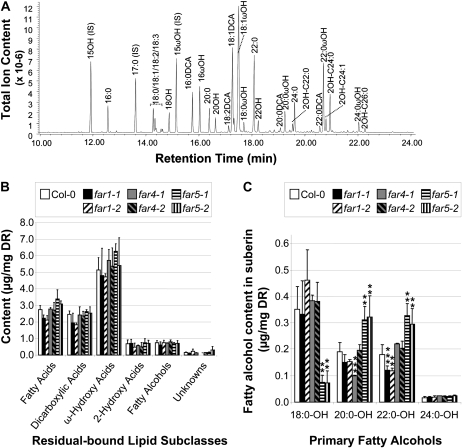
The pairs of far1, far4, and far5 T-DNA mutant alleles described above were first analyzed for suberin monomer composition in roots of 6-week-old plants grown in soil. Gas chromatography-mass spectrometry (GC-MS) analysis of residual bound lipids after extensive chloroform/methanol extraction of whole roots allowed the identification and quantification of 41 different compounds, of which the major ones are indicated in Figure 5A. The major lipid classes usually found in root suberin (i.e. fatty acids, α,ω-dicarboxylic acids, ω-hydroxy acids, and primary fatty alcohols) were detected. 2-Hydroxy acids were also detected, but these have been speculated to come from sphingolipids rather than representing suberin monomers (Molina et al., 2006). No significant differences in the total quantity of these lipid classes were observed in the different far mutant lines (Fig. 5B). However, inspection of the fatty alcohol subclass revealed significant variations among the chain length distributions depending on the mutant (Fig. 5C; Supplemental Table S2). The most pronounced phenotype was observed in both far5-1 and far5-2, where the C18:0-OH content was 80% decreased. This sharp reduction in C18:0-OH was accompanied by a 60% increase in the amounts of both C20:0-OH and C22:0-OH. In the far1 lines, the amounts of C20:0-OH and C22:0-OH were reduced by 20% and 25%, respectively, while C18:0-OH content remained about the same. The far4-2 line, which contains a T-DNA insertion in the 3′ untranslated region and only a slight reduction in FAR4 transcript abundance (Fig. 4B), had a fatty alcohol composition that was not significantly different from the wild type. In contrast, the far4-1 line, which is a transcriptional null of FAR4, had a 50% decrease of C20:0-OH. The levels of C18:0-OH and C22:0-OH were not significantly altered in far4-1.
Figure 5.
Aliphatic suberin composition in roots of soil-grown wild-type, far1, far4, and far5 plants. A, Gas chromatogram of suberin monomers released by depolymerization of solvent-extracted roots of wild-type Arabidopsis plants. Suberin monomers were released by transmethylation and hydroxyl groups silylated before separation by GC and detection of total ion content with a mass spectrometer. The peaks corresponding to the internal standards (IS) pentadecanol (C15:0-OH), ω-pentalactone (C15ωOH), and heptadecanoic acid (C17:0) are indicated, as are each of the major suberin monomers identified by GC-MS. B, Suberin monomer composition, where mean values are shown in μg mg−1 delipidated dry residue (DR) ± sd of three replicates. Monomers are sorted by compound class along the x axis. C, Fatty alcohol content of suberin sorted into individual chain lengths, where mean values are shown in μg mg−1 delipidated dry residue of three replicates. Errors bars indicate sd, and significance was assessed by Student's t test: ** P < 0.05, *** P < 0.01.
There was some variability in the suberin composition data obtained from soil-grown plants. Therefore, we analyzed root suberin composition of wild-type and mutant plants grown on tissue culture medium. Large amounts of roots were easily recovered from plants grown for 5 weeks on vertical plates containing Murashige and Skoog (MS) medium and 1.5% (w/v) agar. We restricted this analysis to the strongest mutant allele of each FAR: far1-1, far4-1, and far5-1 (Table I; Supplemental Fig. S2). As previously observed with soil-grown plants, no significant differences could be detected in the levels of any fatty acids, α,ω-dicarboxylic acids, or ω-hydroxy acids or in the total suberin load between the three far mutants and the wild type (Table I; Supplemental Fig. S2). The absolute amounts of all suberin monomers were generally increased in roots grown in tissue culture in comparison with the amounts measured in roots grown in soil, with the exception of 2-hydroxy acids, which were less abundant (Table I; Fig. 5B). This discrepancy may come from stresses induced by growing the plants in vitro for 5 weeks under continuous light or by soil components that caused artifactual increases in the dry residue weight, thus resulting in reduced total suberin load. Nevertheless, as observed with soil-grown plants, the primary alcohol chain length distributions varied significantly between the different mutants and wild-type plants grown in tissue culture (Table I). Similar to the results obtained from roots grown on soil, the content of C20:0-OH in far4-1 was reduced by 55%, while C22:0-OH was increased by 36%. In far5-1, there was an 80% decrease in C18:0-OH and a 20% increase of C20:0-OH. Finally, in far1-1, the amount of C22:0-OH was reduced by 30%, while the other fatty alcohols were not affected. As for soil-grown plants, the total fatty alcohol content was not significantly affected in far1-1 and far4-1 and was only slightly reduced in far5-1 (Table I).
Table I. Aliphatic suberin composition in roots of tissue culture-grown wild-type, far1-1, far4-1, and far5-1 plants.
Each value is the mean shown in μg mg−1 delipidated dry residue ± sd of three to five biological replicates. The amount of each individual chain length of primary alcohol is reported below the total amount of primary alcohols in each genetic background.
| Chemical Class | Col-0 (Wild Type) | far1-1 | far4-1 | far5-1 |
| Fatty acids | 4.66 ± 0.71 | 4.77 ± 0.58 | 5.20 ± 0.12 | 4.43 ± 0.15 |
| α,ω-Dicarboxylic acids | 4.07 ± 0.71 | 3.55 ± 0.65 | 3.89 ± 0.49 | 3.43 ± 0.58 |
| ω-Hydroxy acids | 6.64 ± 0.68 | 6.42 ± 1.16 | 6.72 ± 0.52 | 6.96 ± 0.28 |
| 2-Hydroxy acids | 0.12 ± 0.03 | 0.10 ± 0.03 | 0.11 ± 0.03 | 0.12 ± 0.03 |
| Primary alcohols | 1.45 ± 0.11 | 1.37 ± 0.24 | 1.51 ± 0.22 | 0.99 ± 0.19 |
| C18:0-OH | 0.68 ± 0.07 | 0.75 ± 0.11 | 0.75 ± 0.11 | 0.15 ± 0.07 |
| C20:0-OH | 0.31 ± 0.02 | 0.29 ± 0.06 | 0.14 ± 0.02 | 0.37 ± 0.07 |
| C22:0-OH | 0.44 ± 0.04 | 0.31 ± 0.08 | 0.60 ± 0.09 | 0.46 ± 0.07 |
| C24:0-OH | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.02 ± 0.01 |
| Total load | 16.81 ± 2.12 | 16.26 ± 2.27 | 17.50 ± 1.38 | 15.97 ± 0.90 |
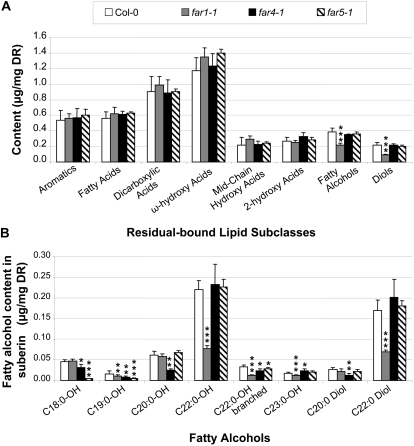
We also analyzed the lipid polyester composition of Arabidopsis wild-type and far mutant seeds (Fig. 6; Supplemental Table S3). Suberin monomers are mostly deposited in the outer integument of the seed coat (Molina et al., 2008). In addition to the components usually found in root suberin, the polyester of seeds contains higher relative amounts of aromatics (mainly trans-ferulic and trans-sinapic acids), midchain hydroxylated fatty acids (10,16-dihydroxy-hexadecanoic and 9,10,18-trihydroxy-octadecenoic acids), diols (1,20-eicosane and 1,22-docosane diols), as well as traces of branched chain compounds (Molina et al., 2006). We did not observe significant variations in the total polyester load of any mutant when compared with the wild type (Supplemental Table S3). Similar amounts of the different subclasses were found in all four lines, with the exception of primary alcohol and diol levels, which were 42% and 56% reduced, respectively, in the far1-1 line relative to the wild type (Fig. 6A; Supplemental Table S3). Although the total fatty alcohol and diol levels were not affected in far4-1 and far5-1, the amount of C18:0-OH was reduced to nearly undetectable levels in far5-1, while the amounts of C20:0-OH and C20:0-diol were reduced by 68% and 50%, respectively, in far4-1 (Fig. 6B). In the far1-1 line, the amounts of C22:0-OH and C22:0-diol were reduced by 65% and 59%, respectively (Fig. 6B). In addition, the amounts of branched chain C22:0-OH and C23:0-OH were significantly reduced in far1-1. Since C18:0-OH, C20:0-OH, and C20:0-diol are minor components of the polyester of wild-type seeds compared with C22:0-OH and C22:0-diol, far1-1 was the only mutant with significantly altered total levels of fatty alcohols and diols (Fig. 6A). The levels of all other suberin components, including the aromatics, remained about the same between the wild type and the three far mutants.
Figure 6.
Lipid polyester composition in seeds of wild-type, far1, far4, and far5 plants. A, Suberin monomer composition, where mean values are shown in μg mg−1 delipidated dry residue (DR) ± sd of five replicates. Monomers are sorted by compound class along the x axis. B, Suberin-associated fatty alcohol content sorted into individual chain lengths, where mean values are shown in μg mg−1 delipidated dry residue of five replicates. Errors bars indicate sd, and significance was assessed by Student's t test: * P < 0.1, ** P < 0.05, *** P < 0.01.
Alterations of Fatty Alcohol Composition in Leaves of far Mutants after Wounding
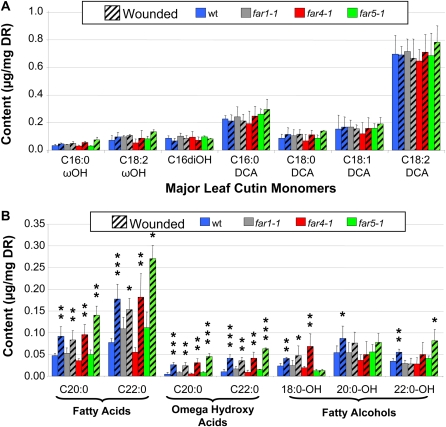
Since the transcript levels of FAR1, FAR4, and FAR5 were increased after wounding (Fig. 3A), we analyzed the levels of primary alcohols in residual bound lipids of leaves before and after wounding. These lipids would collectively represent cutin of the cuticle and possibly suberin deposited in response to wounding. We injured fully developed leaves of 10-week-old plants with a metallic brush and analyzed 5 d later the content of the residual bound lipids. For each line, unwounded leaves from the same plant were used as the control. This method of wounding generated small holes all along the leaf, but in most cases it did not result in dead tissue. After 5 d, total residual bound lipids were slightly increased in wounded leaves of all the lines compared with unwounded leaves, but these differences were not significant according to Student's t test (Supplemental Table S4). Some variability came from the method of wounding itself, as it was difficult to precisely control the number of holes per square centimeter of tissue. In addition, our residual bound lipids contained high amounts of palmitic acid (C16:0) and polyunsaturated C18 fatty acids (C18:2 and C18:3) in comparison with reports in the literature (Bonaventure et al., 2004; Franke et al., 2005), which may have been due to our growing conditions or to incomplete extraction of soluble lipids. Nevertheless, our procedure enabled quantification of the characteristic monomers found in cutin and suberin (residual bound lipids). Wounding treatment did not increase the levels of most major-abundance residual bound lipid components, but the content of some minor-abundance components was significantly increased (Fig. 7; Supplemental Table S4). The C20:0 and C22:0 very-long-chain fatty acids and their corresponding ω-hydroxy fatty acids were significantly induced in wild-type and far mutant lines (Fig. 7B). Similarly, the amounts of C18:0-C22:0 primary fatty alcohols were clearly induced by the wounding treatment in wild-type plants. In contrast, C18:0-OH and C22:0-OH were not increased by wounding in the far5-1 and far1-1 lines, respectively.
Figure 7.
Composition of residual bound lipids in unwounded and wounded leaves of wild-type, far1-1, far4-1, and far5-1 plants. A, Amounts of major monomers in residual bound lipids of unwounded and wounded leaves. B, Amounts of fatty acids, very-long-chain ω-hydroxy acids, and fatty alcohols present in residual bound lipids in unwounded and wounded leaves. All values are means shown in μg mg−1 delipidated dry residue (DR) ± sd of three to four replicates. Errors bars indicate sd, and significance was assessed by Student's t test: * P < 0.1, ** P < 0.05, *** P < 0.01. [See online article for color version of this figure.]
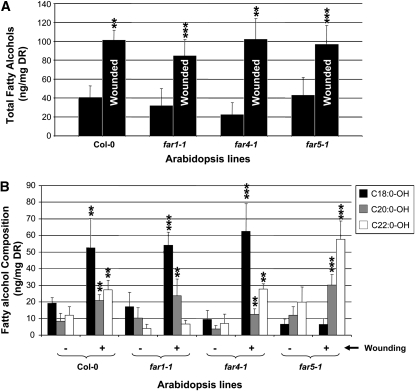
Since the mass spectra of the C20:0 and C22:0 primary alcohols indicated that other compounds coeluted with these alcohols, we only considered the major molecular ion of each fatty alcohol molecule for quantification [m-15, CH3-(CH2)n-O-Si(CH3)2+; Supplemental Fig. S3], rather than total ion content. With this method, contamination from coeluting products was eliminated, allowing for more accurate quantification of each fatty alcohol. Using this method of quantification, the total fatty alcohol content was more than doubled by the wounding treatment in the wild type and in each of the far mutants (Fig. 8A). In the wild type, the content of each chain length of fatty alcohol was more than doubled (Fig. 8B), with C18:0-OH being the most affected (175% increase). A similar increase of C18:0-OH and C22:0-OH was observed in far4-1, but the amount of C20:0-OH was lower than in the wild type with or without wounding treatment. In far1-1, the amounts of C18:0-OH and C20:0-OH primary alcohols increased after wounding as in the wild type, but that of C22:0-OH remained unchanged. The amount of C22:0-OH in unwounded tissues was lower in far1-1 compared with the wild type. Finally, in far5-1, the C18:0-OH content was not increased by wounding, whereas those of C20:0-OH and C22:0-OH increased significantly after wounding, each to a much higher level than that of the wild type. Analogous to the decrease of C22:0-OH and C20:0-OH in unwounded leaves of far1-1 and far4-1, respectively, the level of C18:0-OH in unwounded leaves of far5-1 was significantly decreased relative to the wild type.
Figure 8.
Fatty alcohol content of residual bound lipids in unwounded and wounded leaves of wild-type, far1-1, far4-1, and far5-1plants. A, Total amounts of fatty alcohols in unwounded and wounded leaves. B, Amounts of fatty alcohols sorted by chain length in unwounded and wounded leaves. Quantification of fatty alcohols was based on the major molecular ion of each primary fatty alcohol molecule. All values are means shown in ng mg−1 delipidated dry residue (DR) ± sd of four to six replicates. Errors bars indicate sd, and significance was assessed by Student's t test: ** P < 0.05, *** P < 0.01.
Heterologous Expression of FAR1, FAR4, and FAR5 in Yeast
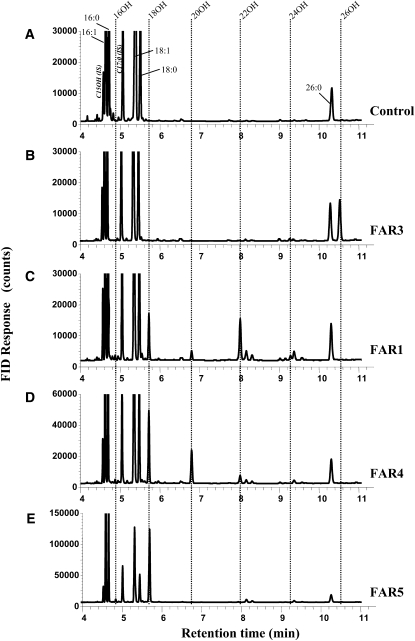
Since C18:0 to C22:0 primary fatty alcohols are detected in suberin of Arabidopsis roots and seed coats, we examined the activities and substrate specificities of the enzymes encoded by the FAR1, FAR4, and FAR5 genes using Saccharomyces cerevisiae as a heterologous system. The open reading frames were subcloned into the yeast expression vector pVT102U, which places the coding sequences under the control of a constitutive promoter, and the resulting constructs were transformed into yeast. The empty vector and pVT102-FAR3 were used as negative and positive controls, respectively. Expression of FAR3/CER4 in yeast results in the production of C24:0-OH and C26:0-OH, in agreement with Rowland et al. (2006), whereas no fatty alcohols were detected in the empty vector control (Fig. 9; Table II). Transgenic yeast expressing FAR1, FAR4, or FAR5 produced C18:0 to C22:0 fatty alcohols, indicating that the three proteins are active alcohol-forming fatty acyl-CoA reductases (Fig. 9; Table II). The major fatty alcohols produced when expressing FAR1 were C18:0-OH and C22:0-OH, but C16:0-OH, C20:0-OH, and C24:0-OH were also detected. Expression of FAR4 resulted mainly in the production of C20:0-OH and C18:0-OH, while FAR5 expression led nearly exclusively to the production of C18:0-OH. The identities of the different fatty alcohols produced were confirmed by GC-MS analyses (Supplemental Fig. S3). In no case were unsaturated fatty alcohols detected. Quantification of internal lipids in the yeast strains revealed that expression of FAR5 generated the most fatty alcohol, with 2.14 μg per unit optical density (OD; Table II). FAR1 and FAR4 each generated much less, with 0.90 and 0.97 μg of fatty alcohol produced per unit OD, respectively, while FAR3 generated 0.60 μg of fatty alcohol per unit OD. In contrast, all strains had similar total fatty acid contents (Table II). The high production of C18:0-OH observed in the case of FAR5 resulted in a lower C18 fatty acid content and a higher C16 fatty acid content.
Figure 9.
Fatty acyl chain profiles of transgenic yeast expressing Arabidopsis FARs. Yeast were transformed with empty vector control pVT102U (A) or with this vector harboring FAR3 (B), FAR1 (C), FAR4 (D), or FAR5 (E). Fatty acyl chains were extracted from yeast cells and analyzed by GC. Major fatty acid peaks are indicated in A, and retention times of the saturated primary alcohols are indicated by dashed lines with chain lengths indicated above the chromatograms. The peaks corresponding to the internal standards (IS) pentadecanol (C15OH) and heptadecanoic acid methyl ester (C17:0) are indicated. FID, Flame ionization detection.
Table II. Internal fatty acyl chain composition of the different FAR-expressing yeast strains.
Fatty acyl chains from whole cells were prepared, derivatized, and analyzed by GC. Data are expressed as μg per unit OD600, and each value is the mean ± sd from five biological replicates. n.d., Not detected.
| Fatty Acyl Chain | Vector Control | FAR1 | FAR3 | FAR4 | FAR5 |
| Fatty acids | |||||
| 16:0 | 1.23 ± 0.01 | 1.24 ± 0.05 | 1.36 ± 0.09 | 1.56 ± 0.10 | 1.76 ± 0.18 |
| 16:1 | 2.85 ± 0.14 | 2.53 ± 0.17 | 2.90 ± 0.28 | 3.28 ± 0.09 | 3.34 ± 0.30 |
| 18:0 | 1.05 ± 0.15 | 1.12 ± 0.07 | 1.15 ± 0.12 | 1.01 ± 0.07 | 0.77 ± 0.06 |
| 18:1 | 4.09 ± 0.24 | 3.62 ± 0.27 | 3.60 ± 0.21 | 3.38 ± 0.16 | 2.84 ± 0.27 |
| 26:0 | 0.18 ± 0.01 | 0.18 ± 0.03 | 0.29 ± 0.05 | 0.15 ± 0.03 | 0.11 ± 0.03 |
| Total fatty acids | 9.40 ± 0.81 | 8.69 ± 0.77 | 9.30 ± 0.97 | 9.39 ± 0.84 | 8.82 ± 0.99 |
| Fatty alcohols | |||||
| 16:0-OH | n.d. | 0.02 ± 0.00 | n.d. | 0.01 ± 0.00 | 0.06 ± 0.01 |
| 18:0-OH | n.d. | 0.28 ± 0.03 | n.d. | 0.52 ± 0.07 | 2.07 ± 0.37 |
| 20:0-OH | n.d. | 0.07 ± 0.01 | n.d. | 0.32 ± 0.05 | 0.01 ± 0.00 |
| 22:0-OH | n.d. | 0.49 ± 0.10 | n.d. | 0.11 ± 0.02 | n.d. |
| 24:0-OH | n.d. | 0.04 ± 0.01 | 0.02 ± 0.01 | n.d. | n.d. |
| 26:0-OH | n.d. | n.d. | 0.59 ± 0.04 | n.d. | n.d. |
| Total fatty alcohols | n.d. | 0.90 ± 0.10 | 0.60 ± 0.04 | 0.97 ± 0.13 | 2.14 ± 0.37 |
DISCUSSION
Suberin is a protective biopolyester that consists of ferulic acid, glycerol, and aliphatic moieties. In this study, we identified three Arabidopsis FAR enzymes, FAR1, FAR4, and FAR5, which are responsible for generating primary fatty alcohols associated with suberin. Evidence for this conclusion is as follows: (1) the gene expression patterns of these three FARs are associated with known sites of suberin deposition; (2) the chain lengths of primary alcohols produced by heterologous expression of these FARs in yeast are consistent with the chain lengths of primary alcohols found in Arabidopsis suberin; and (3) mutants of FAR1, FAR4, and FAR5 are each differentially affected in primary alcohol levels associated with suberin in various tissues.
FAR1, FAR4, and FAR5 Are Expressed at Sites of Suberin Deposition
Suberin is constitutively deposited in the cell walls of root endodermal cells. The Casparian strip of the root endodermis, which is made up of suberin, lignin, and other primary cell wall components, has a major role in the prevention of apoplastic movement of ions and water from the root cortex to the stele (Enstone et al., 2003). Therefore, we focused our attention on root-expressed FARs to identify those producing fatty alcohols associated with suberin. FAR1, FAR4, and FAR5 were found to be root expressed, consistent with the developmental expression patterns reported using DNA microarrays (Schmid et al., 2005). Expression of the GUS reporter gene under control of the promoters of these root-expressed FARs in the endodermal cells is similar to that of six other Arabidopsis suberin-associated genes: GPAT5, AFST, KCS2/DAISY, KCS20, CYP86A1/HORST, and CYP86B1/RALPH (Beisson et al., 2007; Hofer et al., 2008; Compagnon et al., 2009; Lee et al., 2009; Molina et al., 2009). The walls of Arabidopsis root endodermal cells are characterized by suberin lamellae (Franke et al., 2005) and are stained by lipophilic dyes (Franke et al., 2005; Beisson et al., 2007; Compagnon et al., 2009). In aerial tissues, the receptacle of mature siliques, where abscission of petals takes place, is also stained by these lipophilic dyes and is coincident with KCS2/DAISY and CYP86B1/RALPH gene expression (Compagnon et al., 2009; Franke et al., 2009). Likewise, GUS reporter gene activity driven by all three FAR promoters was observed in silique receptacles. The deposition of suberin in this region is likely important to seal off the area that was formerly attached to the petals. Espelie et al. (1980) showed that the micropyle-chalazal region of grapefruit (Citrus paradisi) seed is also sealed by suberized cell walls. FAR1 and FAR4 promoter-driven GUS activity was observed in the micropyle region of the Arabidopsis seed coat, similar to that found for GPAT5, KCS2/DAISY, and CYP86B1/RALPH (Beisson et al., 2007; Compagnon et al., 2009; Franke et al., 2009). FAR5 promoter-driven GUS activity was not observed in the micropyle region, suggesting that FAR5 does not generate suberin-associated fatty alcohols at this location. We had difficulties obtaining consistent GUS staining patterns in seed coats, but all three FARs are likely to be expressed in the seed coat at some developmental stage, because we observed alterations in fatty alcohol levels in the seed coat lipid polyester in each of the three far mutants, with far1 being the most dramatically affected.
Suberization of cell walls occurs in response to pathogen attack or mechanically induced stress (Dean and Kolattukudy, 1976; Bernards and Lewis, 1998). The polymer likely serves to seal off the tissue to prevent further opportunistic pathogen infections and also to prevent prolonged water or solute leakage. The wound induction kinetics of FAR1, FAR4, and FAR5 were roughly similar, peaking at around 2 d and with induced transcripts remaining high even after 4 d. This gene induction correlated with increased amounts of fatty alcohols in the residual bound lipids of leaves 5 d after wounding. The KCS2/DAISY transcript is similarly up-regulated at 24 h after wounding (Franke et al., 2009), which likely accounts for the increases in very-long-chain fatty acids at 5 d post wounding. Salt (e.g. NaCl) stress leads to increased suberization of endodermal and hypodermal root cell walls in castor bean (Ricinus communis; Schreiber et al., 2005a), and a 2-fold increase in total root suberin occurs in Arabidopsis roots in response to high levels of NaCl (Franke et al., 2009). Although it was not shown specifically that fatty alcohols are increased by salinity in Arabidopsis roots, this is likely to be the case, since FAR1, FAR4, and FAR5 gene expression levels were increased by exposure to high salt.
The gene expression patterns of FAR4 and FAR5 were largely restricted to sites of suberin deposition, whether constitutive or inducible, and it is thus likely that they serve only to produce fatty alcohols of suberin. FAR1 was found to be more widely expressed, such as in flower petals, cotyledons, and young leaves, and may have nonsuberin functions. FAR1 is unlikely to have a role in generating fatty alcohols of cuticular wax, because null mutants of far3/cer4 are almost completely deficient of all chain-length fatty alcohols normally found in this aerial hydrophobic barrier. The contribution of other FARs to suberin-associated fatty alcohol biosynthesis is likely to be minimal, based on their gene expression patterns, which are not at known sites of suberin deposition. FAR3/CER4, which generates cuticular C24-C30 fatty alcohols in epidermal cells, is expressed at low levels in roots but not specifically in endodermal cells (Rowland et al., 2006). FAR3/CER4 is thus unlikely to significantly influence root suberin composition. Altogether, these data suggest that FAR1, FAR4, and FAR5 represent the three members of the Arabidopsis FAR multigene family that are involved in suberin synthesis.
Substrate Specificities of FAR1, FAR4, and FAR5
Fatty alcohols ranging in length from C18:0-OH to C22:0-OH have been found in the suberin polyester of Arabidopsis roots and seed coats (Franke et al., 2005; Molina et al., 2006; Beisson et al., 2007; this study). In each of the far mutants in this study, their distribution was differentially affected. These analyses strongly suggest that, in planta, FAR1 is most important for generating C22:0 primary alcohol, FAR4 is most important for generating C20:0 primary alcohol, and FAR5 is most important for generating C18:0 primary alcohol. Expression in yeast confirmed that FAR1, FAR4, and FAR5 produce fatty alcohols ranging from C18:0 to C22:0. These three enzymes are 67% to 73% identical and 80% to 86% similar at the amino acid level (Rowland et al., 2006). In yeast, the apparent substrate specificity of FAR5 was particularly stringent for C18 acyl chains, with the other two FARs able to accept a couple of chain lengths. FAR1 mostly generated C22:0 and C18:0 primary alcohols in yeast, while FAR4 mostly generated C20:0 and C18:0 primary alcohols. When FAR1 and FAR3 were expressed in Escherichia coli, both led to the accumulation of C16:0-OH and C18:1-OH (Doan et al., 2009). This does not fit the plant biochemical phenotype and is in contrast to what was observed here using the yeast system. However, the acyl chains in E. coli are linked to acyl carrier protein, rather than CoA, and this may affect substrate specificity. Expression in yeast, therefore, appears to be a better indicator of in planta substrate specificity than the E. coli system. For example, bacterial expression of FAR1 yielded C16:0-OH and C18:1-OH in similar proportions (Doan et al., 2009), whereas these are not produced when FAR1 is expressed in yeast and instead C18:0-OH and C22:0-OH mainly accumulate (this study). The production of these latter primary alcohols in yeast better reflects the role of FAR1 in producing C22:0 fatty alcohols associated with root, seed coat, and wound-induced suberin. Nevertheless, even in yeast, the available acyl-CoA substrates need to be considered when evaluating FAR substrate specificities. FAR5 preferentially used C18:0-CoA, which is abundant in the acyl-CoA pool of yeast cells (Domergue et al., 2003). In that context, the detection of C18:0-OH in yeast when expressing FAR1 or FAR4 may be due to the relatively high abundance of C18:0-CoA, since there was no effect on C18:0-OH in the far1 and far4 mutants. Nevertheless, the fact that no unsaturated fatty alcohols could be detected, despite monounsaturated fatty acids being highly abundant in the yeast acyl-CoA pool (Domergue et al., 2003), strongly suggests that these enzymes only accept saturated acyl-CoA chains as substrates. In addition, FAR5 effectively reduced C18:0-CoA while displaying nearly no activity on C16:0-CoA, which is more abundant than C18:0-CoA in the acyl-CoA pool (Domergue et al., 2003). It thus appears that FAR enzymes have strong chain length specificities. Ultimately, measurements of substrate specificity using purified enzymes in vitro will be required for a quantitative assessment of relative substrate specificities.
Production of Primary Alcohols Associated with Suberin Is Affected in far1, far4, and far5 Mutants
Individual chain lengths of primary alcohols of root and seed coat suberin were reduced in the far mutants: far1 had a decrease in C22:0-OH, far4 had a decrease in C20:0-OH, and far5 had a decrease in C18:0-OH. With the exception of C20:0 and C22:0 diols, which were reduced in the seed coat polyester of far4 and far1, respectively, the other suberin chemical classes were not affected in the different far mutants, indicating that perturbations in primary alcohol accumulation, at least at the levels altered in these mutants, do not affect other constituents. Reciprocally, the suberin mutants gpat5 (Beisson et al., 2007), afst (Molina et al., 2009), and cyp86b1 (Compagnon et al., 2009), have wild-type fatty alcohol levels, despite being significantly affected in at least one other suberin monomer component. Fatty alcohols have a limited ability to be incorporated into polymers when compared with bifunctional monomers, which may account for the lack of alterations in other polymeric components. Also, a decrease in one chain length of fatty alcohol in a far mutant was sometimes accompanied by an increase in another fatty alcohol(s), such that the total fatty alcohol content of the far mutant was similar to the wild type (e.g. increases of C20:0-OH and C22:0-OH in far5 mutants). Although we observed compensatory effects, we did not detect major increases in the transcript levels of the other two suberin-associated FARs when one was mutated, but there could be changes in protein activities. Alternatively, there may be a certain amount of total fatty alcohols that can be incorporated into or associated with the suberin polymer, such that reduction in one fatty alcohol component allows for the increased accumulation of other chain lengths. That is, more sites of incorporation become available. Suberin-associated waxes of roots, which are soluble in organic solvents, contain components that are chemically related to suberin monomers, and in particular alkyl hydroxycinnamate esters, in which the alkyl moieties are C18-C22 saturated primary alcohols (Li et al., 2007b; Molina et al., 2009). Although we attempted to extract these waxes from roots of plants grown either on soil or in tissue culture, the only compounds we could detect by GC-MS after rapid chloroform dipping of roots were sterol derivatives; therefore, we could not assess the relative alterations in abundance of primary alcohols in this fraction using our far mutants.
Wounded plant tissue develops a wound periderm (Yang and Bernards, 2006). We have demonstrated here that wounding increases the levels of some polyester monomer components of Arabidopsis leaves. The components characteristic of cutin were not wound induced, whereas the characteristic very-long-chain components of suberin (i.e. C20:0 and C22:0 chain length fatty acids, ω-fatty acids, and primary alcohols) were increased 5 d after wounding. In most plants, α,ω-dicarboxylic acids are aliphatics unique to suberized tissue and can be used as diagnostic markers to differentiate between suberized and cutinized tissues (Matzke and Riederer, 1991). However, Arabidopsis cutin is atypical in that high amounts of α,ω-dicarboxylic acids are present in cutin (Bonaventure et al., 2004; Franke et al., 2005). Although C18:1 ω-hydroxy (coeluting with C18:0 dicarboxylic acid in our procedure) and α,ω-dicarboxylic acids are major components of suberin, we did not observe any significant increase in these compounds after wounding. This may be due to the fact that in leaf cutin, the metabolism of polyester biosynthesis is different, with C18:2 α,ω-bifunctional components being far more abundant than the corresponding C18:1 molecules. Alternatively, only certain suberin monomers may be increased in wounded leaf tissues of Arabidopsis. As discussed above, the increase in primary alcohols present in the residual lipids of leaves after wounding correlates well with the wound induction of the FAR1, FAR4, and FAR5 genes. Basal levels of C18:0-C22:0 primary alcohols were found in leaves, and these levels were differentially affected in the far mutants, according to the same pattern that was observed in their root suberin compositions. We did not detect transcripts for these FARs in unwounded leaves, but they could be present at low levels. Alternatively, these genes could be active at earlier stages, which was apparent for FAR1, and the fatty alcohols present are due to a small amount of suberin deposited early in plant development or in response to unknown stresses.
Functions of Fatty Alcohols Associated with Suberin
The primary role of suberized cells appears to be that of controlling water and solute transport (Bernards, 2002), but it has also been postulated that suberized cells act as preformed, as well as wound-induced, antimicrobial barriers (Kolattukudy, 2001). An Arabidopsis mutant called enhanced suberin1 is characterized by increased root suberin, and this was correlated with reduced daytime transpiration rates and increased water use efficiency (Baxter et al., 2009). The Arabidopsis gpat5-1 mutant has an approximately 50% decrease in aliphatic suberin in roots and seed coats, and this is correlated with increases in both root and seed coat permeabilities (Beisson et al., 2007). Since primary fatty alcohols constitute less than 9% of the total aliphatic content in Arabidopsis and as each mutant was affected in only one primary alcohol chain length with the overall amount of fatty alcohol remaining basically unchanged (Table I), it is unlikely that far mutants would have such a dramatic effect on permeability. We carefully monitored for developmental abnormalities in the different far mutants, especially in the development of their roots, but we did not detect any obvious developmental differences when compared with the wild type. We also did not detect major reductions in germination rates or seed coat permeability to tetrazolium salts, even with far1-1, which had the largest reduction in fatty alcohol levels associated with seed coat suberin (data not shown). This absence of phenotype raises the question of the importance of fatty alcohols in root suberin. However, if the role of fatty alcohols in suberin is limited, why then are fatty alcohols consistently found to be associated with suberin? Also, why does the plant require three complementary proteins when just a single FAR with broader substrate specificity would be sufficient? Although the absence of one FAR enzyme had no major effect on plant growth and development, the presence of three genes having similar spatiotemporal gene expression patterns, shared transcriptional regulation, and coding for enzymes with complementary catalytic activities strongly suggests that fatty alcohols have fundamental functions in suberin polyester structure and/or function. A structural role for fatty alcohols is supported by the recent finding that pollen grains of ms2/far2 mutants, which are affected in fatty alcohols associated with sporopollenin, have a “zebra” phenotype. This phenotype is a major defect in exine architecture characterized by a striped appearance on the pollen surface (Dobritsa et al., 2009). The same phenotype is observed with cyp704B1 mutants, indicating that fatty alcohols are as important as ω-hydroxy fatty acids for sporopollenin integrity (Dobritsa et al., 2009). Future studies with double or triple mutants (far1, far4, and/or far5) may unravel important functions for primary alcohols in suberin assembly and suberin permeability characteristics. Also, long-chain fatty alcohols have been found to have antibacterial action (Masao et al., 1987), and thus fatty alcohols may have an important role in pathogen resistance.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants were in the Columbia (Col-0) ecotype background. The following T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org): SALK_068605 (far1-1), SALK_149469 (far1-2), SALK_000229 (far4-1), SALK_147493 (far4-2), SALK_152963 (far5-1), and SALK_070363 (far5-2). For experimental analyses, sterilized seeds were sown on MS medium supplemented with 0.7% agar. The seeds were stratified in the dark for 3 to 4 d at 4°C and then transferred to long-day conditions (16-h-light/8-h-dark cycle) at 22°C or continuous light conditions at 20°C. Seedlings were transferred 10 d to 3 weeks later to a soil-vermiculite mixture and further grown at 20°C to 22°C under long-day conditions.
Tissue-Specific Gene Expression Analysis by Quantitative RT-PCR
RNA was extracted from Arabidopsis tissues using the RNeasy Plant mini kit (Qiagen), and purified RNA was treated with DNase I. First-strand cDNA was prepared from 1 μg of total RNA with SuperScript Reverse Transcriptase II (Invitrogen) and oligo(dT)18, according to the manufacturer's instructions. A 0.2-μL aliquot of the total reaction volume (20 μL) was used as a template in real-time (quantitative) RT-PCR. Amplification by PCR was performed using the gene-specific primers labeled as FARx-qPCR-x listed in Supplemental Table S5. To ensure that transcript amplification was specific, each set of PCR primers was designed from CATMA sequences (www.catma.org) and specificity was verified by amplification of a single PCR product of the correct size from each set of primers. Quantitative PCR was performed on an iCycler (Bio-Rad) using SYBR Green Master Mix (Bio-Rad) as described previously (Joubès et al., 2008). The relative transcript abundance of ACT2, EF-1α, eIF-4A-1, UBQ10, and PP2A in each sample was used to normalize for differences of total RNA amounts (Joubès et al., 2008).
Promoter:GUS Fusions, Transformation of Arabidopsis, and GUS Histochemical Assay
Upstream sequences of approximately 2.5 kb that included the first three codons of FAR1, FAR4, and FAR5 were amplified by PCR using as templates bacterial artificial chromosomes (BACs) containing the genomic regions of interest. A 2,550-bp fragment of FAR1 upstream sequence was amplified using BAC MIQJ16 as template and primers FAR1-Pro-F and FAR1-Pro-R. A 2,560-bp fragment of FAR4 upstream sequence was amplified using BAC F14L2 as template and primers FAR4-Pro-F and FAR4-Pro-R. A 2,600-bp fragment of FAR5 upstream sequence was amplified using BAC F14L2 as template and primers FAR5-Pro-F and FAR5-Pro-R. The forward (F) and reverse (R) primers contained SalI and BamHI recognition sites, respectively, and the sequences are found in Supplemental Table S5. The amplified products were digested with SalI and BamHI and cloned between the corresponding sites of pBI101 (Clontech) fusing the first few codons from each DNA fragment in frame with the coding region of the GUS gene. These constructs were introduced into wild-type Col-0 plants by Agrobacterium tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998).
GUS staining solution was composed of 50 mm sodium phosphate, pH 7.0, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.1% (v/v) Triton X-100, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. Aerial tissues were preincubated in 90% chilled acetone for 5 min and then vacuum infiltrated with GUS staining solution. Tissues were incubated in GUS staining solution at 37°C for 2 h (root tissue) or 12 h (aerial tissues). For whole tissues, samples were cleared in 70% (v/v) ethanol following staining and imaged using a stereomicroscope (Zeiss SteREO Discovery V20). For tissue sectioning, GUS-stained root tissues were incubated with FAA fixative containing formaldehyde (3.7%), glacial acetic acid (5%), and ethanol (50%) for 4 h at room temperature. Following fixation, samples were rinsed for 2 min in 50% ethanol and dehydrated in an ethanol series of 50%, 60%, 70%, 85%, 95%, and three changes of 100% ethanol for 30 min at each concentration. Dehydrated samples were then treated with xylene:ethanol solution of 25%:75%, 50%:50%, and 75%:25% strength for 30 min each, followed by 100% xylene solution for 1 h. The tissue samples were slowly infiltrated with paraffin wax overnight at room temperature. After several wax changes at 6-h intervals incubating at 60°C, the wax molds were allowed to solidify and stored at 4°C. Twelve-micrometer-thick sections were cut on a microtome, dewaxed, and imaged using a compound microscope (Zeiss Axio Imager M2).
Wounding of transgenic promoter:GUS plants was carried out on stems and leaves of 6-week-old plants grown under long-day conditions (16-h-light/8-h-dark cycle). Using a needle, injury was made of approximately 1 cm longitudinally between internodes of the mature stem regions or by puncturing leaves at multiple positions. The plants were returned to the growth cabinet, and the samples were collected for staining after 24 h.
Gene Expression Analysis by RT-PCR after Wounding and Salt Treatment
Wounding of stems of 6-week-old wild-type plants (stage 6.3 according to Boyes et al., 2001) was carried out as described above, and tissues were harvested after 1, 24, 48, 72, and 96 h. As a control, a sample was taken before wounding. For the salt stress study, seedlings were first grown on MS agar plates for 7 d and then transferred to MS agar plates containing 0, 50, 100, or 200 mm NaCl. Samples were harvested immediately, after 4 h, and after 24 h. From tissues harvested in both the wounding and salt stress experiments, RNA was extracted using a phenol-guanidine thiocyanate extraction procedure according to the manufacturer's instructions (Trizol reagent; Invitrogen). First-strand cDNA was prepared from 1 μg of total RNA using SuperScript Reverse Transcriptase III (Invitrogen) and oligo(dT)18 according to the manufacturer's instructions. Two microliters of a five-times diluted RT reaction (cDNA) was used as template in a 20-μL PCR using gene-specific primers labeled as xxxx-RT-x in Supplemental Table S5. The gene-specific primers were designed to span introns to differentiate between products amplified from cDNA and PCR products amplified from contaminating genomic DNA. The sizes of the amplified products for FAR1, FAR4, FAR5, RD29A, and GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT (GAPC) were 490, 950, 420, 1,000, and 250 bp, respectively.
Characterization of FAR Activity in Saccharomyces cerevisiae
The coding sequences for FAR1, FAR3, FAR4, and FAR5 were amplified from cDNA using primers labeled as FARx-ORF-x in Supplemental Table S5. The corresponding PCR fragments were cloned into pDONR221 ENTRY vector by the Gateway recombinational cloning technology using the attB × attP (BP) recombination sites. Fragments containing open reading frame sequences were subsequently transferred into the pVT102-U-GW Destination vector by LR cloning. To obtain the pVT102-U-GW Destination vector, the Gateway cassette (1,711 bp; Karimi et al., 2002) was amplified by PCR, phosphorylated with T4 polynucleotide kinase, and inserted into the yeast expression vector pVT102-U (Vernet et al., 1987), which had been digested with XhoI and blunt ended with Klenow enzyme.
The S. cerevisiae strain INVSc1 (Invitrogen) was transformed with yeast expression vectors containing the different open reading frames by a polyethylene glycol/lithium acetate protocol (Ausubel et al., 1995). After selection on minimal medium agar plates without uracil, cells harboring the yeast plasmid were cultivated in liquid minimal medium at 30°C. Starting from precultures with an OD600 of 0.05, expression cultures were grown for 5 d at 30°C and directly used for fatty acid analysis. Fatty acid methyl esters were obtained by transmethylation at 80°C for 1 h of yeast cell sediments with 0.5 m sulfuric acid in methanol containing 2% (v/v) dimethoxypropane and 50 μg of heptadecanoic acid (C17:0) as well as 20 μg of pentadecanol (C15:0-OH) as internal standards. After cooling, 1 mL of NaCl (2.5%, w/v) was added, and fatty acyl chains were extracted twice in hexane. Extracts were dried under a gentle stream of nitrogen and dissolved into 100 μL of N,O-bis(trimethylsilyl)trifluoroacetamide:trimethylchlorosilane (BSTFA-TMCS; 99:1), and free hydroxyl groups were derivatized at 80°C for 1 h. Surplus BSTFA-TMCS was evaporated under nitrogen, and samples were dissolved in hexane for analysis using a Hewlett-Packard 5890 Series II gas chromatograph equipped with an HP-1 column (30 m × 0.32 mm × 0.25 μm) and a flame ionization detector. The initial temperature of 50°C was held for 1 min, increased at 50°C min−1 to 200°C, held for 1 min at 200°C, increased again at 10°C min−1 to 320°C, and held for 8 min at 320°C. Quantification of fatty acids and fatty alcohols was based on peak areas and the respective internal standard, C17:0 or C15:0-OH. Qualitative analyses were performed using an Agilent 6850 gas chromatograph equipped with an HP-5MS column (30 m × 0.25 mm × 0.25 μm) and an Agilent 5975 mass spectrometric detector (70 eV, mass-to-charge ratio 50–750). The same GC program was used, with helium (1.5 mL min−1) as carrier gas.
Characterization of T-DNA Insertion Lines
T-DNA insertion mutant lines for FAR1, FAR4, and FAR5 were identified from the Sequence-Indexed Library of Insertion Mutations using the Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress). Homozygous insertion lines were identified from among the segregating plants by PCR screening for both the presence of a T-DNA insertion at the locus and the absence of an intact gene. Genomic DNA was isolated from individual plants and genotyped using gene-specific primers labeled as SALK_xxxxxx-TDNA-xx in Supplemental Table S5 and LBb1, which is a T-DNA left border primer. The exact positions of the T-DNA insertions were determined by sequencing the PCR products. For transcript analysis in the mutants, RNA was extracted from root tissues and analyzed by RT-PCR using the same primers and conditions described above for transcript analysis after wounding and salt stress.
Analysis of Lipid Polyester Composition (Suberin and Cutin)
Roots from 6-week-old plants (stage 6.3 according to Boyes et al., 2001) grown on soil were carefully separated from the soil, washed thoroughly in water, and dried on paper towels before delipidation. Roots from mutant plants grown for 5 weeks in tissue culture on vertical plates (MS medium) containing 1.5% (w/v) agar were easily separated from the growing medium and dried on paper towels before delipidation. For seed coat analysis, all the lines were grown together, dry seeds were collected, and about 50 mg of dry seeds was used per replicate. For wounding experiments, four to five leaves from fully developed plants grown on soil for about 10 weeks (stage 8.0 according to Boyes et al., 2001) were injured with a metallic brush, and small holes were produced all along the leaf. Five days later, these leaves, as well as four to five unwounded leaves from the same plant, were harvested and directly used for delipidation.
For delipidation, roots and leaf tissues freshly collected as described above were immersed in hot isopropanol for 10 min at 80°C. For seed coat analysis, seeds were ground in cold isopropanol using a mortar and pestle, and then the mixture was transferred to tubes and heated for 30 min at 80°C. After cooling, samples were extensively delipidated by extracting the soluble lipids successively for 24 h with CHCl3:CH3OH (2:1, v/v), CHCl3:CH3OH (1:1, v/v), CHCl3:CH3OH (1:2, v/v), and CH3OH, all performed at 4°C on a wheel rotating at 33 rpm. Each extraction was conducted once for roots and leaves but twice for seeds. Samples were dried in a fume hood at room temperature for 2 d and then in a desiccator for another 2 d. The dried residues were weighed, and 10 to 30 mg of each sample was depolymerized by transmethylation at 85°C for 3 h using 1 m sulfuric acid in methanol containing 2% (v/v) dimethoxypropane and spiked with 10 μg of heptadecanoic acid (C17:0), 10 μg of ω-pentalactone (C15ωOH), and 10 μg of pentadecanol (C15:0-OH) as internal standards. After cooling, 3 mL of NaCl (2.5%, w/v) was added, and the released fatty acyl chains were extracted twice in hexane for residual lipids derived from roots and leaves and twice in dichloromethane for residual lipids derived from seeds. Extracts were washed once with 3 mL of saline solution (200 mm NaCl and 200 mm Tris, pH 8.0), dried under a gentle stream of nitrogen, and dissolved in 150 μL of BSTFA-TMCS. Free hydroxyl groups were derivatized at 110°C for 30 min, surplus BSTFA-TMCS was evaporated under nitrogen, and samples were dissolved in hexane for analysis using GC-MS under the same conditions described above for yeast acyl chain analysis except that the first temperature ramp (50°C–150°C) was done at 25°C min−1 instead of 50°C min−1. Quantification of fatty acids, hydroxyl acids, and fatty alcohols was based on peak areas, which were derived from total ion content, and using the respective internal standards (C17:0, ω-pentalactone, or C15:0-OH).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression patterns of FAR1 (A), FAR4 (B), and FAR5 (C) detected in transgenic promoter:GUS lines.
Supplemental Figure S2. Aliphatic suberin composition in roots of tissue culture-grown wild-type, far1-1, far4-1, and far5-1 plants.
Supplemental Figure S3. Identification by GC-MS of primary fatty alcohols produced in transgenic yeast expressing Arabidopsis FAR1.
Supplemental Table S1. FAR genes in Arabidopsis.
Supplemental Table S2. Aliphatic suberin composition in roots of soil-grown wild-type, far1, far4, and far5 plants.
Supplemental Table S3. Composition of residual bound lipids in seeds of wild-type, far1-1, far4-1, and far5-1 plants.
Supplemental Table S4. Composition of residual bound lipids in unwounded and wounded leaves of wild-type, far1-1, far4-1, and far5-1 plants.
Supplemental Table S5. Sequences of DNA primers used for PCR.
Supplementary Material
Acknowledgments
We thank the Salk Institute for Genomic Analysis Laboratory and the Arabidopsis Biological Resource Center for providing the sequence-indexed Arabidopsis T-DNA insertion lines. O.R. thanks Sarah Amer for assistance with propagation of promoter:GUS transgenic plants. F.D. thanks Larissa Zita Nyamba Mendome and Sabine Scandola for participating in this work and Fred Beisson as well as Yonghua Beisson-Li for helpful discussions.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen DM, Varki A. (1995) Current Protocols in Molecular Biology. John Wiley & Sons, New York [Google Scholar]
- Baxter I, Hosmani PS, Rus A, Lahner B, Borevitz JO, Muthukumar B, Mickelbart MV, Schreiber L, Franke RB, Salt DE. (2009) Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet 5: e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA. (2002) Demystifying suberin. Can J Bot 80: 227–240 [Google Scholar]
- Bernards MA, Lewis NG. (1998) The macromolecular aromatic domain in suberized tissue: a changing paradigm. Phytochemistry 47: 915–933 [DOI] [PubMed] [Google Scholar]
- Bernards MA, Lopez ML, Zajicek J, Lewis NG. (1995) Hydroxycinnamic acid-derived polymers constitute the polyaromatic domain of suberin. J Biol Chem 270: 7382–7386 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M. (2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octdeca-cis-6,cis-9-diene-1,18-dioate as the major component. Plant J 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JB, Russell DW. (2004) Mammalian wax biosynthesis. I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem 279: 37789–37797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F. (2009) CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol 150: 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean BB, Kolattukudy PE. (1976) Synthesis of suberin during wound-healing in jade leaves, tomato fruit, and bean pods. Plant Physiol 58: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan TTP, Carlsson AS, Hamberg M, Low L, Stymne S, Olsson P. (2009) Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J Plant Physiol 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. (2009) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Abbadi A, Ott C, Zank TK, Zähringer U, Heinz E. (2003) Acyl carriers used as substrates by the desaturases and elongases involved in very long-chain polyunsaturated fatty acids biosynthesis reconstituted in yeast. J Biol Chem 278: 35115–35126 [DOI] [PubMed] [Google Scholar]
- Enstone DE, Peterson CA, Ma F. (2003) Root endodermis and exodermis: structure, function, and responses to the environment. J Plant Growth Regul 21: 335–351 [Google Scholar]
- Espelie KE, Davis RW, Kolattukudy PE. (1980) Composition, ultrastructure and function of the cutin- and suberin-containing layers in the leaf, fruit peel, juice-sac and inner seed coat of grapefruit (Citrus paradisi Macfed.). Planta 149: 498–511 [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. (2005) Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Franke R, Hofer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L. (2009) The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J 57: 80–95 [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L. (2007) Suberin: a biopolyester forming apoplastic plant interfaces. Curr Opin Plant Biol 10: 252–259 [DOI] [PubMed] [Google Scholar]
- Gao JY, Yu XH, Liu CJ. (2009) A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc Natl Acad Sci USA 106: 18855–18860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça J, Pereira H. (2000) Suberin in potato periderm: glycerol, long chain monomers, and glyceryl and feruloyl dimers. J Agric Food Chem 48: 5476–5483 [DOI] [PubMed] [Google Scholar]
- Graça J, Santos S. (2006) Glycerol-derived ester oligomers from cork suberin. Chem Phys Lipids 144: 96–107 [DOI] [PubMed] [Google Scholar]
- Hofer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. (2008) The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot 59: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R. (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. (1970) Reduction of fatty acids to alcohols by cell-free preparation of Euglena gracilis. Biochemistry 9: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. (2001) Polyesters in higher plants. Adv Biochem Eng Biotechnol 71: 1–49 [DOI] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC. (2009) Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge JB. (2007a) Identification of acyltransferases required for cutin synthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Ohlrogge JB, Pollard M. (2007b) Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol 144: 1267–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma FS, Peterson CA. (2003) Current insights into the development, structure, and chemistry of the endodermis and exodermis of roots. Can J Bot 81: 405–421 [Google Scholar]
- Masao H, Kumi M, Sumitra H. (1987) Effect of long chain fatty acids and fatty alcohols on Streptococcus mutants. Chem Pharm Bull (Tokyo) 35: 3507–3510 [DOI] [PubMed] [Google Scholar]
- Matzke K, Riederer M. (1991) A comparative study into the chemical constitution of cutins and suberins from Picea abies (L.) Karst., Quercus robur L., and Fagus sylvatica L. Planta 185: 233–245 [DOI] [PubMed] [Google Scholar]
- Metz JG, Pollard MR, Anderson L, Hayes TR, Lassner MW. (2000) Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol 122: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moire L, Schmutz A, Buchala A, Yan B, Stark RE, Ryser U. (1999) Glycerol is a suberin monomer: new experimental evidence for an old hypothesis. Plant Physiol 119: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Beisson-Li Y, Beisson F, Ohlrogge JB, Pollard M. (2009) Identification of an Arabidopsis feruloyl-CoA transferase required for suberin synthesis. Plant Physiol 151: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge JB, Pollard M. (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M. (2008) Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J 53: 437–449 [DOI] [PubMed] [Google Scholar]
- Moto K, Yoshiga T, Yamamoto M, Takahashi S, Okano K, Ando T, Nakata T, Matsumoto S. (2003) Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc Natl Acad Sci USA 100: 9156–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Pollard M, McKeon T, Gupta LM, Stumpf PK. (1979) Studies on biosynthesis of waxes by developing jojoba seed. II. The demonstration of wax biosynthesis by cell-free homogenates. Lipids 14: 651–662 [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohman JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schreiber L, Franke R, Hartmann K. (2005a) Effects of NO3 deficiency and NaCl stress on suberin deposition in rhizo- and hypodermal (RHCW) and endodermal cell walls (ECW) of castor bean (Ricinus communis L.) roots. Plant Soil 269: 333–339 [Google Scholar]
- Schreiber L, Franke R, Hartmann KD, Ranathunge K, Steudle E. (2005b) The chemical composition of suberin in apoplastic barriers affects radial hydraulic conductivity differently in roots of rice (Oryza sativa L. cv. IR64) and corn (Zea mays L. cv. Helix). J Exp Bot 56: 1427–1436 [DOI] [PubMed] [Google Scholar]
- Serra O, Hohn C, Franke R, Prat S, Molinas M, Figueras M. (2010) A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. Plant J 62: 277–290 [DOI] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. (2009a) Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J Exp Bot 60: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Sauveplane V, Pinot F, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. (2009b) CYP86A33-targeted gene silencing in potato tuber alters suberin composition, distorts suberin lamellae, and impairs the periderm's water barrier function. Plant Physiol 149: 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY. (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52: 225–233 [DOI] [PubMed] [Google Scholar]
- Vioque J, Kolattukudy PE. (1997) Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch Biochem Biophys 340: 64–72 [DOI] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Dumonceaux T, Zou J, Datla R, Selvaraj G. (2002) Male gametophyte development in bread wheat (Triticum aestivum L.): molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J 30: 613–623 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (1993) Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol 101: 1119–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bernards MA. (2006) Wound-induced metabolism in potato (Solanum tuberosum) tubers. Plant Signal Behav 1: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J, Ruel K, Ryser U, Schreiber L. (1999) Chemical analysis and immunolocalisation of lignin and suberin in endodermal and hypodermal/rhizodermal cell walls of developing maize (Zea mays L.) primary roots. Planta 209: 1–12 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.