Abstract
Although the primary structure of proteins, nucleic acids, and carbohydrates can be readily determined, no sequencing method has been described yet for the second most abundant biopolymer on earth (i.e. lignin). Within secondary-thickened plant cell walls, lignin forms an aromatic mesh arising from the combinatorial radical-radical coupling of monolignols and many other less abundant monomers. This polymerization process leads to a plethora of units and linkage types that affect the physicochemical characteristics of the cell wall. Current methods to analyze the lignin structure focus only on the frequency of the major monomeric units and interunit linkage types but do not provide information on the presence of less abundant unknown units and linkage types, nor on how linkages affect the formation of neighboring linkages. Such information can only be obtained using a sequencing approach. Here, we describe, to our knowledge for the first time, a sequencing strategy for lignin oligomers using mass spectrometry. This strategy was then evaluated on the oligomers extracted from wild-type poplar (Populus tremula × Populus tremuloides) xylem. In total, 134 lignin trimers to hexamers were observed, of which 36 could be completely sequenced. Interestingly, based on molecular mass data of the unknown monomeric and dimeric substructures, at least 10 unknown monomeric units or interunit linkage types were observed, one of which was identified as an arylglycerol end unit.
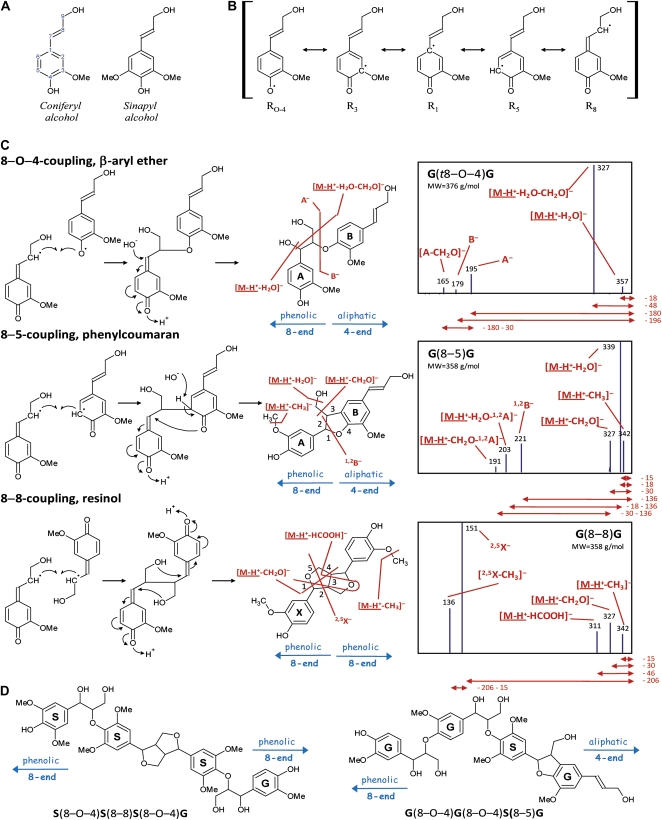
Lignin is an aromatic heteropolymer that is mainly present in secondary-thickened plant cell walls, allowing the transport of water and nutrients and providing the necessary strength for the plant to grow upwardly (Boerjan et al., 2003; Vanholme et al., 2008, 2010). In angiosperms, lignin is predominantly composed of guaiacyl (G) and syringyl (S) units that are derived from combinatorial radical-radical coupling of the monolignols coniferyl and sinapyl alcohol, respectively (Ralph et al., 2004; Fig. 1A). Following oxidation of the monolignols by peroxidase and/or laccase, the resulting electron-delocalized radical has unpaired electron density at its 1-, 3-, O–4-, 5-, and 8-positions (Fig. 1B); note that much of the lignin literature uses Greek letters for the side chain, α, β, and γ for the 7-, 8-, and 9-positions. As radical coupling at the 8-position is favored, coupling with another monolignol radical affords, after rearomatization, a mixture of dehydrodimers with 8–8-, 8–5-, and 8–O–4-linkages (Fig. 1C). Following dimerization, polymerization will continue by the coupling of the 8-position of an incoming monolignol radical to the O–4-position of the dimer's phenolic end. In the case of a G dimer, coupling can also occur, albeit at a lower frequency, to the 5-position. Thus, chain elongation creates 8–5- and 8–O–4-linkages (Adler, 1977). Besides the monolignols and other monomers that are present in minor amounts (Boerjan et al., 2003), the plasticity of lignin polymerization permits the incorporation of any phenolic that enters the lignification site, subject to its chemical cross-coupling propensity. This property might be exploited for the production of more easily degradable lignins (Ralph et al., 1997, 2001, 2004; Sederoff et al., 1999; Baucher et al., 2003; Ralph, 2006, 2010; Grabber et al., 2008), allowing more efficient processing of plant biomass toward fermentable sugars in the route to bioethanol and other biobased products.
Figure 1.
Radical-radical coupling during lignin polymerization. A, Main angiosperm monolignols. B, Delocalized radical following monolignol oxidation. C, Main types of monolignol dimerizations and their MS2 spectra. Below the spectra, the neutral loss for each first product ion is noted in D. For phenylcoumarans and resinols, the pathway II first product ions are specified by superscripts indicating the bonds that are cleaved. This numbering system has been suggested previously to annotate the first product ions upon CID of flavonoids (Ma et al., 1997; Fabre et al., 2001; Morreel et al., 2006) and upon CID of oligosaccharides (Domon and Costello, 1988). Most first product ions are due to charge-driven fragmentations in which the charge center initiates the dissociation, yet the pathway I-associated formaldehyde loss upon CID of β-aryl ethers as well as the pathway I-associated methyl radical loss upon CID of phenylcoumarans and resinols are examples of charge-remote fragmentations (i.e. where the fragmentating center occurs remote from the charge center; Bowie, 1990). D, Examples of tetralignols observed in poplar xylem (Morreel et al., 2004a). [See online article for color version of this figure.]
To date, the lignin structure has been mainly studied by degradation of the polymer into smaller fragments, which can then be quantified. Several methods, such as nitrobenzene oxidation, permanganate oxidation, hydrogenolysis, ozonolysis, and acidolysis, have been developed (Adler, 1977), with the most informative for lignin structure being thioacidolysis (Lapierre et al., 1985) and derivatization followed by reductive cleavage (DFRC; Lu and Ralph, 1997). Both methods cleave the β-ether bonds, and the released units are then measured by gas chromatography (GC) or GC-mass spectrometry (MS). Thioacidolysis followed by Raney-Ni desulfurization and DFRC also allow the analysis of dimers that have a (so-called condensed) carbon-carbon linkage, such as an 8–8- or 8–5-linkage. The latter methods thus only analyze a fraction of the lignin polymer. A nondegradative method is based on the NMR analysis of extracted lignins or even whole cell walls (Lu and Ralph, 2003; Ralph and Landucci, 2010). However, whereas NMR is the preferred method for the elucidation of unknown compounds, it is a low-sensitivity method that works best on purified compounds. Because extracted lignins or cell walls are a complex mixture of different components, NMR does not easily discern which units are attached to which, yet the method is very powerful for examining the frequencies of the different building blocks and the different bond types.
Thioacidolysis, DFRC, and NMR analysis also have the limitation that only those peaks are quantified that are a priori known to correspond with the main types of units or linkages. Peaks corresponding to new units or linkage types are difficult to annotate, as they are only highlighted in comparative analyses between wild-type and transgenic/mutant plants. Additionally, purifying these peaks is cumbersome. Their structural elucidation and authentication, therefore, is solely based on the spiking of chemically synthesized standards. Furthermore, no trimeric or higher order oligomeric structures can be sequence analyzed by these methods. Hence, hardly any information is available concerning the extent to which linkage formation is affected by the adjacent linkage type. An exception was obtained from softwood lignin analyses, where it was apparent that most of the resinols (8–8-linked units) seemed to be 5–O–4-linked (Önnerud and Gellerstedt, 2003), underscoring the importance of such conditional linkage frequencies that are needed to correctly model the lignin structure (van Parijs et al., 2010). It would thus be very informative to have a strategy that allows determining the sequence of monomers and the bonds connecting them in individual lignin oligomers.
Over the past few years, a series of lignin oligomers (i.e. dilignols, trilignols, and tetralignols [Fig. 1D]) were detected in poplar (Populus spp.) and tobacco (Nicotiana tabacum) xylem with liquid chromatography (LC) coupled to ion trap (IT) MS (Morreel et al., 2004a, 2004b; Damiani et al., 2005; Dauwe et al., 2007; Leplé et al., 2007). Their structures were resolved using mass spectral information and further authenticated by chemical synthesis. The structures of all oligolignols were in agreement with the theory of combinatorial coupling. Interestingly, oligolignol profiling also allowed the detection of less abundant units, derived, for example, from sinapyl p-hydroxybenzoate in poplar (SP; Morreel et al., 2004a), from feruloyl tyramine in tobacco (Dauwe et al., 2007), or from feruloyl malate in Arabidopsis (Arabidopsis thaliana; Rohde et al., 2004), and even new units and linkage types were revealed, such as the 5-hydroxyguaiacyl unit in benzodioxane structures in caffeic acid 3-O-methyltransferase-deficient poplar (Morreel et al., 2004b). Additionally, LC-MS analysis of the dilignols enabled a study of the gas-phase fragmentation pathways of the different linkage types (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data) and allowed characteristic MS2 first product ions to be annotated.
Here, we combine the MS-based fragmentation rules into a method that allows (1) sequencing of individual lignin oligomers, (2) detecting new units and linkages, and (3) obtaining conditional frequencies for the different linkage types. Based on these rules, 134 oligolignols present in the xylem of poplar (Populus tremula × Populus tremuloides), ranging from dimers up to hexamers, more than half of which possessed likely unknown linkage types or units, were sequenced. This is, to our knowledge, the first paper presenting a sequencing strategy for lignin oligomers.
RESULTS
Characteristic Fragmentations of Lignin Interunit Linkage Types
To develop a MS-based tool for the sequencing of lignin oligomers, the characteristic gas-phase fragmentations for each of the main linkage units (i.e. 8–O–4, 8–5, and 8–8) were elucidated by analyzing a series of lignin dimers using IT-MS in the negative ion mode (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data). Below, the annotation of the characteristic first product ions is presented. These were then used to design the sequencing strategy.
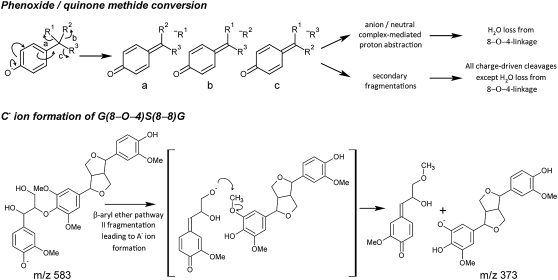
Phenoxide anions were generated in the ionization source, and all charge-driven collision-induced dissociations (CID) were initiated by the conversion of the phenoxide anion to a quinone methide with the concomitant elimination of the chemical entity linked to the 7-position (Fig. 2). This entity is then lost as a neutral fragment via an anion/neutral complex-mediated proton abstraction (water loss from the β-aryl ether linkage unit; Fig. 1C; Bowie, 1990), or it induces a secondary fragmentation pathway that might be mediated via an anion/neutral complex (all remaining charge-driven fragmentations in Fig. 1C; K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data). For each of the three main linkage unit types in lignin, the β-aryl ethers (8–O–4), phenylcoumarans (8–5), and resinols (8–8), this phenoxide/quinone methide conversion yields two groups of characteristic MS2 first product ions: (1) small neutral losses and (2) fragmentations leading to the cleavage of the linkage. Hereafter, these first product ions are called pathway I and pathway II first product ions, respectively.
Figure 2.
CID pathways. Phenoxide anion/quinone methide conversion with charge migration that was shown to induce all charge-driven fragmentation pathways of the lignin linkage types (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data). A new β-aryl ether-associated pathway II fragmentation was observed upon CID of X(8–8)X-containing trilignols, leading to the formation of the C− ion at m/z 373.
The pathway I small neutral losses yield information on the linkage unit (Fig. 1C). A β-aryl ether [Fig. 1C; G(t8–O–4)G] is recognized by the loss of water (−18 D, [M-H+-H2O]−) and the combined loss of water and formaldehyde (−48 D, [M-H+-H2O-CH2O]−). Upon CID of phenylcoumarans [Fig. 1C; G(8–5)G], in addition to formaldehyde loss (−30 D, [M-H+-CH2O]−), water loss (−18 D, [M-H+-H2O]−) occurs also due to the primary alcohol function. As a primary alcohol group is absent in resinols [Fig. 1C; G(8–8)G], no water loss is observed, although a formaldehyde loss (−30 D, [M-H+-CH2O]−) is always present. Furthermore, a rearrangement occurs upon which formic acid (−46 D, [M-H+-HCOOH]−) is expelled. Except for these charge-driven losses, a charge-remote formaldehyde loss (−30 D, [M-H+-CH2O]−data not shown) occurs upon CID of β-aryl ethers; both phenylcoumarans [Fig. 1C; G(8–5)G] and resinols [Fig. 1C; G(8–8)G] expel a methyl radical (−15 D, [M-H+-CH3]−) due to homolytic fragmentation when methoxylated units are present (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data).
After annotating the linkage unit, the G or S moiety involved is deduced from the molecular masses of the precursor ion and first product ions resulting from the cleavage of the linkage unit (Fig. 1C). These pathway II fragmentations include the A−[A-CH2O]−and B− ions for the β-aryl ethers, the 1,2B−[M-H+-H2O-1,2A]−and [M-H+-CH2O-1,2A]− ions for the phenylcoumarans, and the 2,5X−[2,5X-CH3]− ions for the resinols. Their corresponding fragmentation pathways are the topic of another paper (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data).
Sequencing of Trimers Based on Their Negative Ion MS2 Spectra
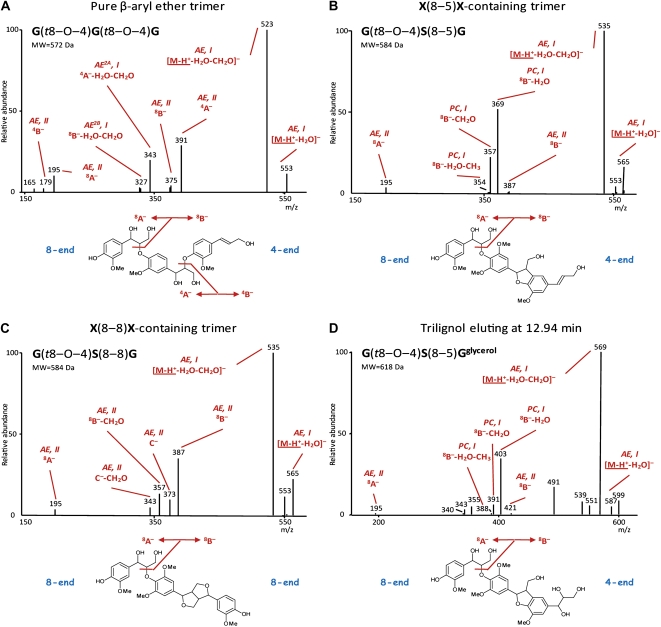
The pathway I and II first product ions for the different linkage units should be consistently observed in the spectra of higher order lignin oligomers to be retained in the sequencing model. In a first step, this was verified by analyzing previously synthesized lignin trimers using LC-IT-MS (Fig. 3; Table I).
Figure 3.
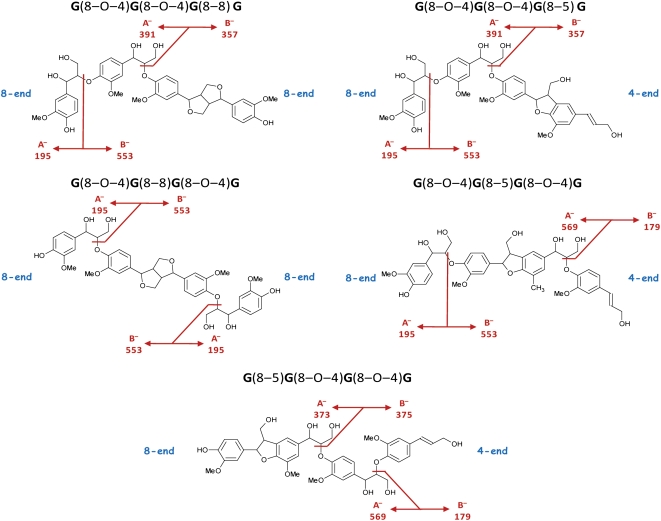
MS2 spectra of trilignols. A, Pure β-aryl ether [G(8–O–4)G(8–O–4)G]. B, X(8–5)X-containing β-aryl ether [G(8–O–4)S(8–5)G]. C, X(8–8)X-containing β-aryl ether [G(8–O–4)S(8–8)G]. D, G(8–O–4)S(8–5)Gglycerol. Characteristic first product ions (see also Fig. 1 and “Materials and Methods”) are indicated. [See online article for color version of this figure.]
Table I. MS fragmentation patterns of trilignols.
Relative intensity of the first product ions as compared with the base peak is given in parentheses. Asterisks indicate that neutral loss of this fragmentation depends on the mass of the involved units.
| First Product Ion | Type | Fragmentation Pathway | Neutral Loss | G(t8–O–4)G(t8–O–4)G | G(t8–O–4)S(8–5)G | S(t8–O–4)S(8–5)G | S(t8–O–4)G(8–5)G | G(t8–O–4)S(8–5)G′ | G(e8–O–4)S(8–5)G′ | G(t8–O–4)S(8–8)G | G(t8–O–4)S(8–8)S | G(e8–O–4)S(8–8)S | S(t8–O–4)S(8–8)S |
| D | |||||||||||||
| Collision energy (%) | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | |||
| [M-H+]− | 571 (0) | 583 (0) | 613 (0) | 583 (0) | 581 (0) | 581 (0) | 583 (0) | 613 (0) | 613 (0) | 643 (0) | |||
| [M-H+-H2O]− | AE | I | 18 | 553 (2) | 565 (3) | 595 (10) | 565 (4) | 563 (8) | 563 (3) | 565 (12) | 595 (12) | 595 (18) | 625 (5) |
| [M-H+-CH2O]− | AE | I | 30 | 541 (2) | 553 (5) | 583 (7) | 553 (2) | 551 (35) | 551 (8) | 553 (19) | 583 (22) | 583 (16) | 613 (12) |
| [M-H+-H2O-CH2O]− | AE | I | 48 | 523 (100) | 535 (100) | 565 (100) | 535 (100) | 533 (94) | 533 (5) | 535 (100) | 565 (100) | 565 (13) | 595 (100) |
| 8A− | AE | II | * | 195 (1)a | 195 (10) | 225 (17) | 195 (3) | 195 (4) | 195 (6) | 195 (4) | 195 (6) | 225 (20) | |
| 8A−-CH2O | * | 165 (5) | 195 (8) | 165 (1) | 165 (2) | 165 (1) | 165 (5) | ||||||
| 8B− | AE | II | * | 375 (1) | 387 (2) | 387 (4) | 357 (3) | 385 (5) | 385 (10) | 387 (41) | 417 (52) | 417 (64) | 417 (27) |
| 4A− | AE | II | * | 391 (35) | |||||||||
| 4B− | AE | II | * | 179 (1)a | |||||||||
| 8B−-H2O | PC | I | * | 369 (70) | 369 (38) | 339 (28) | 367 (100) | 367 (100) | |||||
| 8B−-CH2O | PC/AEb | I/IIb | * | 357 (24) | 357 (15) | 327 (21) | 355 (36) | 355 (15) | 357 (24) | 387 (33) | 387 (51) | 387 (27) | |
| 8B−-H2O-CH2O | AE2B | I | * | 327 (8) | |||||||||
| 4A−-H2O | * | 373 (1) | |||||||||||
| 4A−-H2O-CH2O | AE2A | I | * | 343 (36) | |||||||||
| A−|8B− | * | 195 (1)a | |||||||||||
| B−|8B− | * | 179 (1)a | |||||||||||
| A−|4A− | * | 195 (1)a | |||||||||||
| B−|4A− | * | 195 (1)a | |||||||||||
| 8B−-H2O-CH3 | PC | I | * | 354 (1) | 354 (4) | 324 (4) | 352 (8) | 352 (4) | |||||
| C− | AE | II | * | 373 (7) | 403 (68) | 403 (100) | 403 (28) | ||||||
| C−-CH2O | AE | II | * | 343 (7) | 373 (40) | 373 (33) | 373 (9) |
These first product ions arise from multiple fragmentation pathways.
This first product ion was classified as PC, I or AE, II in the case of X(8–5)X- or X(8–8)X-containing trimers, respectively. Y−|nX− with X, Y = A or B and n = 4 or 8 indicates the β-aryl ether-associated A− or B− ion formation resulting from the cleavage of the 8B− or 4A− dimeric first product ion. Type and fragmentation pathway are given for those first product ions mentioned in Figure 3.
As the main interunit linkage type in lignin is 8–O–4 (Fig. 3A), evaluating the MS2 spectra for those that contain its pathway I (AE, I; [M-H+-H2O]−[M-H+-H2O-CH2O]−for nomenclature of the product ions, see “Materials and Methods”) characteristic ions can be used to annotate candidate oligolignols and to start the sequencing of trimers. The 8–O–4-associated pathway II fragmentations, yielding the A−B− first product ions (AE, II), result in ions corresponding to a dimeric substructure and a monomeric unit.
If the β-aryl ether is present at the 8-end of the trimer as defined in Figure 3, the 8A− first product ion will represent the monomeric unit (Fig. 3A). Because it contains an additional oxygen as compared with the original monolignol it was derived from, its mass will be 16 D larger. In the case of coniferyl alcohol (180 D) or sinapyl alcohol (210 D), this will yield a first product ion at mass-to-charge ratio (m/z) 195 or 225, respectively. The dimeric first product ion resulting from the pathway II cleavage at the 8-end is referred to as 8B− (Fig. 3A). However, a β-aryl ether at the 4-end will eliminate a 4B− monomeric first product ion (Fig. 3A) at m/z 179 or 209 in the case of a G or S unit, respectively, and a 4A− dimeric first product ion (Fig. 3A) bearing an additional oxygen as compared with the original dilignol, hence having a m/z value that is 16 units higher. Evidently, cleavage of the β-aryl ether at the 4-end is a charge-remote process.
MS2 Spectra of Pure β-Aryl Ether Trimers
Most of the first product ions present in the MS2 spectra (Fig. 3A) of these trimers are associated with the β-aryl ether pathway I and II fragmentations of either interunit linkage type (see above). The pathway II-derived first product ions possess sufficient internal energy to induce a secondary fragmentation leading to “second product” ions within the MS2 spectrum (annotated with AE2A or AE2B in Fig. 3A). The pathway II first product ions corresponding with the dimeric substructures (i.e. the 4A−8B− ions) still contain an 8–O–4-linkage and, thus, will be subjected to secondary pathway I and II dissociations. The pathway I fragmentations yield the 4A−-H2O-CH2O and 8B−-H2O-CH2O “second product” ions, whereas the ions of the pathway II fragmentations will contribute to the intensity of already existing m/z peaks (Table I, A−|8B−B−|8B−A−|4A−and B−|4A−). The MS3 spectrum of the dimeric 8B− ion (m/z 375) is similar to the MS2 spectrum of G(t8–O–4)G.
MS2 Spectra of X(8–5)X-Containing Trimers (X = G or S)
The 8–5-linkages originate from monolignol dimerization as well as from the coupling of a monolignol to the G end of a lignin polymer (Ralph et al., 2004). Thus, the phenylcoumaran linkage can appear at either end of a trilignol. After pathway II cleavage of the β-aryl ether in an X(8–5)X-containing trimer, G(t8–O–4)S(8–5)G (Fig. 3B; Table I), the first product ion representing the phenylcoumaran dimeric substructure [i.e. the 4A− or the 8B− ion depending on the position of the 8–5-linkage; MS2 spectra of X(8–5)X-containing trimers in Fig. 3B] is subjected to phenylcoumaran-associated pathway I fragmentations (PC, I), resulting in a typical m/z peak triplet (8B−-H2O, 8B−-CH2O, and 8B−-H2O-CH3 ions) separated by 12 and 3 D. The 8B−/4A− first product ion itself is only present in minor amounts. Clearly, the residual internal energy of such a first product ion is high enough to favor further dehydration or, to a somewhat lesser extent, formaldehyde loss via one of the primary alcohol groups (8B−-H2O and 8B−-CH2O in Table I). A combined dehydration and demethylation explains the limited formation of the third m/z peak (8B−-H2O-CH3 in Table I) within the triplet.
Phenylcoumaran-associated pathway II fragmentations are not observed in the MS2 spectrum but occur upon MS3 fragmentation of the 8B−/4A− ion. For example, MS3 of the G(t8–O–4)S(8–5)G-derived 8B− first product ion at m/z 387 yields minor ions at m/z 221, 203, and 191, corresponding with the 1,2B−[M-H+-H2O-1,2A]−and [M-H+-CH2O-1,2A]− ions in the spectrum of phenylcoumaran dimers (Fig. 1).
Other MS2 first product ions of G(t8–O–4)S(8–5)G were also subjected to MS3 fragmentation. Interestingly, the [M-H+-CH2O]− ion at m/z 553 fragmented mainly by losing 48 D, yielding a MS3 second product ion at m/z 505. This indicates that the MS3 fragmentation affords the combined water/CH2O loss typical of β-aryl ethers, indicating that the MS2-generated [M-H+-CH2O]− ion originates at least partly from a charge-remote fragmentation at the phenylcoumaran.
MS2 Spectra of X(8–8)X-Containing Trimers
Although it has been hypothesized that 8–8-coupling of a monolignol to a growing polymer might occur (Zhang et al., 2003), the 8–8-linkages are mainly, if not exclusively, produced during monolignol dimerization. As two phenolic end groups are retained, this interunit linkage type has only 8-ends (Fig. 3C; Table I). Because, for the formation of a trimer, a β-aryl ether can be formed at either end of the 8–8-linked dimer, the β-aryl ether-associated pathway II cleavage of the trimer affords only 8B−8A− first product ions. The MS2 spectra (Fig. 3C; Table I) of these trimers show prominent 8B− first product ions. In addition, a new pathway II cleavage of the β-aryl ether is observed leading to the C−C−-CH2O first product ions (Figs. 2 and 3C). Therefore, the 8B−C−8B−-CH2O, and C−-CH2O first product ions afford a characteristic quartet of m/z peaks separated by 14, 16, and 14 D. A charge-driven pathway (Fig. 2) can be envisioned when the β-aryl ether-associated 8A− first product ion attacks the methoxy group in a neutral/anion complex-mediated reaction. Such a SN2 mechanism has been shown to occur by the attack of various anions on alkyl phenyl ethers in the gas phase using labeled reactants (Houriet and Tureček, 1994). Plausibly, the C−-CH2O ion originates from the C− ion via a charge-remote reaction rather than via a charge-driven pathway.
Resinol-associated pathway II fragmentations are not observed upon MS2 of the trimers. However, the MS3 spectrum of the G(8–O–4)S(8–8)G-derived 8B− first product ion at m/z 387 shows low-abundance second product ions at m/z 372, 341, 181/151, and 166/136 similar to the [M-H+-CH3]−[M-H+-HCOOH]−2,5X−and [2,5X-CH3]− first product ions observed in the MS2 spectra of resinol dimers (Fig. 1). However, the base peak at m/z 218 in the MS3 spectrum of the 8B− ion at m/z 387 results from a homolytic fragmentation that does not occur upon MS2 fragmentation of resinol dimers. These results indicate that the internal energy of the MS2-produced resinol dimer B− ion differs from that of a resinol dimer ion produced in the ionization source.
As was observed for X(8–5)X-containing β-aryl ether trimers, MS3 dissociation of the [M-H+-CH2O]− ion yielded a base peak due to a combined water/CH2O loss (−48 D). Again, this demonstrates that the CH2O loss in the MS2 spectrum is partly due to a charge-remote process, this time associated with the resinol linkage unit in the trimer.
Pathway I fragmentations can be traced in the MS2 spectra of all three linkage unit types, but only 8–O–4-linkage units will yield pathway II-associated first product ions in the MS2 spectra of lignin trimers, whereas pathway II fragmentations of 8–5- and 8–8-linkage units are only revealed upon MS3 fragmentation. Nonetheless, observation of the presence of an 8–5- or 8–8-linkage unit using the MS2 spectrum is possible due to the appearance of a characteristic m/z triplet or quartet, respectively.
Sequencing of Tetramers and Higher Order Oligomers
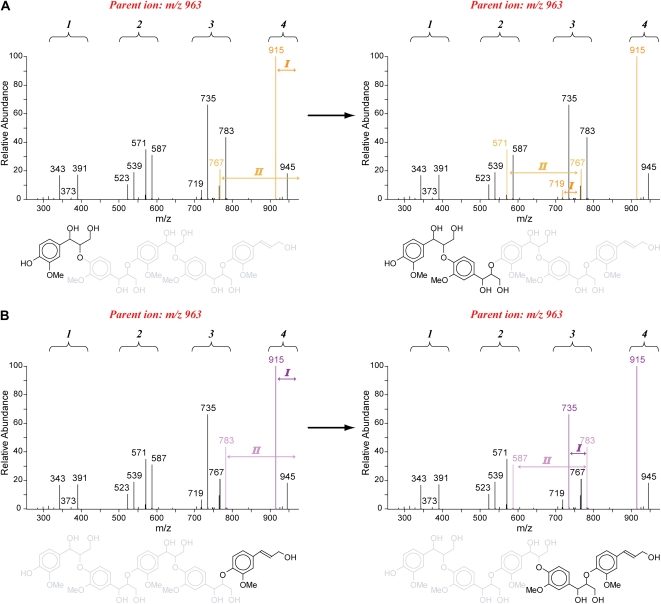
The pathway I and II fragmentation rules can equally be applied for the sequencing of higher order oligomers. Upon annotation of the β-aryl ether-associated pathway I first product ions, the pathway II ions are traced. These pathway II-associated 8B−/4A− first product ions are then starting points to search again for the pathway I and II ions indicative of a subsequent 8–O–4-linkage unit. Iteration of this process resolves the structure of pure β-aryl ether oligomers (Fig. 4). This nested strategy is based on the assumption that the β-aryl ether-associated pathway II cleavage is a charge-driven process. However, trimer sequencing showed that this fragmentation also occurs as a charge-remote process, indicating that cleavage can also happen randomly along the oligomer. As a consequence, a regular spacing between groups of first product ions (i.e. m/z clusters) is visible in the MS2 spectrum, of which the number of m/z clusters represents the number of 8–O–4-linkage units. Actually, in Figure 4, these m/z clusters arise because the charge-remote pathway II cleavages yield A−A−-H2O-CH2O (AE2A, I) ions for the dimeric, trimeric, and tetrameric substructures. In addition to this A ion series (light and dark purple for the A−A−-H2O-CH2O ions in Fig. 4), also the B−B−-H2O-CH2O (AE2B, I) ions for the dimeric, trimeric, and tetrameric substructures, called the B ion series (light and dark yellow for the B−B−-H2O-CH2O ions in Fig. 4), are observed. Surveying the mass differences between the ions belonging to the A or B series will also lead to the oligomer sequence. From Figure 4, it is also clear that the A ion series is more abundant than the B ion series. It should be mentioned, though, that first product ions with a mass that is less than one-third of the parent mass are no longer observed in the MS2 spectrum. Thus, for longer oligomers, MSn or tandem MS is necessary.
Figure 4.
Sequencing approach illustrated on the pentamer G(8–O–4)G(8–O–4)G(8–O–4)G(8–O–4)G. The number of linkages can be deduced from the number of ion clusters. A, Sequencing starting from the 8-end using the B ion series (yellow). First product ions due to pathway I and II fragmentations are shown (dark and light yellow). B, Annotation of first product ions (dark and light purple, A ion series) after sequencing from the 4-end.
Both the nested and the survey approach aid in sequencing higher order oligomers that contain a condensed linkage unit. In tetramers, the condensed linkage unit might be present at two (in the case of an 8–8-linkage unit) or at three (in the case of an 8–5-linkage unit) different positions. The 8–8-linkage unit can be the central linkage unit or can occupy one of the end positions (both are 8-ends), giving two different MS2 spectra. However, the end-wise positions are not identical in the case of an 8–5-linkage unit, hence providing three different MS2 spectra.
A tetramer with an 8–8-linkage unit will yield the resinol-characteristic m/z quartet. If the tetramer has an 8–8-end-linkage unit, first product ions representing the dimeric β-aryl ether substructure will be present in both the A and B ion series (Fig. 5). These latter first product ions will be absent in the case of an internal 8–8-linkage unit. In addition, the presence of two 8–O–4-linked phenolic end groups will yield new types of first product ions in the MS2 spectrum due to their combined pathway I losses; that is, neutral losses of 36 (2H2O) and 96 (2H2O + 2CH2O) D (Morreel et al., 2004a).
Figure 5.
Typical A and B ion series for G β-aryl ether tetramers containing one condensed linkage unit. Two configurations are possible in the case of an 8–8-linkage unit and three in the case of an 8–5-linkage unit. [See online article for color version of this figure.]
The presence of an 8–5-linkage unit is evident from the characteristic m/z triplet within the MS2 spectrum. The A and B ion series within the spectrum of a tetramer with an internal 8–5-linkage unit will lack first product ions representing the dimeric β-aryl ether substructure (Fig. 5). When the tetramer has an 8–5-end-linkage unit, it occurs at the 4- or 8-end. If this condensed linkage unit is present at the 8-end, the A ion series representative of the monomeric substructure and a B series first product ion associated with a trimeric substructure are lacking in the MS2 spectrum. Conversely, an A series first product ion for a trimeric substructure and a B series peak for a monomeric substructure are not observed in the spectrum of a 4-end-located 8–5-linkage unit. Thus, the absence of certain ions in the A and B series is indicative of the location of the condensed linkage unit.
For the sequencing of higher order oligomers, the nested and survey approaches might be combined. The nested approach necessitates the use of MSn, whereas the survey approach might be difficult, as the MS2 spectra are often complicated with peaks that hide the m/z clusters. Moreover, the smaller the substructure, the less abundant the corresponding A and B ions are, especially when condensed linkage units are present. With both sequencing approaches, the presence of the G and S moieties linked by 8–O–4-, 8–5-, and 8–8-bonds can be revealed. As the fragmentation pathways of these linkage units might alter when they connect to other units, such as hydroxycinnamaldehyde- (G′ and S′) or SP-derived units, these units were not yet included in the sequence strategy.
Detection of Oligolignols with Unknown Units in Wild-Type Poplar Xylem
As a proof of concept, the “nested” sequencing approach was applied to sequence trilignols and higher order oligolignols that are present in poplar xylem. Previously, a wide variety of oligolignols has been profiled using reverse-phase LC-MS (Morreel et al., 2004a, 2004b). In this study, accurate masses were obtained using ion cyclotron MS, whereas MSn was obtained in parallel via IT-MS or by direct infusion following LC fractionation. In cases where the sequence could not be resolved based on the G and S units, we also considered the presence of G′, S′, and SP units, as all three were previously observed in poplar oligolignols (Morreel et al., 2004a).
Based on the pathway I peak pattern for β-aryl ethers, 134 oligolignols, from trimers up to hexamers, were detected (Supplemental Table S1). Often, multiple isomers were present [e.g. isomers of G(8–O–4)S(8–8)S and S(8–O–4)S(8–5)G eluted at four different retention times]. This is not surprising, as they correspond to physically distinct isomers of each of these compounds. Twenty-five of the 134 oligolignols could be completely sequenced by our approach taking only G and S units into account. Of the remainder, a complete sequence was obtained for nine oligolignols when taking the presence of a G′, S′, or SP unit into account, and two structures were resolved after taking a hexose or methyl group into account. Still, 98 remained only partially sequenced. In 11 cases, this arose from the inability to determine the correct position of some of the units. Following a more elaborate MSn analysis, two of these 11 oligolignols were shown to contain a putative reduced phenylcoumaran or resinol linkage unit. Preliminary sequence terminations became more frequent as sequences were longer (18 cases), requiring that MSn of particular first product ions had to be performed. However, the majority of the cases (69 cases) could not be sequenced due to the presence of as yet unknown units. Surveying Supplemental Table S1 for unknown units or dimeric substructures indicates at least 10 unknown units or linkages.
Unraveling the Dimeric Substructure of 422 D
One frequently observed unknown dimeric substructure had a molecular mass of 422 D. The structure of this moiety was further resolved by a more in-depth analysis of the MSn spectra obtained for the oligolignol with m/z 617.2244 eluting at 12.94 min (Fig. 3D; Supplemental Fig. S1). The molecular formula obtained for this accurate mass, C31H37O13 (Δppm = 0.71), indicated a trilignol with two G units and one S unit. The MS2 spectrum showed the β-aryl ether-associated pathway I losses at m/z 599 and 569. The pathway II fragmentation led to an 8A− first product ion at m/z 195 and a reduced abundance of a 8B− ion at m/z 421. Phenylcoumaran-associated pathway I losses occurred from the 8B− first product ion, leading to the characteristic triplet at m/z 403, 391, and 388. Therefore, this structure was partially elucidated as an analog of G(8–O–4)S(8–5)G. The mass indicated the presence of two more hydroxyl groups on this structure. The MS3 spectra of the [M-H+-H2O-CH2O]−8B− first product ions afforded both a combined loss of water and CH2O as a base peak. This supports the presence of a 1,3-diol function on the side chain of the aliphatic end group. Finally, in the MS2 spectrum, a second phenylcoumaran-associated triplet at m/z 355, 343, and 340 was observed, exactly 66 atomic mass units less than the m/z 403, 391, and 388 triplet, implying the presence of another hydroxyl function besides the 1,3-diol group on the side chain of the aliphatic end group. In conclusion, this oligolignol is G(8–O–4)S(8–5)Gglycerol, with a guaiacylglycerol (Gglycerol) unit as the aliphatic end group (Fig. 3D). The MS2-initiated 422-D neutral loss corresponds with the S(8–5)Gglycerol dimeric substructure. Additional support was obtained from MSn analysis of a synthesized phenylcoumaran with a similar structure: G(8–5)Gglycerol. MS3 analyses of the S(8–5)Gglycerol-associated 8B− ion at m/z 421 showed the same neutral losses (−18, −30, and −48 D) as observed in the MS2 spectrum of G(8–5)Gglycerol. Furthermore, the MS3 spectrum of the MS2-generated ion at m/z 343, derived from the combined loss of water and CH2O from the glycerol moiety of G(8–5)Gglycerol, yielded a second product ion at m/z 207 due to the loss of the 8-end-located G unit. Similarly, MS3 fragmentation of the ion at m/z 373 in the MS2 spectrum of G(8–O–4)S(8–5)Gglycerol (i.e. the 8B−-H2O-CH2O first product ion) yielded a major ion at m/z 207 upon expelling the 8-end-located S unit. The latter fragmentation occurs similarly as described for the 1,2B− first product ion obtained upon CID of phenylcoumaran dilignols (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data). In total, five oligolignols could be identified: four isomers of G(8–O–4)S(8–5)Gglycerol and one isomer of S(8–O–4)S(8–5)Gglycerol (Supplemental Table S1). Oligolignols containing a (G8–5)Gglycerol moiety were not observed.
DISCUSSION
Lignin is the last plant biopolymer defying sequencing. Of course, a primary sequence of polymer ensembles has no meaning if the polymerization is combinatorial and there is no defined biosynthetic “sequence” (Ralph et al., 2008). Nevertheless, being able to determine the actual structural sequence in lignin oligomers is of enormous value: it provides information on how the polymer is made, and it also reflects the conditions of monolignol coupling during polymerization in the cell wall. It is known from experiments with dehydrogenation polymers that the conditions during polymerization have a large impact on lignin structure. Similarly, in planta, the structures of the polymers also vary among cell types and individual cell wall layers (Ruel et al., 2009). Furthermore, the sequencing method allows oligolignols that do not fit with our current knowledge of lignin structure to be identified, thus pinpointing those that deserve further research to characterize novel units and bonds. No unambiguous methods to determine the primary structure of even moderately complex oligomers, other than by synthesizing authentic compounds for comparison, have been published to date. Here, and together with another study focusing on dimers only (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data), we were able to deduce the gas-phase fragmentation behavior of the different lignin units using CID of the negative ions from a variety of synthesized dilignols, trilignols, and tetralignols. Applying these rules to the MS2 spectra of poplar oligolignols allowed sequencing of up to hexamers. Previously, even heptamers and octamers have been observed in oligolignol profiles of poplar xylem (G. Goeminne, K. Morreel, and W. Boerjan, unpublished data); such oligomers appear to be close to the estimated average polymer length of extremely S-rich lignins (i.e. approximately 10 units; Stewart et al., 2009). However, the longer an oligolignol, the more isomers exist, in minute individual amounts, that cannot be fully resolved by reverse-phase LC.
The MS-based sequencing of all our lignin oligomers was possible mainly via a nested strategy: following the characterization of the linkage unit using the specific pathway I neutral losses, the attached units or oligomeric moieties are identified using the pathway II first product ions. This is in contrast with the MS-based sequencing of peptides and nucleic acids that are both characterized by only one type of bond between the units. For these biopolymers, sequencing is solely based on first product ions that result from the cleavage of the bonds linking the units. For peptides measured in the positive ionization mode (Biemann, 1988), cleavage of the alkyl-carbonyl bond, the peptide-amide bond, and the amino-alkyl bond leads to the an, bn, and cn ion series when the charge is present on the N terminus on the one hand, and otherwise to the C terminus-associated xn, yn, and zn ion series, respectively. Cleaving the phosphoester and phosphorus-oxygen bonds of positively ionized nucleic acids by CID yields the a, b, c, and d ion series associated with the 5′ terminus and the w, x, y, and z ion series when the charge is on the 3′ terminus (McLuckey et al., 1992). A similar nomenclature was also proposed for the dissociation of negatively charged nucleic acids (McLuckey and Habibi-Goudarzi, 1993). As these cleavages occur randomly throughout the peptide or nucleic acid oligomer, the annotation of such a series of first product ions reveals the primary structure. Such a “survey” approach can also be employed for lignin oligomers and might even more easily indicate the location of a condensed linkage unit than the nested approach, but when a condensed bond was present, the spectra were rapidly blurred by the multitude of ions, preventing an efficient use of the survey method. Sequencing oligosaccharides is similarly performed via a survey method using the first product ions arising from glycosidic bond cleavage (i.e. the nonreducing end B and C ion series and the reducing end Y and Z ion series; Domon and Costello, 1988). Nevertheless, branching occurs frequently and may occur at any of the different alcohol groups of the carbohydrate unit. Although the size of the branch can be deduced from the B, C, Y, and Z ion series, the linkage position can only be determined using the nonreducing end A and reducing end X series that originate from ring cleavage. Lignin is a largely linear polymer, yet branching does occur via 5–5- (resulting in dibenzodioxocin units) and 4–O–5-linkages. In this study, the sequencing approach was based on linear oligomers of p-hydroxyphenyl, G, and S units that are connected by 8–O–4-, 8–5-, and 8–8-bonds, yielding the β-aryl ether, phenylcoumaran, and resinol linkage units, respectively. The MS fragmentation behavior of branched linkage units and of the less abundant 8–1-linkages still has to be investigated.
To date, the lignin structure has been investigated by degradative methods, such as thioacidolysis (Lapierre et al., 1985) or DFRC (Lu and Ralph, 1997), or by NMR of purified lignin or whole cell walls (Lu and Ralph, 2003). These methods only address a priori known monomeric and dimeric moieties in the polymer and provide little information on the actual sequence of individual polymers. Here, we present an MS-based sequencing approach for lignin oligomers. By targeting the oligolignols, no further degradation or derivatization is needed, and only a mild extraction step is involved. LC-MS peaks that represent oligolignols can be quickly annotated by searching for β-aryl ether-characteristic fragmentations in their MS2 spectra. The subsequent MS-based sequencing of the lignin oligomers then enables new units or linkages to be pinpointed. It should be mentioned that a whole series of peaks was found with masses ranging from trimers to tetramers, for which the MS2 spectra clearly indicated the presence of an 8–8- or an 8–5-linkage unit. However, because no β-aryl ether linkage unit was present and no MS2 pathway II fragmentations occur for condensed linkage units, we were not able to sequence these compounds. Interestingly, as oligolignols containing unknown units or linkage units can fairly readily be purified by reverse-phase HPLC for further identification, they offer an alternative way to identify the unknown NMR peaks that are associated with lignin. To date, annotating these unknown NMR peaks is solely based on synthesizing candidate compounds that are then used for NMR comparison or spiking experiments.
Oligolignol profiling not only provides information about the lignin oligomers but also applies to the (neo)lignans: the secondary metabolites for which the biosynthesis also starts with the radical-radical (cross-)coupling of monolignols (Umezawa, 2003). (Neo)lignan biosynthesis might be more elaborate than previously thought. For example, in poplar xylem, we detected two trilignol hexosides, one of which we were only able to partially sequence. As there are no glycosyltransferases present in the cell wall, the detection of the two trilignol hexosides in this study suggests that they are more likely to be sesquineolignans and that oligomerization might occur also within the cell, likely within the vacuole. To verify their vacuolar localization, oligolignol profiling of isolated vacuoles should be performed. The optical activities of the aglycones should also be determined; lignans are optically active, whereas lignins are racemic (Ralph et al., 1999).
The oligolignol profiling of poplar xylem extracts unleashed 134 β-aryl ether-containing oligolignols, the majority of which possessed an unknown unit and/or linkage structure. Based on all sequence information, the synthesis of all these oligolignols was in agreement with combinatorial coupling expectations (Ralph et al., 2004): multiple isomers were often detected, and no oligolignol sequence violated the cross-coupling propensities of monomers and oligomers, as was discussed previously (Morreel et al., 2004a). This study also confirms the SP unit as an authentic lignin unit. In addition to G, S, G′, and S′ units, SP units were readily incorporated: six, one, and four peaks were noted corresponding to SP-containing trilignols, a tetralignol, and pentalignols, respectively. Five of the six SP-containing trilignols were isomers. In all of these oligolignols, the SP unit was connected via its 8-position in an 8–O–4-, 8–8-, or 8–5-linkage unit. An SP-containing dilignol and a trilignol were previously observed in poplar xylem (Morreel et al., 2004a), but the presence of SP units here in five trilignol isomers underscores the incorporation of sinapyl p-hydroxybenzoate also via combinatorial coupling.
Using high-resolution MS and MSn spectra, the structure of a more frequently encountered dimeric moiety in the total list of oligolignols was unraveled: S(8–5)Gglycerol. Arylglycerol end units such as Gglycerol are known to occur as minor structures in lignin (Kilpeläinen et al., 1994). They were suspected to arise from acid hydrolysis of β-aryl ether linkage units during lignin isolation, but this could not be confirmed upon hydrolysis of model compounds (Higuchi et al., 1974). Alternatively, they might be caused upon side chain homolysis/oxidation invoked by the ball-milling step (Ralph et al., 2004). However, all three arylglycerols (p-hydroxyphenyl, guaiacyl, and syringyl) could be isolated, albeit in low yields, from enzymic dehydrogenation of their respective monolignols (Higuchi et al., 1974). Furthermore, p-hydroxyphenylglycerol end units were observed in a dehydrogenation polymer (that was never subjected to ball milling) obtained using p-coumaryl alcohol as monomer (Ralph et al., 2006). Finally, arylglycerol end units were shown to be increased in lignins rich in p-hydroxyphenyl units and in lignins that were obtained by down-regulating p-coumarate 3-hydroxylase in alfalfa (Medicago sativa; Ralph et al., 2006) or hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata lignifying cell cultures (Wagner et al., 2007). These results indicate that arylglycerol end units are a feature of natural lignins, as suggested previously (Higuchi et al., 1974). Arylglycerol end units might result from the oxidative conditions during lignification. Gellerstedt and Pettersson (1975) have shown that they are formed when singlet oxygen adds to the double bond, providing a peroxirane intermediate. Water addition then yields a glycol structure. The same authors showed that the main venue of the peroxirane intermediate is its rearrangement to a dioxetane, which then cleaves to give two aldehydes. Therefore, postcoupling oxidation of G(8–O–4)S(8–5)G would yield G(8–O–4)S(8–5)Gglycerol but predominantly G(8–O–4)S(8–5)V′ (V′ is the unit derived from vanillin). However, the latter oligolignol was not observed in our oligolignol profiles, despite the fact that the V′ unit has been detected previously in poplar oligolignols (Morreel et al., 2004a), suggesting that G(8–O–4)S(8–5)Gglycerol might not be entirely due to oxidation of G(8–O–4)S(8–5)G by singlet oxygen. Alternatively, guaiacylglycerol could come directly from coniferyl alcohol via hydrogen peroxide/peroxidase, but this would not explain the detection of four G(8–O–4)S(8–5)Gglycerol isomers but only one S(8–O–4)S(8–5)Gglycerol isomer and the apparent absence of arylglycerol end units within the pool of pure β-aryl ether trilignols. However, this selective oxidation of the phenylcoumaran aliphatic side chain may originate from the higher abundance of X(8–5)X-containing trilignols compared with pure β-aryl ether-containing trilignols. Also, the dilignols were too low in abundance, preventing the detection of G(8–5)Gglycerol or S(8–5)Gglycerol. Together, these results question whether the Gglycerol units are solely due to oxidation. Alternatively, guaiacylglycerol might be synthesized intracellularly, as it was isolated as only one enantiomeric form, the d-threo-isomer, from Zantedeschia aethiopica (Della Greca et al., 1998), and guaiacylglycerol glucosides were detected in leaves of Juniperus phoenicea (Comte et al., 1997), in roots and rhizomes of Sinopodophyllum emodi (Zhao et al., 2001), in stems of Hydnocarpus annamensis (Shi et al., 2006) and Xylosma controversum (Xu et al., 2008), and in leaves and branches of Camellia amplexicaulis (Tung et al., 2009).
MATERIALS AND METHODS
Chemicals
Synthesized oligolignols, G(t8–O–4)G(t8–O–4)G, G(t8–O–4)S(8–5)G, S(t8–O–4)S(8–5)G, S(t8–O–4)G(8–5)G, G(t8–O–4)S(8–5)G′, G(e8–O–4)S(8–5)G′, G(t8–O–4)S(8–8)G, G(t8–O–4)S(8–8)S, G(e8–O–4)S(8–8)S, and S(t8–O–4)S(8–8)S, have been reported previously (Morreel et al., 2004a).
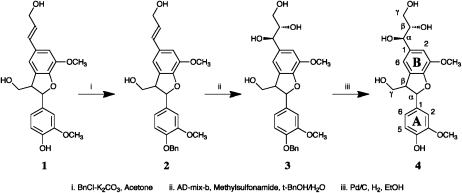
Synthesis of G(8–5)Gglycerol Model 4
Starting with G(8–5)G dimer 1, benzylation with benzyl bromide-K2CO3 in acetone at room temperature for 2 d produced compound 2 in about 20% yield. Following the standard Sharpless asymmetric dihydroxylation method with AD-mix-β, compound 2 was converted to compound 3 in 64% yield after thin-layer chromatography purification. Finally, debenzylation of compound 3 with palladium/carbon in ethanol under a hydrogen balloon at room temperature for 1.5 h gave the G(8–5)Gglycerol dimer 4 quantitatively. Since asymmetric dihydroxylation catalyst AD-mix-β was used to generate the glycerol side chain, the configuration of hydroxyls at the α- and β-positions of the B-unit in compound 4 is as shown in Figure 6. Therefore only two diastereomers were produced for compound 4 synthesized in this way. NMR data of compound 4 are as follows: δH: 3.44 (1H, m, Bγ1), 3.53 (1H, m, Bγ2), 3.55 (1H, m, Aβ), 3.67 (1H, m, Bβ), 3.83 (3H, s, OMe), 3.84 (1H, m, Aγ1), 3.85 (3H, s, OMe), 3.88 (1H, m, Aγ2), 4.60 (2H, d, J = 6.13 Hz), 5.56 (1H, d, J = 6.62) [two close doublets appear representing two isomers], 6.82 (1H, d, J =8.2 Hz, A5), 6.90 (1H, dd, J = 8.2, 1.85 Hz, A6), 6.95 (2H, br-s, B2/6), 7.05 (1H, d, J =1.85 Hz); δC: 54.83, 54.85 (Aβ), 56.16, 56.28 (OMe), 63.89, 63.94 (Bγ), 64.5 (Aγ), 74.82, 74.87 (Aβ), 77.23, 77.26 (Bβ), 110.34 (A2), 112.08, 112.17 (B2), 115.59 (A5), 116.09, 116.18 (B6), 119.47 (A6), 129.58, 129.60 (B5), 134.39 (A1), 136.58, 136.60 (B1), 144.66, 144.69 (B3), 147.12 (A4), 148.27 (A3), 148.37 (B4).
Figure 6.
Synthesis of G(8–5)Gglycerol. Note that the α-, β-, and γ-positions are also referred to as the 7-, 8-, and 9-positions.
Poplar Growth Conditions
Poplar trees (Populus tremula × Populus tremuloides, clone T89) were obtained by in vitro propagation of shoots followed by growth on Murashige and Skoog medium (Duchefa) for 3 months. No hormones or other compounds were added. Rooted plantlets were transferred to the greenhouse (21°C, 60% humidity, 16-h-light/8-h-dark regime, 40–60 μmol m−2 s−1 photosynthetic photon flux) and grown for 3 months until harvest, reaching a height of approximately 2 m.
Oligolignol Extraction
Following debarking, xylem tissue (approximately 600 mg fresh weight) was scraped with a scalpel from a 40-cm-long, debarked stem cut at 50 cm above the ground. After grinding in liquid nitrogen, one-third of the tissue was extracted with 1 mL of methanol. After lyophilization of the methanol, the pellet was dissolved in 200 μL of cyclohexane and 200 μL of water. For the generation of MSn spectra, 25 μL of the water phase was analyzed with reverse-phase LC, and fractions were postcolumn collected as described below.
Direct Infusion MSn Analysis of Standards
A 100 μm solution of each standard, flowing at a rate of 10 μL min−1, was mixed with a flow of 300 μL min−1 (water:methanol, 50:50 [v/v], 0.1% acetate) before entering a LCQ Classic IT-MS device upgraded to a LCQ Deca (Thermo Fisher Scientific). Analytes were negatively ionized by atmospheric pressure chemical ionization using the following parameter values: capillary temperature, 150°C; vaporizer temperature, 350°C; sheath gas, 25 (arbitrary); auxiliary gas, 3 (arbitrary); source current, 5 μA. MSn analysis was performed by CID using helium as the collision gas. The MSn spectra were analyzed with Xcalibur version 1.2.
MSn Spectra Generation of Poplar Oligolignols
For reverse-phase LC, an Acquity UPLC BEH C18 column (150 × 2.1 mm, 1.7 μm; Waters) was serially coupled to an Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 μm) and mounted on an ultrahigh-performance LC system consisting of an Accela pump (Thermo Electron) and Accela autosampler (Thermo Electron). The Accela LC system was hyphenated to a LTQ FT Ultra (Thermo Electron) via an Advion Triversa NanoMate (Advion BioSciences). The following gradient was run using 0.1% aqueous acetic acid (solvent A) and acetonitrile:water (99:1, v/v) acidified with 0.1% acetic acid (solvent B): 0 min, 5% B; 30 min, 45% B; 35 min, 100% B. The loop size, flow, and column temperature were 25 μL, 300 μL min−1, and 80°C, respectively. Full loop injection was applied. An additional postcolumn flow of 10 μL min−1 2-propanol was delivered by an Agilent Technologies 1200 Series capillary pump (G1376A) coupled to a 1200 Series G1379B degasser. The NanoMate was operated in the LC Chip Coupling with Fraction Collection mode, in which 500 nL min−1 was infused via an A-Chip (Advion BioSciences) into the LTQ FT Ultra, and the remaining flow was collected into fractions of 80 μL. Negative ionization was obtained with a spray voltage of 1.7 kV. Using a capillary temperature of 180°C, full Fourier transform-MS spectra between m/z 120 and 1,400 were recorded at a resolution of 100,000.
The collected fractions were subsequently negatively ionized with a NanoMate spray voltage and pressure of 1.7 kV and 3 p.s.i. and a LTQ FT Ultra capillary temperature of 180°C. MSn spectra (maximum breadth, 50; maximum depth, 4) were generated with the LTQ IT-MS device using helium as the collision gas and a collision energy of 35%. Spectra were assembled and analyzed with Mass Frontier version 5.0 (Thermo Electron).
Shorthand Naming of Oligolignols
The oligolignols were named as described by Morreel et al. (2004a). The linkage type (i.e. 8–O–4, 8–8, and 8–5) is denoted as (8–O–4), (8–8), and (8–5), respectively. Isomers (threo and erythro) of a β-aryl ether linkage unit are indicated between parentheses as t and e. The threo/erythro configuration was only noted for the standards and not for the extracted oligolignols from poplar, where it was not always possible to deduce the configuration. Units are written outside the parentheses (Table II). Derivatizations of units are noted via superscripts; in the case of a lariciresinol or secoisolariciresinol linkage unit, as red (for reduced) for one or both units of the 8–8-linkage types, respectively. Therefore, lariciresinol and secoisolariciresinol are indicated as Gred(8–8)G or G(8–8)Gred and Gred(8–8)Gred, respectively.
Table II. Shorthand names of lignin oligomer units.
| Shorthand Name | Unit Type |
| G | Guaiacyl unit, derived from coniferyl alcohol |
| S | Syringyl unit, derived from sinapyl alcohol |
| G′ | Unit derived from coniferaldehyde |
| S′ | Unit derived from sinapaldehyde |
| V′ | Unit derived from vanillin |
| SP | Unit derived from sinapyl p-hydroxybenzoate |
| Gglycerol | Guaiacylglycerol unit |
For convenience throughout the text, the phenolic end of an oligolignol, which is involved in further coupling with new monomer radicals, is termed the 8-end, and the aliphatic end is termed the 4-end, to conform with the order of the numbers representing the bond type in G(8–O–4)S.
Additionally, the aromatic nucleus present in the polymer (e.g. G or S) is called a unit, whereas the interunit linkage structure (e.g. β-aryl ether, phenylcoumaran, or resinol) is referred to as a linkage unit.
Naming Convention for Product Ions
For the naming of the product ions observed upon CID of dilignols, the fragmentations of the different linkage units were grouped into those that are indicative for the presence of a linkage unit (pathway I) and those that cleave it (pathway II). For pathway II ions of phenylcoumarans and resinols, numbers in superscript refer to the bonds that are broken (K. Morreel, H. Kim, F. Lu, T. Akiyama, O. Dima, C. Niculaes, R. Vanholme, G. Goeminne, D. Inzé, E. Messens, J. Ralph, and W. Boerjan, unpublished data).
For trilignols and higher order oligolignols, the pathway II fragmentations are not completely the same as those observed in the MS2 spectra of dilignols [e.g. the β-aryl ether pathway II-associated C− ion occurs only upon CID of X(8–8)X-containing trimers; Fig. 2]. Additionally, the presence of multiple linkage units provides multiple pathway I and II m/z peak groups in the MS2 spectrum. Therefore, additional nomenclature is necessary to annotate all product ions: AE, β-aryl ether linkage unit; PC, phenylcoumaran linkage unit; I, pathway I; II, pathway II. Pathway II first product ions (i.e. due to MS2 fragmentation) were observed for β-aryl ether linkages but not for the other linkage types. Product ions derived from a β-aryl ether-associated pathway II fragmentation (i.e. the A−B− product ions) often possess sufficient internal energy to provoke a secondary fragmentation, leading to fragmentations of the remaining bond. In the case of a pure β-aryl ether, this is annotated as AE2X, with X either A or B when the secondary fragmentation originates from an A− or B− product ion, respectively. The A−/B− product ions resulting from a β-aryl ether cleavage are further annotated with a 4 or 8 in superscript to indicate whether they derive from the 8-end- or 4-end-located 8–O–4-linkage unit. This superscript should not be confused with the superscripts used for pathway II product ions of dilignols.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MSn spectra of G(8–O–4)S(8–5)Gglycerol.
Supplemental Table S1. Oligolignols present in poplar xylem.
Supplementary Material
Acknowledgments
We thank Martine De Cock for help in preparing the manuscript.
References
- Adler E. (1977) Lignin chemistry: past, present and future. Wood Sci Technol 11: 169–218 [Google Scholar]
- Baucher M, Halpin C, Petit-Conil M, Boerjan W. (2003) Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Biemann K. (1988) Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom 16: 99–111 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bowie JH. (1990) The fragmentations of even-electron organic negative ions. Mass Spectrom Rev 9: 349–379 [Google Scholar]
- Comte G, Vercauteren J, Chulia AJ, Allais DP, Delage C. (1997) Phenylpropanoids from leaves of Juniperus phoenicea. Phytochemistry 45: 1679–1682 [Google Scholar]
- Damiani I, Morreel K, Danoun S, Goeminne G, Yahiaoui N, Marque C, Kopka J, Messens E, Goffner D, Boerjan W, et al. (2005) Metabolite profiling reveals a role for atypical cinnamyl alcohol dehydrogenase CAD1 in the synthesis of coniferyl alcohol in tobacco xylem. Plant Mol Biol 59: 753–769 [DOI] [PubMed] [Google Scholar]
- Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, Van Beeumen J, Ralph J, Boudet AM, Kopka J, Rochange SF, et al. (2007) Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Della Greca M, Ferrara M, Fiorentino A, Monaco P, Previtera L. (1998) Antialgal compounds from Zantedeschia aethiopica. Phytochemistry 49: 1299–1304 [Google Scholar]
- Domon B, Costello CE. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J 5: 397–409 [Google Scholar]
- Fabre N, Rustan I, de Hoffmann E, Quetin-Leclercq J. (2001) Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J Am Soc Mass Spectrom 12: 707–715 [DOI] [PubMed] [Google Scholar]
- Gellerstedt G, Pettersson EL. (1975) Light-induced oxidation of lignin: the behaviour of structural units containing a ring-conjugated double bond. Acta Chem Scand A 29: 1005–1010 [Google Scholar]
- Grabber JH, Hatfield RD, Lu F, Ralph J. (2008) Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9: 2510–2516 [DOI] [PubMed] [Google Scholar]
- Higuchi T, Nakatsubo F, Ikeda Y. (1974) Enzymic formation of arylglycerols from p-hydroxycinnamyl alcohols. Holzforschung 28: 189–192 [Google Scholar]
- Houriet R, Tureček F. (1994) Stereochemical effects in ion-molecule reactions studied by ion-cyclotron-resonance spectroscopy. Splitter JS, Tureček F, , Applications of Mass Spectrometry to Organic Stereochemistry. Methods in Stereochemical Analysis Series. Wiley-VCH, New York, pp 483–508 [Google Scholar]
- Kilpeläinen I, Sipilä J, Brunow G, Lundquist K, Ede RM. (1994) Application of two-dimensional NMR spectroscopy to wood lignin structure determination and identification of some minor structural units of hard- and softwood lignins. J Agric Food Chem 42: 2790–2794 [Google Scholar]
- Lapierre C, Monties B, Rolando C. (1985) Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol 5: 277–292 [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Ralph J. (1997) Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J Agric Food Chem 45: 2590–2592 [Google Scholar]
- Lu F, Ralph J. (2003) Non-degradative dissolution and acetylation of ball-milled plant cell walls: high-resolution solution-state NMR. Plant J 35: 535–544 [DOI] [PubMed] [Google Scholar]
- Ma YL, Li QM, Van den Heuvel H, Claeys M. (1997) Characterization of flavone and flavonol aglycones by collision-induced dissociation tandem mass spectrometry. Rapid Commun Mass Spectrom 11: 1357–1364 [DOI] [PubMed] [Google Scholar]
- McLuckey SA, Habibi-Goudarzi S. (1993) Decompositions of multiply charged oligonucleotide anions. J Am Chem Soc 115: 12085–12095 [Google Scholar]
- McLuckey SA, Van Berkel GJ, Glish GL. (1992) Tandem mass spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom 3: 60–70 [DOI] [PubMed] [Google Scholar]
- Morreel K, Goeminne G, Storme V, Sterck L, Ralph J, Coppieters W, Breyne P, Steenackers M, Georges M, Messens E, et al. (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J 47: 224–237 [DOI] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W. (2004a) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Lu F, Goeminne G, Busson R, Herdewijn P, Goeman JL, Van der Eycken J, Boerjan W, Messens E. (2004b) Phenolic profiling of caffeic acid O-methyltransferase-deficient poplar reveals novel benzodioxane oligolignols. Plant Physiol 136: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önnerud H, Gellerstedt G. (2003) Inhomogeneities in the chemical structure of hardwood lignins. Holzforschung 57: 255–265 [Google Scholar]
- Ralph J. (2006) What makes a good monolignol substitute? Hayashi T, , The Science and Lore of the Plant Cell Wall Biosynthesis, Structure and Function. BrownWalker Press, Boca Raton, FL, pp 285–293 [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ralph J, Brunow G, Harris PJ, Dixon RA, Schatz PF, Boerjan W. (2008) Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? Daayf F El Hadrami A Adam L Balance GM, , Recent Advances in Polyphenol Research, Vol 1 Wiley-Blackwell Publishing, Oxford, pp 36–66 [Google Scholar]
- Ralph J, Landucci LL. (2010) NMR of lignins. Heitner C, Dimmel DR, , Lignins. Marcel Dekker, New York, pp 137–234 [Google Scholar]
- Ralph J, Lapierre C, Marita JM, Kim H, Lu F, Hatfield RD, Ralph S, Chapple C, Franke R, Hemm MR, et al. (2001) Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Ralph J, MacKay JJ, Hatfield RD, O'Malley DM, Whetten RW, Sederoff RR. (1997) Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Ralph J, Peng J, Lu F, Hatfield RD, Helm RF. (1999) Are lignins optically active? J Agric Food Chem 47: 2991–2996 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP. (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Sederoff RR, MacKay JJ, Ralph J, Hatfield RD. (1999) Unexpected variation in lignin. Curr Opin Plant Biol 2: 145–152 [DOI] [PubMed] [Google Scholar]
- Shi HM, Wen J, Jia CQ, Jin W, Zhang XF, Yao ZR, Tu PF. (2006) Two new phenolic glycosides from the barks of Hydnocarpus annamensis and their anti-inflammatory and anti-oxidation activities. Planta Med 72: 948–950 [DOI] [PubMed] [Google Scholar]
- Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD. (2009) The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol 150: 621–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung NH, Ding Y, Choi EM, Minh CV, Kim YH. (2009) New neolignan component from Camellia amplexicaulis and effects on osteoblast differentiation. Chem Pharm Bull (Tokyo) 57: 65–68 [DOI] [PubMed] [Google Scholar]
- Umezawa T. (2003) Diversity in lignan biosynthesis. Phytochem Rev 2: 371–390 [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- van Parijs FRD, Morreel K, Ralph J, Boerjan W, Merks RMH. (2010) Modeling lignin polymerization. I. Simulation model of dehydrogenation polymers. Plant Physiol 153: 1332–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L. (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZR, Chai XY, Bai CC, Ren HY, Lu YN, Shi HM, Tu PF. (2008) Xylocosides A–G, phenolic glucosides from the stems of Xylosma controversum. Helv Chim Acta 91: 1346–1354 [Google Scholar]
- Zhang L, Henriksson G, Gellerstedt G. (2003) The formation of β-β structures in lignin biosynthesis: are there two different pathways? Org Biomol Chem 1: 3621–3624 [DOI] [PubMed] [Google Scholar]
- Zhao C, Cao W, Nagatsu A, Ogihara Y. (2001) Three new glycosides from Sinopodophyllum emodi (Wall.) Ying. Chem Pharm Bull (Tokyo) 49: 1474–1476 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.