Abstract
At the core of the circadian network in Arabidopsis (Arabidopsis thaliana), clock genes/proteins form multiple transcriptional/translational negative feedback loops and generate a basic approximately 24-h oscillation, which provides daily regulation for a wide range of processes. This temporal organization enhances the fitness of plants only if it corresponds to the natural day/night cycles. Light, absorbed by photoreceptors, is the most effective signal in synchronizing the oscillator to environmental cycles. Phytochrome B (PHYB) is the major red/far-red light-absorbing phytochrome receptor in light-grown plants. Besides modulating the pace and phase of the circadian clock, PHYB controls photomorphogenesis and delays flowering. It has been demonstrated that the nuclear-localized amino-terminal domain of PHYB is capable of controlling photomorphogenesis and, partly, flowering. Here, we show (1) that PHYB derivatives containing 651 or 450 amino acid residues of the amino-terminal domains are functional in mediating red light signaling to the clock, (2) that circadian entrainment is a nuclear function of PHYB, and (3) that a 410-amino acid amino-terminal fragment does not possess any functions of PHYB due to impaired chromophore binding. However, we provide evidence that the carboxyl-terminal domain is required to mediate entrainment in white light, suggesting a role for this domain in integrating red and blue light signaling to the clock. Moreover, careful analysis of the circadian phenotype of phyB-9 indicates that PHYB provides light signaling for different regulatory loops of the circadian oscillator in a different manner, which results in an apparent decoupling of the loops in the absence of PHYB under specific light conditions.
Circadian clocks regulate many rhythmic cellular and physiological processes and allow a wide range of organisms to adapt to the predictable daily changes in the environment (e.g. day/night cycles; Harmer, 2009). At the core of the clock, components of the central oscillator (clock genes/proteins) mutually regulate their expression/activity via multiple feedback loops, which results in an autonomous, self-sustained approximately 24-h oscillation. The core oscillation is relayed to diverse clock-controlled processes (gene expression, physiology, behavior) via the output pathway (Mas and Yanovsky, 2009). As the period length of the core oscillation always deviates from 24 h, the clock must be resynchronized to the environmental cycles regularly in order to provide precise temporal information. In nature, this resetting occurs on a daily basis in response to periodic environmental cues (e.g. changes in temperature and light conditions). Light is absorbed by specialized photoreceptors, and signals are forwarded by the input pathway to modulate the pace and the phase of the oscillator (Kozma-Bognar and Kaldi, 2008). The Arabidopsis (Arabidopsis thaliana) circadian oscillator is supposed to consist of three interlocked feedback loops (Locke et al., 2006). In the first loop (central or coupling loop), the morning-expressed CIRCADIAN CLOCK ASSOCIATED1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY) transcription factors inhibit the expression of the TIMING OF CAB EXPRESSION1 (TOC1) gene; conversely, the evening-expressed TOC1 positively regulates the transcription of CCA1/LHY (Alabadi et al., 2001). In the second loop (evening loop), the predicted factor Y induces TOC1 expression during the afternoon/evening, while TOC1 represses Y during the night (Locke et al., 2005). It has been demonstrated that GIGANTEA (GI) functions as a component of Y (Locke et al., 2006). In the so-called morning loop, CCA1/LHY up-regulate the PSEUDO RESPONSE REGULATOR7/9 (PRR7/9) genes (homologs of TOC1) in the morning, and PRR7/9 proteins down-regulate CCA1/LHY expression during the day (Farre et al., 2005). The coordinated function of the three loops is required to generate the approximately 24-h basic oscillations in Arabidopsis (Locke et al., 2006).
Light signals are perceived and transduced to the clock by specialized photoreceptors, including members of the blue light-absorbing cryptochrome (CRY) and the red/far-red light-sensing phytochrome (PHY) families (Lin and Todo, 2005; Franklin and Quail, 2010). The Arabidopsis PHY photoreceptor family consists of five members (PHYA to PHYE), which function as molecular light switches. In the dark, PHYs are present in their inactive red light-absorbing (Pr) form (λmax = 660 nm). After capturing a photon by the covalently bound linear tetrapyrrole chromophore, they are converted to the active far-red light-absorbing conformer (Pfr), which initiates downstream signaling events in the cytosol or in the nucleus. The active Pfr form is converted to Pr by far-red light (λmax = 730 nm; Rockwell et al., 2006). The Pfr conformers of PHYs are translocated to the nuclei, where they form characteristic nuclear bodies (Kircher et al., 2002). The exact composition and function of nuclear bodies are not yet known, but they may represent multiprotein complexes, where PHYs interact with transcription factors and other regulatory proteins to control the expression of light-induced genes (Bae and Choi, 2008).
The involvement of PHYA, PHYB, PHYD, and PHYE has been demonstrated in the function of red light input to the clock (Somers et al., 1998; Devlin and Kay, 2000; Yanovsky et al., 2001). PHY signaling affects transcription rate and mRNA and protein turnover of several clock components, although the signal transduction pathways linking these events with light-activated PHYs are largely unknown (Kozma-Bognar and Kaldi, 2008). On the other hand, several reports have demonstrated that PHY-mediated continuous red light signals inversely affect the free-running period length in a fluence rate-dependent manner (parametric entrainment), and discrete red light pulses elicit characteristic phase shifts of the clock and overt circadian rhythms (nonparametric entrainment; Millar, 2003). The role of PHYB has been seen in both processes. It has been shown by several research groups that the absence of PHYB function (i.e. in phyB mutants) results in a long period of the rhythmic transcription of the CHLOROPHYLL A/B BINDING2 (CAB2) gene at higher fluences of continuous red light (Somers et al., 1998; Devlin and Kay, 2000; Hall et al., 2002), whereas no period alterations were observed in phyB mutants in continuous white light (Hall et al., 2002; Salome et al., 2002). Interestingly, phyB-9 but not phyB-1 displayed an early phase of CAB2:LUC expression under these conditions (Hall et al., 2002; Salome et al., 2002). This could indicate allele- and/or ecotype-specific effects of PHYB function or could reflect the functional interaction of the PHY- and CRY-mediated signaling to the clock in white light, the precise spectral composition and fluence rate of which are difficult to reproduce. Using leaf-movement assays, it also has been shown that PHYB is required for red light-induced phase shifts (Yanovsky et al., 2001). It should be noted that in all the experiments reported so far, the function of PHYB as a circadian input photoreceptor was evaluated on the basis of altered output rhythms (CAB2, PHYB expression, leaf movements), but the rhythmic expression of core clock components has not been tested yet.
PHYB was first identified by the characteristic photomorphogenic phenotype of phyB mutants, which show elongated hypocotyls, the lack of hypocotyl hook opening, and reduced cotyledon expansion in red light (Reed et al., 1993). The molecular pathway by which PHYB controls photomorphogenesis involves the nuclear translocation of PHYB Pfr and its interaction with regulatory proteins (Bae and Choi, 2008; Fankhauser and Chen, 2008). The initiation of flowering is a complex developmental process in Arabidopsis partly controlled by the rhythmically expressed transcriptional activator CONSTANS (CO) protein (Imaizumi and Kay, 2006). Light signals mediated by PHYB destabilize CO protein specifically in the morning (Valverde et al., 2004). As a result, phyB mutants flower earlier in any light conditions tested so far (Endo et al., 2005).
The characteristic domain structure of PHYs and analysis of phy mutants displaying altered light sensing or signaling capabilities suggested that the N-terminal domain is required for light absorption, whereas downstream signaling cascades are activated by the C-terminal domain (Park et al., 2000). However, it has been shown that N-terminal fragments of PHYB containing 651 or 450 amino acids of the receptor fused to the bacterial GUS protein (providing dimerization motifs) and nuclear localization signals (NLS) were biologically active in regulating photomorphogenesis (Matsushita et al., 2003; Oka et al., 2004). These reports have also demonstrated that the 651-amino acid, but not the 450-amino acid, N-terminal fragment retained the function of PHYB in controlling flowering. These results indicated that the His kinase-like subdomain, which is located within the C-terminal part of PHYB, is dispensable for downstream signaling mediating photomorphogenesis and flowering.
However, information on the circadian functions of N-terminal fragments of PHYB of different lengths or the identification of the minimal PHYB fragment required for certain functions of the receptor are still missing. To address this question, a series of fusion proteins using 651, 450, or 410 amino acids of the N-terminal part of PHYB were expressed in the phyB-9 mutant background. Instead of the relatively large GUS protein, a well-defined short dimerization domain of the transcription factor CPRF4 (Kircher et al., 1998) was fused to the PHYB fragments to facilitate the formation of homodimers. Subcellular localization was controlled by short NLS or nuclear exclusion signals (NES). All fusion proteins carried the yellow fluorescent protein (YFP) for easy detection of the fusion proteins in vitro and in vivo. Here, we demonstrate that both the 651- and 450-amino acid N-terminal fragments of PHYB mediate entrainment of the circadian clock. We show that this function of PHYB requires nuclear localization of the receptor. We provide evidence that the 410-amino acid N-terminal fragment lacks any physiological functions of PHYB, because it fails to autoligate the chromophore required for light absorption. Moreover, detailed quantitative analysis of core clock gene expression in phyB-9 indicates that PHYB affects the pace of the morning and the evening loops to different extents, which results in the apparent decoupling of the two loops under specific light conditions.
RESULTS
Structure, Expression, and Subcellular Localization of N-Terminal PHYB Fragments in Transgenic Plants
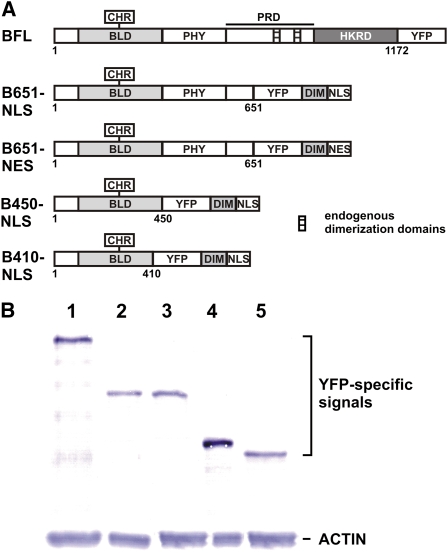
To test the functions of PHYB derivatives of different lengths, truncated versions containing 651, 450, and 410 amino acid residues were expressed in fusion with the YFP, a dimerization domain, and a NLS in the phyB-9 mutant background (B561-NLS, B450-NLS, and B410-NLS, respectively). The wild-type full-length PHYB protein tagged with YFP was used as a positive control (BFL), whereas B651-NES, which contains a NES instead of the NLS, was generated to test the effect of subcellular localization of PHYB on its function. Figure 1A shows the structures of the fusion proteins and the characteristic domain structure of the wild-type PHYB receptor. The approximately 120-kD PHYB folds into two major domains, the N-terminal and the C-terminal parts, which are separated by a flexible hinge region. The very N-terminal end of phytochromes plays a role in regulating the stability of the Pfr conformer (Sweere et al., 2001; Trupkin et al., 2007). The bilin-lyase motif carries the covalently bound tetrapyrrole chromophore phytochromobilin (Wu and Lagarias, 2000). The phytochrome domain (PHY) contributes to the integrity of Pfr (Montgomery and Lagarias, 2002). The first half of the C-terminal part, called the PAS-related domain, contains two PAS-like subdomains, a core regulator region termed the Quail box, and dimerization motifs. A His kinase-related domain showing homology to bacterial two-component sensor kinases is located at the C terminus of the protein (Rockwell et al., 2006). Expression of the different PHYB fragments was tested by western-blot analysis (Fig. 1B). B450-NLS was expressed at a slightly higher level; BFL, B651-NLS, B651-NES, and B410-NLS showed comparable expression levels in transgenic lines selected for further experiments. Subcellular localization of the fusion proteins was tested by confocal laser scanning and epifluorescence microscopy. Confocal images shown in Figure 2 report YFP fluorescence data from 1-μm optical sections of cells and convincingly demonstrate that the vast majority of NLS- or NES-tagged PHYB derivatives are localized within or outside the nucleus, respectively. The NLS-tagged forms showed diffuse distribution in the nucleus, whereas BFL formed nuclear bodies in the nuclei of etiolated plants illuminated with red light for 24 h, in agreement with previously published results. Epifluorescence images shown in Supplemental Figure S1 demonstrate that nuclear translocation and the formation of nuclear bodies by BFL are light dependent, whereas the intracellular distribution pattern of B651-NLS or B651-NES was unaffected by the light conditions. These results verify the correct expression and subcellular compartmentalization of the truncated PHYB fragments.
Figure 1.
Structure and expression of fusion proteins containing truncated derivatives of PHYB. A, Schematic illustration of the truncated PHYB constructs. BLD, Bilin lyase domain; CHR, chromophore; PHY, phytochrome domain; PRD, PAS-related domain; HKRD, His kinase-related domain; DIM, dimerization domain. Expression of all constructs was driven by the 35S promoter from Cauliflower mosaic virus. B, Western-blot analysis of PHYB protein expression levels. Seedlings expressing BFL (lane 1), B651-NLS (lane 2), B651-NES (lane 3), B450-NLS (lane 4), or B410-NLS (lane 5) were grown in 12-h-white-light/12-h-dark cycles for 7 d before collecting samples for protein extraction. A GFP-specific antibody was used to detect PHYB fusion proteins. [See online article for color version of this figure.]
Figure 2.
Subcellular localization of PHYB derivatives. Panels on the left show confocal laser scanning microscopy images of hypocotyl epidermal cells expressing the indicated PHYB derivatives. Four-day-old etiolated plants were irradiated with red light for 24 h prior to microscopic analysis. Confocal images of YFP fluorescence represent optical sections of about 1 μm. Panels on the right show the corresponding light transmission images. Each frame represents an area of 48 × 48 μm. nu, Nucleus.
Differential Complementation of Photomorphogenic and Flowering Time Phenotypes of phyB-9 by PHYB Derivatives
To test the effectiveness of the different PHYB fusion proteins to inhibit hypocotyl elongation, seedlings were grown at different fluence rates of red light for 4 d and hypocotyl lengths were measured. Figure 3A shows that BFL, B651-NLS, and B450-NLS could rescue the long-hypocotyl phenotype of phyB-9, whereas B651-NES and B410-NLS were ineffective. BFL was hypersensitive compared with ecotype Columbia-0 (Col-0) at all fluence rates. B651-NLS and B450-NLS showed hypersensitivity at medium, but especially at low, fluence rates. Similar results were obtained from plants grown in continuous white light or in light/dark cycles (data not shown). Mutations in the PHYB gene cause early flowering under long-day (Neff and Chory, 1998), short-day, or even continuous light conditions (Endo et al., 2005). To test if the truncated PHYB fragments are able to restore the early-flowering phenotype of phyB-9, plants were grown in long-day (16-h-white light/8-h-dark) conditions and flowering time was determined as the number of rosette leaves at the time of bolting. Figure 3B demonstrates that BFL and B651-NLS flowered as the wild type, but B651-NES, B450-NLS, and B410-NLS were indistinguishable from phyB-9.
Figure 3.
Complementation of photomorphogenic and flowering time phenotypes of phyB-9 by PHYB fragments. A, Fluence rate curve of hypocotyl elongation in continuous red light. Wild-type (Col-0), phyB-9, and transgenic seedlings expressing the truncated versions of PHYB in the phyB-9 background were grown in constant red light at the indicated fluence rates for 4 d, then the hypocotyl length of the seedlings was measured. Values were normalized to the hypocotyl length of the corresponding dark-grown seedlings. B, Flowering time of plants grown under long days (16 h of white light/8 h of dark). Flowering time was determined as the number of rosette leaves at the time of the emergence of inflorescence. Error bars represent se.
These results indicate that the 651-amino acid N-terminal fragment of PHYB acts as a functional photoreceptor if it is localized in the nucleus, whereas B410-NLS is not sufficient to regulate the light-dependent physiological processes tested. Interestingly, B450-NLS could mediate signaling to inhibit hypocotyl elongation, but it was unable to function in flowering time determination.
The phyB-9 Mutation Affects Rhythmic Transcription of Central Clock Components in a Different Manner
It has been demonstrated that loss of PHYB function results in a long-period phenotype of the CAB2:LUC marker at medium and high fluence rates of continuous red light (Somers et al., 1998). However, it was presumed, but not directly tested, that rhythmic expression of core clock components is affected in the same way. To test this, CAB2:LUC+, CCA1:LUC+, TOC1:LUC+, GI:LUC+, and PRR9:LUC+ markers were introduced in Col-0 and phyB-9 backgrounds, and free-running period length and circadian phase of the expression of these markers were tested at different fluence rates of red light. Figure 4 shows that, according to Aschoff's rule, period length of all markers decreased with increasing light intensity and that the phyB-9 mutation did not eliminate this response. phyB-9 plants showed longer periods at most fluence rates tested. However, two classes of genes could be established based on the actual effect of the phyB-9 mutation on the period of their rhythmic transcription. CCA1:LUC+ and PRR9:LUC+ showed relatively large period differences (approximately 1 h) between Col-0 and phyB-9 over the entire fluence rate range (Fig. 4, B and C). In contrast, TOC1:LUC+ and GI:LUC+ displayed smaller, but significant, period differences (approximately 0.5 h), mainly at medium fluence rates (Fig. 4, D and E). Interestingly, in the case of both markers, Col-0 and phyB-9 plants showed identical periods at the lowest fluence rate tested (10 μmol m−2 s−1). Moreover, the period of GI:LUC+ was not lengthened in the phyB-9 mutant at the highest fluence rate. TOC1:LUC+ showed longer, although less significant, periods in phyB-9 under these conditions. The period length of the circadian output marker CAB2:LUC+ showed characteristics similar to those of CCA1:LUC+ and PRR9:LUC+ (Fig. 4A). The phyB-9 mutation also affected the circadian phase of rhythmic expression of marker genes differently. PRR9:LUC+ and TOC1:LUC+ showed 1- to 2-h phase delays over the entire fluence rate range tested (Fig. 4, H and I). In contrast, CAB2:LUC+, CCA1:LUC+, and GI:LUC+ showed no significant phase changes in their expression patterns at any fluence rate (Fig. 4, F, G, and J). These results indicate that PHYB provides different quality and/or quantity of input signals to the morning (CCA1, PRR9) and the evening (TOC1, GI) loops of the plant circadian oscillator in a fluence rate-dependent manner, which results in an apparent decoupling of the two loops at certain light intensities.
Figure 4.
Analysis of the circadian phenotype of phyB-9 in continuous red light. Wild-type Col-0 (white squares) and phyB-9 (black squares) plants expressing CAB2:LUC+ (A and F), CCA1:LUC+ (B and G), PRR9:LUC+ (C and H), TOC1:LUC+ (D and I), or GI:LUC+ (E and J) were grown in 12-h-white-light/12-h-dark cycles for 7 d and transferred to different fluence rates of continuous red light. The free-running period length (A–E) and phase (F–J) of luminescence rhythms were determined and plotted as a function of fluence rate. Phase values were converted to circadian time (CT). Asterisks indicate statistically significant differences between Col-0 and phyB-9 as determined by Student's two-tailed heteroscedastic t test: ** P < 0.01, * P < 0.05.
Complementation of the Circadian Period Phenotype of phyB-9 by PHYB Derivatives in Continuous Red Light
To test the ability of different PHYB fragments to restore the period phenotype of phyB-9, CCA1 and TOC1 mRNA rhythms were monitored by quantitative reverse transcription (qRT)-PCR assays in the different complemented lines. A medium fluence rate of continuous red light (50 μmol m−2 s−1) was used as a condition for free running, since all clock genes showed significant period lengthening in their transcription in this condition (Fig. 4, B–E). Tissue samples were harvested over a 72-h time course starting 60 h after the transfer to continuous red light. Figure 5 clearly shows that CCA1 and TOC1 expression had longer periods in phyB-9 in these conditions, which verifies the previous results (Fig. 4, B and D). The data also demonstrate that loss of PHYB function resulted in a lower amplitude of rhythmic CCA1 expression, but the amplitude of TOC1 expression was apparently unaffected. BFL, B651-NLS, and B450-NLS fully restored the period phenotype for both CCA1 and TOC1, but B651-NES and B410-NLS retained the phyB-9 phenotype. The ectopic expression of the functional PHYB fragments has not resulted in period lengths visibly shorter than that of the wild type. However, the amplitude of CCA1 and TOC1 expression was significantly increased in BFL plants (Fig. 5, A and F), whereas B651-NLS showed a similar increase in the amplitude of CCA1 expression only (Fig. 5B). The amplitude of rhythmic mRNA accumulation was unaffected by other PHYB derivatives. Since it is difficult and inaccurate to estimate period length from rhythmic mRNA data, the CCR2:LUC+ marker was introduced in the various backgrounds in order to obtain quantitative period length estimates in the complementing lines. Supplemental Figure S2 shows period estimates of CCR2:LUC+ expression from plants assayed in continuous red light at 50 μmol m−2 s−1. The period length of CCR2:LUC+ expression was approximately 1 h longer in phyB-9 compared with the wild type. Consistent with the mRNA data, BFL and B651-NLS complemented this phenotype, whereas B410-NLS did not. These results demonstrate that (1) N-terminal PHYB fragments containing the first 450 amino acids of PHYB are fully active in light signaling to the clock in continuous red light conditions, and (2) nuclear localization of PHYB is essential for this function.
Figure 5.
Complementation of the long-period phenotype of phyB-9 by PHYB fragments in continuous red light. Wild-type Col-0, phyB-9, and transgenic seedlings expressing BFL (A and F), B651-NLS (B and G), B651-NES (C and H), B450-NLS (D and I), or B410-NLS (E and J) in the phyB-9 background were grown in 12-h-white-light/12-h-dark cycles for 7 d and transferred to continuous red light at 50 μmol m−2 s−1 fluence rate. Samples were harvested at 3-h intervals starting at 60 h after the transfer for 3 d. CCA1 (A–E) and TOC1 (F–J) mRNA levels were determined by qRT-PCR. Data normalized to TUBULIN2/3 levels are shown. Col-0 (green lines) and phyB-9 (red lines) data are shown on each graph. Data from complementing lines are shown by blue lines.
B651-NLS Mediates Resetting of the Clock by Red Light Pulses
The circadian clock free running in darkness responds with characteristic phase shifts to light pulses. The magnitude and direction (delay or advance) of the phase shift depends on the actual phase of the clock at the time when the light pulse is given. Generally, light pulses applied during the first or second half of the subjective night trigger phase delays or advances, respectively (Covington et al., 2001). The role of PHYB in resetting the phase of leaf-movement rhythms in response to red light pulses has been suggested (Yanovsky et al., 2001). To test the function of PHYB fragments in this particular response, plants expressing CCR2:LUC+ in different genetic backgrounds were transferred to constant darkness at dusk (Zeitgeber time 12 [ZT12]) after 7 d of entrainment, and rhythmic luminescence was monitored. ZT0 is defined as the time of the last dark/light transition before transfer to constant conditions. A single red light pulse (100 μmol m−2 s−1 for 1 h) was applied to half of the plants after 29 h in constant dark, when the largest phase delays are expected (Kevei et al., 2007). Phase shifts were calculated by comparing the phase of CCR2:LUC+ expression in the induced plants with those of the noninduced control plants. Supplemental Figure S3 shows that wild-type plants produced approximately 6-h phase delays under these conditions, whereas the response was significantly reduced, but not eliminated, in the phyB-9 mutant. Similar to complementation of the long-period phenotype, BFL and B651-NLS were able to restore the wild-type response, but B410-NLS was not functional at all. These data suggest that N-terminal fragments of PHYB can mediate not only the effect of continuous light on the pace of the clock but also the effect of short light pulses on the phase of the clock.
The Effect of PHYB on the Clock in Continuous White Light
It has been demonstrated earlier that absence of PHYB function does not change the period of CAB2:LUC(+) expression in continuous white light, which was explained by the contribution of all the other PHY and CRY photoreceptors to entrainment under this condition (Hall et al., 2002; Salome et al., 2002). These reports have also demonstrated that phyB-9 but not phyB-1 mutants show an early phase of CAB2:LUC(+) expression in continuous white light. To investigate the role of PHYB in circadian entrainment in white light, the period and phase of core clock gene expression were analyzed in Col-0 and phyB-9 plants transferred to continuous white light. Surprisingly, with the exception of PRR9, transcription of all clock genes showed a short-period phenotype in phyB-9 (Fig. 6A). The phase of CCA1:LUC+ and TOC1:LUC+ remained unchanged, but GI:LUC+ and PRR9:LUC+ showed an advanced phase of expression in phyB-9 (Fig. 6B). CAB2:LUC+ showed no period change but a slight early-phase phenotype in phyB-9, which is consistent with earlier results (Salome et al., 2002). In contrast, expression of another clock output marker, CCR2:LUC+, displayed significant period shortening and slight phase advance in the mutant plants. These data demonstrate that the absence of PHYB function has markedly different effects on the circadian oscillator in white and red light conditions, which suggests a complex interaction between the red and blue light input pathways to the circadian clock. To identify the particular domain of PHYB responsible for this functional interaction, CCR2:LUC+ rhythms were monitored in transgenic phyB-9 lines expressing different truncated derivatives of PHYB. The data presented in Figure 6C show that only the full-length PHYB, but none of the N-terminal derivatives, was able to restore the wild-type period of CCR2:LUC+. These results indicate that, in contrast to red light, the C-terminal domain of PHYB is required for normal entrainment in white light conditions.
Figure 6.
Circadian phenotypes of phyB-9 in continuous white light. Seedlings expressing different luciferase reporters and/or N-terminal derivatives of PHYB as indicated were grown in 12-h-white-light/12-h-dark cycles for 7 d and transferred to continuous white light at 60 μmol m−2 s−1 fluence rate. A, Period estimates for CCA1:LUC+, TOC1:LUC+, GI:LUC+, PRR9:LUC+, CAB2:LUC+, and CCR2:LUC+ expression in Col-0 (black bars) and phyB-9 mutant (white bars) seedlings. B, Phase estimates for CCA1:LUC+, TOC1:LUC+, GI:LUC+, PRR9:LUC+, CAB2:LUC+, and CCR2:LUC+ expression in Col-0 (black bars) and phyB-9 mutant (white bars) seedlings. C, Period estimates for CCR2:LUC+ expression in Col-0, phyB-9, BFL, B651-NLS, and B410-NLS seedlings. Asterisks indicate statistically significant differences between Col-0 and phyB-9 as determined by Student's two-tailed heteroscedastic t test: ** P < 0.01, * P < 0.05.
DISCUSSION
PHYB is the dominant red/far-red light-absorbing photoreceptor in light-grown Arabidopsis plants. PHYB provides signals to regulate photomorphogenesis, to control flowering, and to entrain the circadian clock. Consequently, phyB mutants display long hypocotyls, accelerated flowering, and long periods of circadian rhythms under specific light conditions (Reed et al., 1993; Somers et al., 1998; Endo et al., 2005). The PHYB receptor folds into two main domains: the chromophore-bearing photosensory N-terminal domain and the C-terminal domain containing a His kinase-like subdomain and protein motifs for dimerization and nuclear translocation. It has been demonstrated that dimers of the N-terminal domain of PHYB possess full PHYB function regarding photomorphogenesis and flowering time determination it they are targeted to the nucleus by added foreign NLS motifs (Matsushita et al., 2003). In other words, the His kinase domain is dispensable for these functions of PHYB, and the main role of the C-terminal part is to provide a platform for dimerization and to control the entry of the protein in the nucleus.
To test the function of the N-terminal domain to provide light signaling to the circadian clock, we expressed the 651-amino acid N-terminal fragment fused to YFP, dimerization domains, and NLS or NES protein motifs (B651-NLS and B651-NES, respectively) in the phyB-9 background, which lacks any PHYB functions (Reed et al., 1993). The fusion proteins, including the YFP-tagged full-length PHYB control (BFL), were expressed at comparable levels (Fig. 1B) and showed the expected subcellular localization (Fig. 2; Supplemental Fig. S1). The light input pathway to the circadian oscillator mediates two modes of entrainment depending on the duration of illumination. In constant light conditions, the activity of the input pathway shortens the free-running period length with increasing light intensity (parametric entrainment). In constant darkness, however, short light pulses elicit discrete phase advances or delays of the oscillator and overt circadian rhythms (nonparametric entrainment; Devlin and Kay, 2001). Red light signals absorbed by PHYB contribute to both types of entrainment (Somers et al., 1998; Yanovsky et al., 2001). Figures 4 and 5 show that rhythmic expression of core oscillator genes has a longer period in phyB-9 plants compared with the wild type in continuous red light at 50 μmol m−2 s−1 fluence rate. Supplemental Figure S3 shows that in the absence of PHYB function, red light pulses trigger smaller phase shifts of the oscillator. Our results demonstrated that B651-NLS was as effective at restoring these phenotypes of the phyB-9 mutant as the full-length PHYB. Interestingly, BFL (and B651-NLS) showed significant increases in the amplitude of mRNA rhythm of CCA1 but did not cause shorter periods. It has been reported that overexpression of PHYB results in a short period of CAB:LUC expression in continuous red light (Somers et al., 1998; Hall et al., 2002) and in early flowering under both short-day and long-day conditions (Bagnall et al., 1995). The lack of circadian and flowering-time phenotypes of BFL can be explained by similar PHYB levels in BFL and wild-type plants, in contrast to lines used in the cited studies, which show at least 15-fold overexpression of the PHYB protein. In contrast to BFL and B651-NLS, B651-NES was unable to restore PHYB function to regulate rhythmic CCA1 or TOC1 expression (Fig. 5, C and H). These results demonstrate that the N-terminal domain of PHYB is able to mediate all aspects of entrainment of the circadian clock, and this function requires nuclear localization of PHYB. The data presented here also verify previous reports, since B651-NLS but not B651-NES restored the photomorphogenic and flowering phenotypes of phyB-9.
The molecular mechanism by which nucleus-localized PHYB mediates entrainment is unclear, but most likely it involves interaction of PHYB with regulatory proteins, which indirectly or directly affect the oscillator. Among such clock-affecting proteins, ZEITLUPE (ZTL) and EARLY FLOWERING3 (ELF3) have been shown to interact with PHYB in yeast or in vitro (Jarillo et al., 2001; Liu et al., 2001). ZTL is an F-box protein that targets TOC1 proteins for ubiquitination by Skp1/Cullin/F-box-type E3 ubiquitin ligases and for subsequent degradation (Mas et al., 2003) and severely affects the free-running period length (Somers et al., 2000; Kevei et al., 2006). ELF3 is a clock-controlled nuclear protein that attenuates light-induced resetting of the clock (Covington et al., 2001). However, both ZTL and ELF3 bind to the C-terminal domain of PHYB, which is dispensable for circadian function in red light according to our data; therefore, these interactions have no relevance to entrainment under these conditions.
In fact, PIF3 has been shown to interact with the N-terminal domain of PHYB Pfr (Shimizu-Sato et al., 2002) and also with TOC1 (Yamashino et al., 2003). Since PIF3 potentially binds to the G-box element located in the promoter of CCA1 and LHY genes, the PHYB-PIF3-TOC1 complex could provide a mechanism for directing regulatory light signals to certain core oscillator genes (Martinez-Garcia et al., 2000). It has been shown that PIF1, PIF4, and PIF5 also interact with TOC1 (Yamashino et al., 2003). However, misexpression of PIF3 or PIF5 does not affect entrainment of the plant circadian clock (Fujimori et al., 2004; Viczian et al., 2005). Although the lack of such phenotypes could be explained by possible redundant coaction of several PIF3-like transcription factors, PIFs are probably not the terminal components of PHYB-mediated red light input to the clock but rather represent components of clock-controlled output processes (e.g. rhythmic hypocotyl elongation; Nozue et al., 2007).
Oka et al. (2004) reported that a short fragment containing 450 amino acids of the N-terminal domain of PHYB mediates photomorphogenic responses in the nucleus. We created phyB-9 plants expressing this PHYB derivative in fusion with dimerization and NLS signals (B450-NLS). Analysis of these transgenic lines verified that the PHY subdomain of the N-terminal part of PHYB is not required for the inhibition of hypocotyl elongation (Fig. 3A) and revealed that this subdomain is also dispensable for the regulation of period length. Figure 5, D and I, demonstrates that B450-NLS rescued the wild-type expression pattern of CCA1 and TOC1. Unlike the photomorphogenic and circadian phenotypes, the early-flowering phenotype of phyB-9 was not restored by B450-NLS. Figure 3B shows that B450-NLS plants grown in long days (16 h of light/8 h of dark) flowered at the same time as phyB-9 mutants. This result is consistent with those reported earlier (Oka et al., 2004). This observation indicates that the PHY subdomain may provide a binding site for a yet unidentified factor that is necessary to transmit signals eventually destabilizing CO protein and delaying flowering. The facts that PHYB acts on CO protein levels and that the flowering phenotype of phyB mutants is independent of photoperiod demonstrate that the early flowering of phyB mutants does not arise from altered clock function. Our results further support this conclusion, because B450-NLS plants generate wild-type circadian rhythms yet display the early-flowering phenotype of phyB-9.
To define the minimal N-terminal PHYB fragment, which is still functional in the nucleus, we created transgenic plants expressing a 410-amino acid derivative of PHYB (B410-NLS). Despite the fact that this version is only 40 amino acids shorter than B450-NLS, B410-NLS was unable to complement any phenotype of phyB-9 in any assays. It must be noted that the effect of both B651-NLS and B450-NLS was light dependent, and phenotypes indicating constitutive light-independent signaling were not observed in these plants in darkness (Matsushita et al., 2003; Oka et al., 2004; data not shown). This means that light-induced conformational change is a prerequisite of the biological function of these derivatives. To undergo light-induced Pr ↔ Pfr conversions, the chromophore must be attached to the PHYB apoprotein. Zinc blots were used to test the chromophore-binding capability of the different PHYB derivatives (Oka et al., 2004). Supplemental Figure S4 demonstrates that B651 and B450 were able to autoligate the chromophore. In contrast, B410 failed to incorporate the chromophore, indicating that the deletion probably affected the function of the bilin lyase domain.
Quantitative analysis of clock gene expression in the phyB-9 mutant revealed an unexpected complexity in the function of the light input to the circadian oscillator. The plant oscillator consists of three interlocked transcriptional/translational feedback loops. The “morning” loop is operated by interactions among CCA1/LHY and PRR7/9 genes, the “evening” loop is based on the cross-regulation between TOC1 and Y/GI, and the two loops are coupled by a central loop formed by CCA1/LHY, TOC1, and a yet unidentified X factor. In wild-type plants, these loops are coupled together, which is illustrated by stable and constant phase relationships of expression of these genes under free-running conditions. Theoretically, the morning and evening loops could be uncoupled from each other in certain conditions, for example, as a result of the misfunction of X. Data in Figure 4 demonstrate that loss of PHYB function affects the pace of the morning and evening loops differently. Expression of CCA1 and PRR9, components of the morning loop, shows significant long-period rhythm in phyB-9 over a wide range of fluence rates of continuous red light, but TOC1 and GI expression displays less significant period lengthening. Importantly, at lower fluences of red light, the pace of the evening loop is not affected by the lack of phyB-9, whereas the morning loop runs at a lower pace in the same conditions. These data demonstrate that, in these conditions, the coupling between the morning and evening loops has been weakened in phyB-9. Although the exact molecular mechanism of this phenomenon and the role of PHYB in the apparent decoupling remain unclear, our results shed light on the complexity of the function of the light input to the plant circadian oscillator. This complexity was further expanded by the analysis of circadian gene expression in the phyB-9 mutant in continuous white light (Fig. 6). Our data showed that PHYB has nearly opposite effects on the pace of the clock in red and white light, since circadian period lengths in the phyB-9 mutant are generally lengthened or shortened in continuous red or white light conditions, respectively. On the other hand, it has been demonstrated that the phyB-1 mutation has no effect on period length in monochromatic blue light (Devlin and Kay, 2000). The cool-white fluorescent tubes used in this experiment provide light enriched in blue light but still emit red light capable of PHYB activation. This is indicated by the long hypocotyls of phyB-9 seedlings under this condition (data not shown). It follows that the short-period phenotype phyB-9 is apparent upon simultaneous activation of the red and blue light signaling pathways to the clock, which could be explained by a negative effect of PHYB on blue light signaling. The unexpected short-period phenotype in continuous white light has been observed for leaf-movement rhythms in the quintuple phy mutant (Strasser et al., 2010), also suggesting that blue light input to the clock is attenuated by the action of phytochrome(s). Since PHYB appears to have a positive effect on CRY2 signaling (Mas et al., 2000), other routes of blue light input may be affected. However, molecular details of this particular functional interaction between red and blue light signaling to the clock remain to be elucidated. We showed that BFL, but not B651-NLS, was able to complement the short-period phenotype of phyB-9 in white light (Fig. 6C). Since the BFL and B651-NLS proteins were expressed at similar levels (Fig. 1B), the lack of complementation by B651-NLS is most probably due to absence of the C-terminal domain of PHYB.
In conclusion, our data suggest that the nucleus-localized N-terminal domain of PHYB is fully functional in regulating photomorphogenesis, flowering time, and red light-dependent entrainment of the circadian clock. However, the C-terminal domain is essential for proper circadian entrainment in white light conditions. To explain the requirement of the C-terminal domain for this process, we propose that this domain mediates integration of the blue and red light signaling pathways to the clock.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type and phyB-9 mutant Arabidopsis (Arabidopsis thaliana) plants in the Col-0 background were used in this study. Surface-sterilized seeds were grown in 12-h-white-light (50 μmol m−2 s−1)/12-h-dark cycles at 22°C (MLR-350; Sanyo) for 7 d before being transferred to continuous darkness at ZT12 or to continuous light at ZT0, and all measurements were carried out at constant 22°C. ZT0 is defined as the time of the last dark/light transition before transfer to constant conditions. Illumination was provided by cool-white fluorescent tubes or monochromatic light-emitting diode red light sources (λmax = 667 nm; Quantum Devices).
Construction of Transgenic Plants
The CAB2:LUC+, CCR2:LUC+, CCA1:LUC+, and TOC1:LUC+ constructs have been described (Toth et al., 2001; Doyle et al., 2002; Locke et al., 2006). GI and PRR9 promoter fragments were obtained by PCR performed on genomic DNA of wild-type plants. Unique restriction sites were designed at the 5′ and 3′ ends of the promoter fragments to facilitate cloning in the pPCV-LUC+ binary vector (Toth et al., 2001). The amplified fragments contained the entire 5′ untranslated region but not the ATG of the corresponding genes. Fragment lengths and restriction sites at the 5′ and 3′ ends were as follows: GI, 2,749 bp, XbaI-BamHI; PRR9, 1,330 bp, SalI-BamHI.
DNA fragments encoding the SV40 NLS motif (Kalderon et al., 1984) and the dimerization domain of CPRF4 (Kircher et al., 1998) were the same as described (Pfeiffer et al., 2009). The DNA fragment encoding the NES motif (LALKLAGLDINKTGG) from the heat-stable inhibitor (PKI; Wen et al., 1995) was generated by annealing the following oligonucleotides: 5′-CAAGCTTAACGAGCTTGCTCTTAAGTTGGCTGGACTTGATATTAACAAGACTGGAGGATAGGAGCT-3′ and 5′-CCTATCCTCCAGTCTTGTTAATATCAAGTCCAGCCAACTTAAGAGCAAGCTCGTTAAGCTTGAGCT-3′. The modified pPCV812 binary vectors containing the 35S promoter and DNA fragments coding for YFP, dimerization domain, and NLS or NES motifs were created as described (Pfeiffer et al., 2009). cDNA fragments encoding 651, 450, or 410 amino acid residues of the N-terminal domain of PHYB were amplified by PCR and cloned in the modified pPCV812 vectors. The final constructs were verified by sequencing and were transferred to Agrobacterium tumefaciens GV3101. The constructs were transformed into Col-0 and/or phyB-9 (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog medium supplemented with 15 μg mL−1 hygromycin. Ten to 15 independent transformants for each construct were self-fertilized, and individuals of the T2 or the homozygous T3 progeny were used for luminescence or complementation assays, respectively. To create lines coexpressing the different PHYB derivatives and the CCR2:LUC+ marker, the CCR2:LUC+ construct was transferred in the pPCVB812 vector, which carries a BASTA resistance marker (Bauer et al., 2004). Selected homozygous lines expressing different N-terminal fragments of PHYB were transformed with the CCR2:LUC+ pPCVB812 construct, and transformants were selected based on their resistance to BASTA herbicide.
Analysis of Luminescence Rhythms
Luciferase activity was determined by measuring single seedlings with an automated luminometer (TopCount NXT; Perkin-Elmer) for 2 to 7 d as described previously (Kevei et al., 2006). For fluence rate curves, circadian periods of luminescence rhythms were measured in seedlings transferred to constant illumination of red or white light at the fluence rates indicated. All rhythm data were analyzed with the Biological Rhythms Analysis Software System (available at http://www.amillar.org) running fast Fourier-transform nonlinear least-squares estimation. Variance-weighted mean periods within the circadian range (15–40 h) and se values were estimated as described from 10 to 36 traces per genotype. Phase values were determined as the time of the first full peak of luminescence rhythms in continuous light. Phase values were normalized to free-running period length and are shown as circadian time (Salome et al., 2002). Statistical analysis was done using Student's two-tailed heteroscedastic t test. Experiments were repeated three or four times, and representative data sets are shown. For each genotype, at least three independent transgenic lines were analyzed.
For phase-shift experiments (Supplemental Fig. S3), plants expressing CCR2:LUC+ were grown in 12-h-white-light/12-h-dark cycles for 7 d and transferred to darkness at the last light-to-dark transition. CCR2:LUC+ luminescence was monitored at 1-h intervals for 5 d. After 29 h in constant dark, half of the plants were treated with a 1-h red light pulse at 100 μmol m−2 s−1 fluence rate. Phase values were determined as the time of the second peak after the light pulse and were normalized to the free-running period as above. Light-induced phase shifts were calculated by comparing the phase of CCR2:LUC+ rhythm in the induced and noninduced plants.
Analysis of Gene Expression
Total RNA extraction, cDNA preparation, and qRT-PCR assays were carried out as described (Kevei et al., 2007). CCA1 and TOC1 qRT-PCR primers have been described (Edwards et al., 2005). All graphs show mRNA levels relative to the TUBULIN2/3 mRNA transcript (Endo et al., 2007). Total protein extraction, western-blot analysis, and detection of YFP fusion proteins were done essentially as described (Kevei et al., 2007). The assays were repeated two or three times, and representative data are shown.
Chromophore-Binding Assay (Zinc Blot)
For in vitro reconstitution, 4 μm phycocyanobilin as chromophore was incubated with purified recombinant GST-B651, GST-B450, or GST-B410 protein as described (Lagarias and Lagarias, 1989). Each apoprotein was expressed in Escherichia coli and purified on a glutathione-Sepharose 4B column. Zn2+-induced fluorescence of chromophore-bound proteins was visualized under UV light after SDS-PAGE separation according to Oka et al. (2004).
Measurement of the Hypocotyl Length
For hypocotyl length measurements, seeds were sown on wet filter paper and incubated in the dark for 48 h at 4°C. Cold-treated seeds were then irradiated with 6 h of white light and transferred to 22°C and dark for an additional 18 h. After this treatment, seedlings were grown at different fluences of light for 4 d. Measurement of the hypocotyl length was performed using MetaMorph Software (Universal Imaging). Hypocotyl lengths of light-grown seedlings were normalized to the corresponding dark-grown hypocotyl length. Fluence rate curves for hypocotyl elongation were obtained by plotting relative hypocotyl lengths against the light intensities used in the experiment displayed on a logarithmic scale. Subcellular distribution of YFP fusion proteins was analyzed in 4-d-old seedlings as described previously (Bauer et al., 2004).
Epifluorescence and Confocal Laser Scanning Microscopy
Seeds were sown on a four-layer filter paper and imbibed in water in the dark for 48 h at 4°C. Cold-treated seeds were then transferred to 25°C, irradiated with 18 h of white light to induce homogeneous germination, and grown for additional days in the dark. Six-day-old dark-grown seedlings were then subjected to various light treatments as described.
For epifluorescence and light microscopy, seedlings were transferred to glass slides under dim-green safelight and analyzed with an Axioskop microscope (Zeiss). Excitation and detection of the YFP fluorophore were performed with YFP filter set (AHF Analysentechnik). Each experiment was repeated at least two times using at least five seedlings by analyzing the upper third of the hypocotyl. Representative cells were documented by photography with a digital Axiocam camera system (Zeiss). Documentation of cells was performed during the first 60 s of microscopic analysis to prevent the impact of microscopic light on localization patterns. Photographs were processed for optimal presentation using the Photoshop 7.0 (Adobe Systems Europe) and MS Office (Microsoft) software packages.
For confocal laser scanning microscopy, seedlings were transferred to glass slides under dim-green safelight and analyzed with an LSM 510 microscope equipped with the META device (Zeiss).
Optical sections of about 1 μm were achieved using a 63× Plan-Apochromat/numerical aperture 1.4 objective (Zeiss) and a pinhole size adjusted to 1 airy unit. For excitation of the YFP fluorophore, a 514-nm laser line was used, and the detection range of YFP emission was set in the META detector to 524 to 578 nm. In parallel to YFP analysis, transmission light images were taken.
Measurement of Flowering Time
Seeds were sown on soil and incubated for 2 d in darkness at 4°C. They were subsequently transferred to long-day (16-h-white-light/8-h-dark) conditions. Light sources were fluorescent (cool-white) tubes producing a fluence rate of 60 μmol m−2 s−1. Flowering time was recorded as the number of rosette leaves at the time when inflorescences reached 1 cm in height. The experiment was repeated twice using 30 to 40 plants per genotype.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of PHYB fusion proteins.
Supplemental Figure S2. Quantitative analysis of complementation of the long-period phenotype of phyB-9.
Supplemental Figure S3. The 651-amino acid N-terminal fragment of PHYB is sufficient to mediate red light-induced resetting of the circadian clock.
Supplemental Figure S4. Chromophore does not incorporate into the recombinant protein GST-B410.
Supplementary Material
Acknowledgments
We are grateful to Andrea Keszthelyi, Katalin Joszai, and Gabriella Veres for their excellent technical assistance.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bagnall DJ, King RW, Whitelam GC, Boylan MT, Wagner D, Quail PH. (1995) Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol 108: 1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. (2001) Circadian photoperception. Annu Rev Physiol 63: 677–694 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A. (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A. (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T. (2004) Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Bastow RM, Nagy F, Millar AJ. (2002) Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J 32: 529–537 [DOI] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR. (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. (1984) A short amino acid sequence able to specify nuclear location. Cell 39: 499–509 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Feher B, Toth R, Viczian A, Kircher S, Rea D, Dorjgotov D, Schafer E, Millar AJ, et al. (2007) Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol 17: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Hall A, Kozma-Bognar L, Kim WY, Eriksson ME, Toth R, Hanano S, Feher B, Southern MM, et al. (2006) Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognar L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adam E, Schafer E, Nagy F. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Ledger S, Hayashi H, Weisshaar B, Schafer E, Frohnmeyer H. (1998) CPRF4a, a novel plant bZIP protein of the CPRF family: comparative analyses of light-dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol Gen Genet 257: 595–605 [DOI] [PubMed] [Google Scholar]
- Kozma-Bognar L, Kaldi K. (2008) Synchronization of the fungal and the plant circadian clock by light. ChemBioChem 9: 2565–2573 [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Lagarias DM. (1989) Self-assembly of synthetic phytochrome holoprotein in vitro. Proc Natl Acad Sci USA 86: 5778–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Todo T. (2005) The cryptochromes. Genome Biol 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ. (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, Kay SA. (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Mas P, Yanovsky MJ. (2009) Time for circadian rhythms: plants get synchronized. Curr Opin Plant Biol 12: 574–579 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A. (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Millar AJ. (2003) A suite of photoreceptors entrains the plant circadian clock. J Biol Rhythms 18: 217–226 [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC. (2002) Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci 7: 357–366 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J. (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Suzuki T, Tokutomi S, Nagatani A. (2004) Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16: 2104–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CM, Bhoo SH, Song PS. (2000) Inter-domain crosstalk in the phytochrome molecules. Semin Cell Dev Biol 11: 449–456 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Kunkel T, Hiltbrunner A, Neuhaus G, Wolf I, Speth V, Adam E, Nagy F, Schafer E. (2009) A cell-free system for light-dependent nuclear import of phytochrome. Plant J 57: 680–689 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR. (2002) The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. (2002) A light-switchable gene promoter system. Nat Biotechnol 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD. (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schafer E, Harter K. (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin SA, Debrieux D, Hiltbrunner A, Fankhauser C, Casal JJ. (2007) The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol Biol 63: 669–678 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Viczian A, Kircher S, Fejes E, Millar AJ, Schafer E, Kozma-Bognar L, Nagy F. (2005) Functional characterization of phytochrome interacting factor 3 for the Arabidopsis thaliana circadian clockwork. Plant Cell Physiol 46: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Wu SH, Lagarias JC. (2000) Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochemistry 39: 13487–13495 [DOI] [PubMed] [Google Scholar]
- Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, Mizuno T. (2003) A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 619–629 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, Whitelam GC, Casal JJ. (2001) Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J Biol Rhythms 16: 523–530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.