Abstract
The barley (Hordeum vulgare) brittle stem mutants, fs2, designated X054 and M245, have reduced levels of crystalline cellulose compared with their parental lines Ohichi and Shiroseto. A custom-designed microarray, based on long oligonucleotide technology and including genes involved in cell wall metabolism, revealed that transcript levels of very few genes were altered in the elongation zone of stem internodes, but these included a marked decrease in mRNA for the HvCesA4 cellulose synthase gene of both mutants. In contrast, the abundance of several hundred transcripts changed in the upper, maturation zones of stem internodes, which presumably reflected pleiotropic responses to a weakened cell wall that resulted from the primary genetic lesion. Sequencing of the HvCesA4 genes revealed the presence of a 964-bp solo long terminal repeat of a Copia-like retroelement in the first intron of the HvCesA4 genes of both mutant lines. The retroelement appears to interfere with transcription of the HvCesA4 gene or with processing of the mRNA, and this is likely to account for the lower crystalline cellulose content and lower stem strength of the mutants. The HvCesA4 gene maps to a position on chromosome 1H of barley that coincides with the previously reported position of fs2.
The mechanical strength of stems of cereal crop species is an important agronomic trait, particularly just prior to harvest, when the stems are laden with developing grain. Mechanical failure of stems at this stage, which might be initiated by environmental conditions such as high winds or heavy rain, can lead to substantial losses of yield. While crop species with brittle stems are undesirable in this context, brittle stem mutants are attracting attention in emerging lignocellulosic bioethanol technologies, insofar as they are likely to require lower energy input during grinding of the crop residues prior to the conversion steps of bioethanol production. As a result, there has been considerable interest in identifying factors that influence stem strength and how these factors might be manipulated, either through traditional breeding or emerging genetic engineering approaches, to generate cereal varieties with altered mechanical strength. Stems of plants are variously referred to as stems, stalks, or culms, depending on the species. Here, we will use stems as a general term to cover each of these possibilities.
Brittle stem mutants of rice (Oryza sativa), barley (Hordeum vulgare), and maize (Zea mays) have become important tools for the identification of factors that contribute to stem strength (Kokubo et al., 1989; Li et al., 2003; Ching et al., 2006). In some mutant lines, reduced stem strength has been correlated with decreased cellulose content of the stems (Kokubo et al., 1991; Appenzeller et al., 2004) and with greatly reduced numbers of terminal rosette cellulose synthase complexes in the plasma membrane (Kimura et al., 1999). The content of lignin, pectic polysaccharides, arabinoxylans, and (1,3;1,4)-β-d-glucans in stems of the barley brittle culm mutants remain largely unchanged (Kokubo et al., 1991). In a few cases, the gene that carries a lesion associated with reduced strength and cellulose content has been identified. For example, the Brittle Stalk2 (bk2) gene of maize and the Brittle Culm1 (bc1) gene of rice encode glycosyl phosphatidylinositol-anchored Cobra-like proteins (Li et al., 2003; Ching et al., 2006; Brady et al., 2007; Sindhu et al., 2007). Although the precise biological function of Cobra-like proteins has not been defined, perturbations of the gene result in reduced levels and abnormal orientation of cellulose microfibrils and abnormal anisotropic cell expansion (Schindelman et al., 2001; Roudier et al., 2005). In another example, the bc7 phenotype in rice mutants generated by 60Co γ-irradiation is believed to result from an aberrant cellulose synthase (CesA) gene (Yan et al., 2007). Overall, a large number of different genes, possibly more than 20, have been implicated in various brittle stem mutants or in mutants with reduced cellulose content (Supplemental Table S1), although in most cases the gene mutations have not been identified unequivocally or characterized. Identification of genes involved in brittle stem mutants in commercially important members of the Triticeae has been further hampered because no genome sequence has been available for these species.
Here, we have examined two brittle stem mutants of barley, for which the “brittle” loci were originally designated “fragile stem” (fs). The single-gene brittle stem mutants are spontaneous mutants derived from the parental barley landraces Ohichi (line J755) and Shiroseto (line J156). The corresponding mutant lines, both of which carried mutations at the fs2 locus, are designated X054 and M245, respectively, and have a different genetic background (Kimura et al., 1999). The growth of the internodes of barley stems follows the same pattern as in other cereals, where dividing and elongating cells in each sequential internode push the apical meristem upward (Evert, 2006). The number of divisions and the extent of elongation together dictate the final length of the internode in the stage of primary growth, where cell walls are thin and plastic. Once cells have ceased elongating, secondary thickening of the walls may occur. During secondary thickening, the walls acquire additional layers of material composed mainly of cellulose, heteroxylans, and lignin. This is seen particularly in specialized cells of the vascular bundles and in some cells of ground tissue, including older collenchyma and sclerenchyma fibers (Evert, 2006). Accordingly, there is a continuum of developmental stages along the length of each internode, where the younger cells at the base, or elongation zone, have the thinnest walls and may be still elongating and even dividing. In contrast, older cells in the upper, maturation zone of the internode may already be undergoing secondary thickening.
Our overall experimental approach was to use microarray techniques to identify genes for which transcript abundance in stems of the mutant lines differed significantly from levels in the parental lines. Although a barley gene microarray was commercially available, it was not used here because examination of its constituent genes revealed that many genes from the large CesA and cellulose synthase-like (Csl) gene families of barley (Hazen et al., 2002; Fincher, 2009) were not represented. Therefore, a custom-designed “barley cell wall microarray” was generated, based on long oligonucleotide (60-bp) technologies. Gene sequences were assembled from various publicly available EST and other databases and from our own unpublished sequences of barley HvCesA and HvCsl genes. Approximately 1,400 genes encoding sugar nucleotide-interconverting enzymes, polysaccharide synthases, glycosyl transferases, expansins, cell wall structural proteins, polysaccharide hydrolases and transferases, and enzymes involved in lignin biosynthesis and degradation were selected from the databases.
Special attention was also directed to the sampling of stem tissues for microarray analysis. Thus, comparisons of parental and mutant lines in the younger cells of the basal, elongation regions of the internode were undertaken in attempts to identify the primary genetic lesion. Analyses of the older cells in the upper, maturation zones of the internodes were expected to reveal pleiotropic responses that occur as the plant tries to compensate for reduced stem strength.
The microarray data pointed to a relatively small number of genes that were differentially transcribed in the lower, elongation regions of internodes of the mutant lines, while transcript levels of many genes changed in the more mature, upper regions of the internode. Following confirmation of changes in transcript levels by real-time quantitative (Q)-PCR, a CesA gene that mapped to the same locus as the fs2 gene on barley chromosome 1H was sequenced. A lesion in the gene that is likely to result in the lower crystalline cellulose content and stem strength of the mutant lines was characterized.
RESULTS
Stem Strength and Crystalline Cellulose Content Are Greatly Reduced in the Brittle Culm Mutant Lines
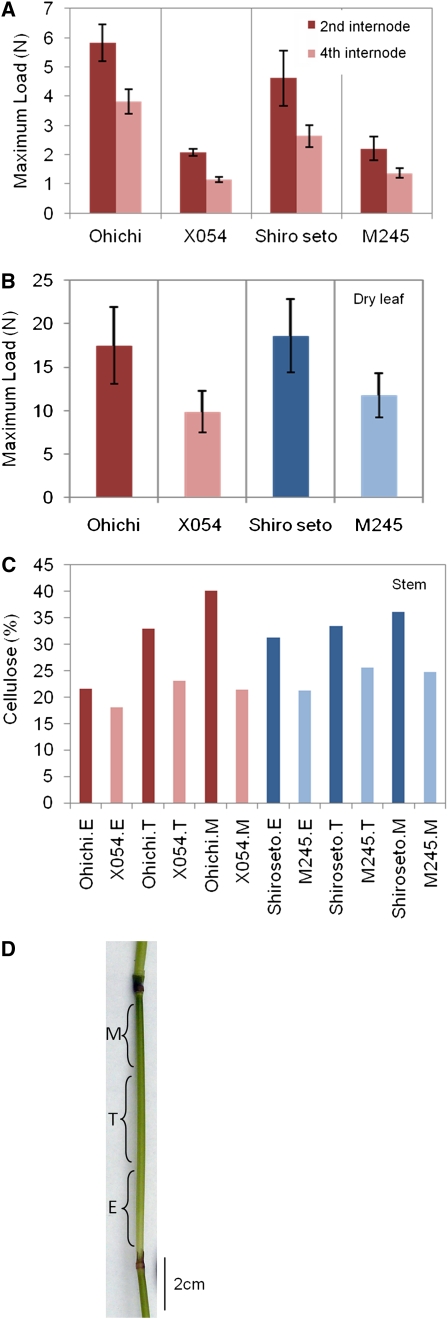
To confirm and quantitate the differences in tissue strength originally reported for the barley brittle stem mutants (Kokubo et al., 1989, 1991), the tensile strength of dry leaves and the flexural strength of fresh stems of the parental and mutant lines were measured. In all cases, the strength parameters of the mutant lines, including the three-point flexural strength of the second and fourth internodes and the “load-to-break” of dry leaves, were significantly lower than those of the corresponding parental lines (Fig. 1, A and B). The crystalline cellulose contents of stems of the two mutant lines, as measured by the acetic acid-nitric acid method of Updegraff (1969), were also reduced to between approximately 60% and 40% of the corresponding parental line (Fig. 1C). The approximate locations of tissues sampled for the subsequent microarray analyses are shown in Figure 1D.
Figure 1.
Comparisons of strength and cellulose content of barley brittle culm mutants and their parental lines. A, Maximum “load-to-bend” of the second and fourth internodes of the barley lines Ohichi (J755) and Shiroseto (J156) compared with the mutant lines X054 and M245, respectively. In both mutant lines and in both internodes, the strength of the stems of the mutant lines is significantly lower than that of the parental lines. B, Maximum load-to-break of dry leaves from the barley lines Ohichi (J755) and Shiroseto (J156) compared with the mutant lines X054 and M245, respectively. Again, the strength of the tissues from the mutant lines is lower than that of the parental lines. C, Crystalline cellulose determination (Updegraff, 1969) of stem cell walls from brittle stem mutants, where E = lower, elongation zone, M = upper, maturation zone, and T = transition zone. Both mutant lines showed reduced cellulose levels relative to their corresponding parental line. For each line, the lower, elongation zone showed the lowest cellulose content of the three zones tested. D, Fourth internode from a barley stem, showing the approximate positions of tissue sampling.
Despite the lower levels of crystalline cellulose observed in stems of the mutant lines (Fig. 1C), a complete linkage analysis of cell wall polysaccharides from leaf and leaf sheath material revealed only minor differences between the mutants and their parental lines (Supplemental Table S2). In particular, levels of the 4-Glcp derivative, which are indicative of (1,4)-linked glucosyl residues, were slightly higher in walls of the mutant lines (Supplemental Table S2). Levels of the other major partially methylated alditol acetate derivatives were also very similar. When we estimated the polysaccharide composition of the walls, based on the structures of well-characterized wall polysaccharides from barley (Gibeaut et al., 2005), we could find no evidence for significant changes in other polysaccharides that contain 4-Glcp residues, namely the (1,3;1,4)-β-d-glucans or xyloglucans (Supplemental Table S3).
These data for wall composition in leaves and crystalline cellulose content in stems suggest that the walls of the barley brittle stem mutant lines contain similar levels of “cellulose,” defined as (1,4)-β-d-glucan, but that the crystallinity or possibly the molecular size of the cellulose is reduced. Given the data of Kokubo et al. (1991), who showed that the degree of polymerization of cellulose was similar between the brittle fs2 lines and corresponding parental lines, it is more likely that the analytical data presented above reflect reduced crystallinity of cellulose in the mutant lines.
Microarray Normalization Procedures Were Evaluated
The barley cell wall microarray chip falls into the category of “boutique array,” because the majority of the features are long oligonucleotides, derived from genes or ESTs, which correspond to fragments of genes involved in one particular function, namely cell wall biology. These oligonucleotide features are referred to here as their corresponding genes. As stated by Oshlack et al. (2007), “there is as yet no widely accepted standard method for normalization of boutique arrays” (p. R2.2). Furthermore, most of the investigations of normalization methods for boutique arrays have focused on two-color cDNA microarrays, not long oligonucleotide arrays (Wilson et al., 2003; Oshlack et al., 2007), and may not be applicable to the one-color, long oligonucleotide array used here. Therefore, three normalization methods were evaluated to account for systematic errors in the data, including the cyclic Loess normalization method (Ballman et al., 2004), 75th percentile scaling, and normalization to the 44 housekeeping genes present on the array. Loess normalization assumes that the majority of the genes are not differentially expressed (Dudoit et al., 2002), and in a study on two-color microarrays, Oshlack et al. (2007) showed that Loess normalization is only robust if less than 20% of genes show asymmetric differential expression.
It appeared from the MA plots of the barley cell wall microarray used here that the number of potentially differentially expressed genes was too large to justify normalizing the data using the fast linear Loess procedure or 75th percentile scaling. Normalization, therefore, was performed using the series of control genes.
Microarray Analyses Reveal Differentially Transcribed Genes
The results of the statistical analysis of the microarray data showed that transcription levels of up to several hundred genes differed between the mutant lines and their near parental lines in the upper, maturation zones of the stem internode (Table I). However, only two features on the microarray were strongly down-regulated in the lower, elongating zones of the internodes in the X054 and the M245 mutants (Table I). The two features, in fact, were both HvCesA4, which had been monitored on the microarray with two separate oligonucleotides. The log2 fold changes of the HvCesA4 transcripts in the mutant lines are shown in Table II, where it is apparent that transcript abundance is approximately 16-fold lower in the mutant lines. Also shown in Table II are the up-regulated genes in the lower, elongation zone, namely a Sasanda retrotransposon of the type I Copia group and genes encoding a putative aquaporin/silicon transporter and a putative ascorbate oxidase. In the case of the Copia retrotransposon, this means that the transcript abundance in the mutant lines is between 48-fold and 630-fold higher than in the parental lines. The sequence of the oligonucleotide probe of the type I Copia retrotransposon corresponded to the gag region, which encodes proteins that form the nucleocapsid core and protease components of the retrotransposon (Grandbastien, 1998).
Table I. Differentially transcribed genes (P ≤ 0.05, FDR adjusted) in internodes from barley stems.
| Genotypes (Wild Type - Mutant) | Position in Internode | Total No. of Differentially Expressed Genes | Down-Regulated in Mutant |
| Ohichi (J755) - X054 | Upper zone | 466 | 212 |
| Lower zone | 4 | 2 | |
| Shiroseto (J156) - M245 | Upper zone | 555 | 176 |
| Lower zone | 4 | 2 |
Table II. Differentially transcribed genes in the lower, elongation zones of internodes of stems from parental lines and the fs2 brittle stem mutants (P ≤ 0.05, FDR adjusted).
| Functional Annotation | Fold Change (log2)a (Parent – Mutant) | P |
| Shiroseto (J156) versus mutant M245 | ||
| Ty1-Copia retrotransposon | −5.84 | 1.02E-07 |
| HvCesA4 (oligonucleotide 1) | 4.28 | 1.79E-05 |
| HvCesA4 (oligonucleotide 2) | 4.55 | 2.57E-05 |
| Putative l-ascorbate oxidase | −3.00 | 0.03 |
| Ohichi (J755) versus mutant X054 | ||
| Ty1-Copia retrotransposon | −6.27 | 2.31E |
| HvCesA4 (oligonucleotide 1) | 3.89 | 9.45E |
| HvCesA4 (oligonucleotide 2) | 4.06 | 0.00017 |
| Putative aquaporin/silicon transporter | −6.97 | 0.00017 |
A negative number here indicates up-regulation, while a positive number indicates down-regulation.
Transcript abundance of differentially transcribed genes in the upper, maturation zones of the barley internodes are shown in Table III, where similar trends are observed for the HvCesA4 gene and the retrotransposon. However, transcripts for a larger number of other genes are also altered in the mutant lines in these upper regions of the internode.
Table III. Selected differentially transcribed genes in the upper zones of internodes from stems of Ohichi (J755) and mutant X054 (P ≤ 0.05, FDR adjusted).
The 12 genes with the highest absolute fold change are listed here.
| Functional Annotation | Fold Change (log2) (J755 – X054) | P |
| Down-regulated in mutant | ||
| Putative cinnamyl alcohol dehydrogenase | 2.96 | 0.0159 |
| Tonoplast membrane integral protein ZmTIP4-2 | 3.17 | 0.0262 |
| Solute carrier family 2, Glc transporter | 3.26 | 0.0078 |
| Putative receptor protein kinase CRINKLY4 | 3.38 | 0.0004 |
| Suc phosphate synthase | 3.51 | 5.42E-05 |
| Cold-regulated protein, complete | 3.72 | 0.0139 |
| Putative peroxidase | 3.95 | 0.0175 |
| Putative cellulose synthase catalytic subunit | 4.23 | 6.81E-06 |
| Putative galactoside 2-α-l-fucosyltransferase | 4.64 | 0.0021 |
| Cellulose synthase catalytic subunit 10 | 4.66 | 4.00E-07 |
| Putative peroxidase | 4.83 | 0.00063 |
| Peroxidase 6 | 7.72 | 0.00027 |
| Up-regulated in mutant | ||
| Putative Pro-rich protein | −8.46 | 0.019 |
| Endoxyloglucan transferase | −7.89 | 1.29E-05 |
| Putative thaumatin protein | −7.33 | 2.18E-05 |
| Floral organ regulator 1 | −7.06 | 0.0132 |
| Putative P450 | −7.03 | 9.00E-06 |
| Retrotransposon protein, putative, Ty1-Copia subclass | −6.79 | 3.52E-10 |
| Putative peroxidase ATP6a | −6.79 | 8.98E-06 |
| Floral organ regulator 1 | −6.47 | 1.10E-05 |
| Expressed protein | −6.39 | 3.51E-05 |
| Putative endo-1,4-β-glucanase | −6.32 | 3.25E-05 |
| Putative (1,4)-β-mannan endohydrolase | −6.22 | 2.51E-06 |
| Putative Pro-rich protein | −6.02 | 0.00049 |
When the levels of HvCesA4 transcripts were compared in the elongation and maturation zones of the parental lines, the HvCesA4 mRNA abundance was generally slightly higher in the maturation zone, consistent with the synthesis of secondary cell walls in that zone (data not shown). For example, in internodes from the Ohichi parent, signal intensities of 14.12 and 12.38 (log2) were observed for the upper, maturation zone and the lower, elongation zone, respectively.
Q-PCR Confirms the Lower Levels of HvCesA4 Transcripts in the Mutant Lines
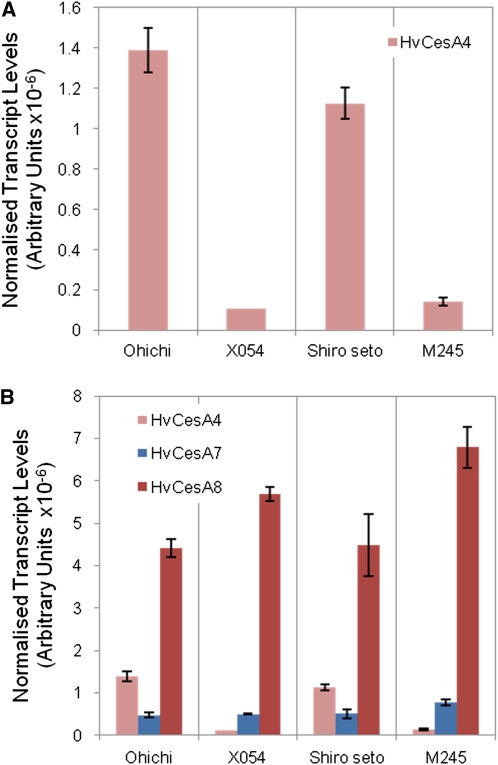
When transcript levels of the HvCesA4 gene were compared quantitatively by Q-PCR in stems of the parental lines and the mutants, 10- to 12-fold decreases were observed in the mutant lines (Fig. 2A). The barley HvCesA4 gene was shown by Burton et al. (2004) to be coexpressed with the HvCesA7 and HvCesA8 genes in a broad range of tissues. However, the lower transcript levels of the HvCesA4 gene in the mutant lines were not matched by lower levels of HvCesA7 and HvCesA8 gene transcripts, which were slightly up-regulated in the mutant lines (Fig. 2B).
Figure 2.
Transcript abundance of cellulose synthase genes. A, Normalized abundance of mRNA levels for cellulose synthase genes in the two barley brittle culm mutants and their parental lines, showing reduced abundance of HvCesA4 transcripts in the mutant lines. B, Normalized abundance of mRNA levels for cellulose synthase genes in the two barley brittle culm mutants and their parental lines, showing a slight up-regulation in HvCesA7 and HvCesA8 transcripts in the mutant lines.
A Retroelement Has Been Inserted into the HvCesA4 Genes of the Mutants
The full genomic sequence of the HvCesA4 gene was obtained from the Morex bacterial artificial chromosome P453O19 clone and used as the basis for primer design during the analysis of the mutant lines. For sequence analysis, several attempts were made to amplify full-length HvCesA4 genes from both the parent and mutant genomic DNA using primers 5UFN and 3UR (Table IV). In each attempt, full-length fragments were generated from the parent lines but never from the mutant genomic DNA preparations. To overcome this, the genes were amplified in two large overlapping fragments from the mutant genomic DNAs, using the primer sets 4MIDF with 3UR or 4REV4 with 5UFN (Table IV). This approach yielded the 3′ sections of the mutant genes, but not the 5′ fragments, and therefore indicated that the problem preventing amplification lay in the 5′ region of the genes. A series of primers flanking each of the first three introns in the 5′ region of the gene were designed (EX1F to EX4R; Table IV), and through systematic PCR across these regions, it became apparent that the region of the gene that was resistant to amplification was close to and within the first intron of the HvCesA4 gene in both the X054 and M245 mutant lines.
Table IV. PCR primers.
| Primer | Sequence (5′–3′) |
| 5UFN | CCCTCCTCCACCACATCATCA |
| 3UR | GATTATACAATGCCCCAAAAGTGC |
| 4MIDF | GAAGCACCGCAAATCGAGCAAGG |
| 4REV4 | CAGCTGATGACGTGGATAGCC |
| EX1F | GAGGATGGCAGCCCCTTCGTG |
| EX2F | ACGGCGACATGGACGACTTAG |
| EX3F | GAGCAGCCGCCGCAGAAGTGG |
| EX2R | CTAAGTCGTCCATGTCGCCGTCC |
| EX3R | CCACTTCTGCGGCGGCTGCTCG |
| EX4R | ATCCTGTCCTTCCACTCCATGCTC |
| PROMF9 | GCTAATCTTTGTGGTGCTGCTTTG |
| PROMR3 | GCACGCAGTGTTGGTATGAAAG |
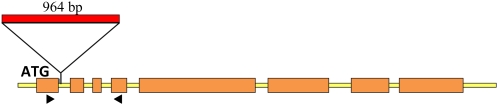
A nested PCR approach across the first intron in the presence of dimethyl sulfoxide eventually yielded a product that was larger than the fragment predicted from the parental sequence. Sequencing of this product revealed a 964-bp insertion in the first intron; the intron was 125 bp in length in the parental lines (Fig. 3). The insertion starts 10 bp inside the 5′ border of the intron and consisted of a 957-bp region that showed 76% sequence identity with the long terminal repeat (LTR) of a Copia-like retrotransposon known as Sasanda (Triticeae Repeat Sequence database, GrainGenes; http://wheat.pw.usda.gov/GG2/blast.shtml). A 7-bp repeat of the upstream 9-bp intron sequence flanking the 5′ end of the insertion was found at the 3′ end of the insert. The sequences and the positions of the insert were identical in the HvCesA4 genes from both X054 and M245.
Figure 3.
Structure of the HvCesA4 gene in the barley brittle stem mutants. The HvCesA4 genes of both mutant lines have a 964-bp insertion in their first intron. The sequence of the insertion corresponds to a solo-LTR of the Sasanda class of type I Copia-like retrotransposons. Orange boxes represent exons, and black arrowheads indicate the positions of the primers used to amplify products from the cDNA (see Fig. 4).
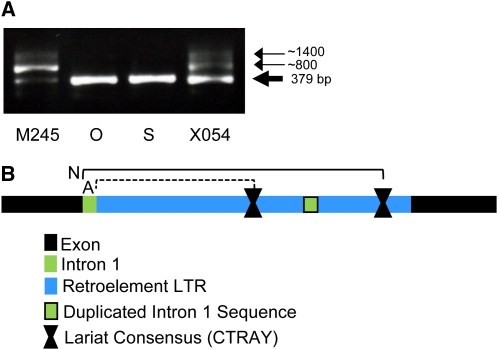
At the mRNA transcript level, PCR amplification across this region of cDNAs synthesized from the two parents and the two corresponding mutants (Table IV) revealed that multiple products were amplified from the X054 and M245 mutant line cDNAs but that a single product of the correctly spliced and predicted size was amplified from cDNAs prepared from parental line RNA (Fig. 4A). For the mutant lines, additional DNA fragments of approximately 380, 500, and 800 bp and 1.4 kb were detected (Fig. 4A). The 1.4-kb fragment has been sequenced and contains the entire retroelement and flanking regions from the HvCesA4 gene; this indicated that no splicing had occurred in this fragment (Supplemental Fig. S1). The smallest fragment (379 bp) was also sequenced and shown to be correctly spliced. Despite frequent attempts, we have been unable to clone the 800-bp cDNA fragment for further analysis, probably due to internal secondary structure.
Figure 4.
Products amplified from barley cDNA preparations using PCR primers spanning the first intron of the HvCesA4 gene. A, The bottom arrow indicates the 379-bp product amplified from cDNA, which was sequenced and confirmed to be the product of a normally spliced mRNA. The other two arrows (approximately 1,400 and 800 bp) indicate products assumed to arise from cDNAs synthesized from incorrectly spliced RNAs that still carry all or part of the LTR insertion in the first intron. The O and S lanes indicate the products from the Ohichi and Shiroseto parent lines, respectively, and M245 and X054 are the PCR products from the corresponding brittle culm mutant lines. B, A schematic diagram (not to scale) of the potential origin of the 503-bp cDNA product that was consistently isolated from PCR of the cDNA corresponding to transcripts of the M245 and X054 HvCesA4 mutants. Note that the 503-bp fragment is not clearly visible in A. Exons 1 and 2 are shown in black, and the normal splicing positions are indicated by N. The aberrant 503-bp product (spliced at position A) retains the first intron (green), but the retroelement (blue) is absent. The positions of the normal (N) and retroelement (A) consensus lariat sequences (CTRAY, where R = A or G and Y = C or T) are shown. In the mutant lines, a lariat could form either between the 5′ end of the intron and the lariat motif sequence in intron 1 during normal splicing (N) or between the 5′ end of the retroelement LTR and the lariat within the retroelement during aberrant splicing (A), leaving intron 1 in the mature mRNA.
The 503-bp fragment contains an intact first intron, but no retrotransposon, in an mRNA in which the downstream introns are all correctly spliced out (Fig. 4B). The retroelement sequence starts with a highly conserved GT intron boundary motif, and there is a branch-point consensus sequence (CTGAT) 671 bp from the 5′ end of the insertion, which would allow the formation of a lariat during splicing. The branch-point consensus sequence for splicing of intron 1 from the wild-type mRNA lies 3′ to the retroelement sequence (Supplemental Fig. S1). Therefore, it seems possible that the splicing mechanism has been perturbed, so that in some cases the lariat forms between the 5′ end of the retroelement sequence and the branch-point consensus within it. In this way, the insert could be spliced out, leaving the original intact intron behind. There is no conserved AG motif at the 3′ end of the retroelement sequence, so a cryptic sequence may be fulfilling this role.
The HvCesA4 Gene and the fs2 Fragile Stem Phenotype Map to the Same Locus on Chromosome 1H
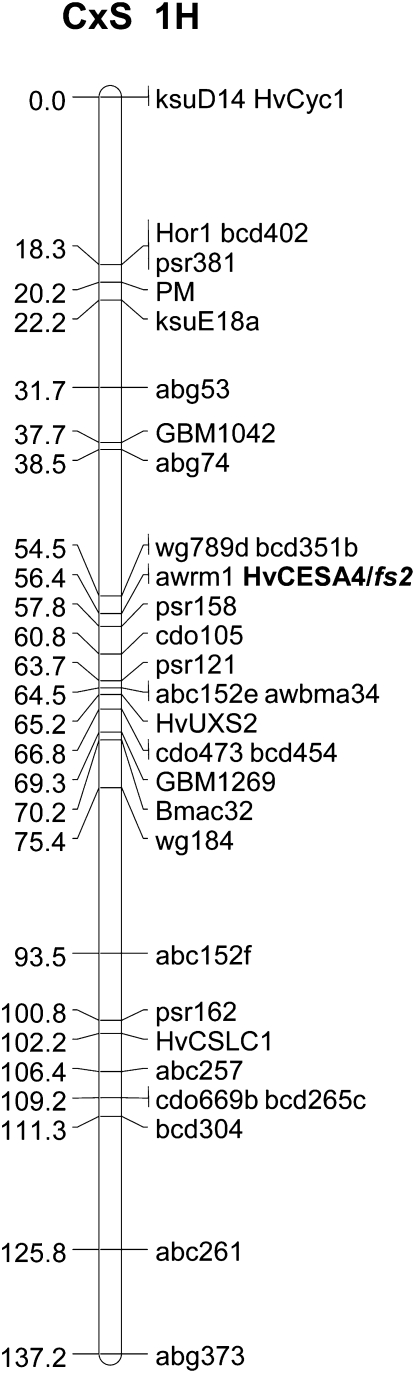
The barley HvCesA4 gene has been previously mapped to the long arm of chromosome 1H using wheat (Triticum aestivum)-barley addition lines (Islam et al., 1981; Burton et al., 2004), but the absence of a polymorphism for the partial HvCesA4 cDNA prevented the gene being mapped to a specific locus (Burton et al., 2004). To enable more precise mapping of the gene, the full genomic sequence of a Morex bacterial artificial chromosome clone (P453O19) was used. The HvCesA4 gene contains eight exons and seven introns, and the full sequence, including the promoter and 3′ untranslated regions, has been submitted to GenBank under accession number HM222644. Acquisition of the full genomic sequence enabled PCR amplification of the gene from the mapping parents Clipper and Sahara. Comparison of the sequenced fragments amplified using the primer set PROMF9 and PROMR3 (Table IV) led to the identification of a polymorphism between cv Clipper and the Sahara landrace. The polymorphism is located in the promoter region 921 bases upstream of the start codon. In the HvCesA4 gene from Clipper, the sequence is AAAAAAATTT, while in Sahara, the corresponding sequence is AAATTTTTTT. Sequencing and scoring of the PCR products amplified from 20 Clipper × Sahara double haploid lines from the mapping population (Karakousis et al., 2003) that were known to carry recombination events on the long arm of chromosome 1H placed the HvCesA4 gene between markers awrm1 and psr158, which are separated by a genetic distance of 1.4 centimorgan (Fig. 5). The mapped position of the HvCesA4 gene is coincident with the previously reported position of fs2 (Takahashi et al., 1966; Tsuchiya, 1972; Tsuchiya and Singh, 1973), as shown in Figure 5.
Figure 5.
Genetic map location of the HvCesA4 gene on chromosome 1H of barley. The genetic map of chromosome 1H of barley shows the colocation of the HvCesA4 cellulose synthase gene and the locus for the fragile stem phenotype (fs2), which are flanked by the markers awrm1 and psr158. Selected double haploid lines from the Clipper × Sahara (CxS) mapping population (Karakousis et al., 2003) were used to map the HvCesA4 gene.
The HvCesA4 Gene Is Not Responsible for the fs3 Fragile Stem Phenotype
As mentioned earlier, lesions in many different genes have been assigned to the brittle stem and low cellulose mutants of cereals and other plants (Supplemental Table S1). Here, we repeated the microarray analyses with extracts from stem internodes from the fs3 fragile stem mutant of barley (M382) and its parental line Kobinkatagi 4 (J066; Kimura et al., 1999). The crystalline cellulose contents of internode sections and leaves of the M382 mutant and its parental line were measured using the acetic-nitric acid method (Updegraff, 1969). In the M382 mutant line, crystalline cellulose contents were reduced by 25% to 50% compared with the J066 wild-type line, depending on the tissue assayed (data not shown), and these reductions were similar to those observed in the two fs2 mutant lines (Fig. 1C).
The microarray analyses revealed completely different transcription patterns in the M382 fs3 mutant compared with those observed in the two fs2 mutants. In contrast to the small number of genes that were differentially transcribed in the lower, elongation zones of stem internodes in the fs2 mutants (Tables I and II), 167 genes (141 up-regulated and 26 down-regulated) potentially related to wall metabolism were differentially transcribed (P ≤ 0.05) in the lower, elongation zones of internodes (Supplemental Table S4). Of these genes, only two were cellulose synthases and both were up-regulated in the M382 fs3 mutant line. In the upper, maturation zones of the internodes, 95 genes (57 up-regulated and 38 down-regulated) were differentially transcribed (P ≤ 0.05; Supplemental Table S5); one cellulose synthase gene was slightly down-regulated in the mutant line. The latter gene, designated OsCesA5 in Supplemental Table S5, is most closely related to the barley HvCesA2 gene, which is believed to be involved in the biosynthesis of cellulose in primary walls and is located on chromosome 5H (Burton et al., 2004).
The fs3 phenotype maps to the short arm of chromosome 7H (data not shown). There are no HvCesA genes on chromosome 7H (Burton et al., 2004). While it is not possible at this stage to identify the gene responsible for the fragile stem phenotype in this fs3 mutant, it may be concluded from these data that transcriptional changes in the fs3 mutant line are quite different from those observed for the fs2 mutants and that the primary genetic lesion is not in a cellulose synthase gene. These data further confirm that mutations that interfere with cellulose synthesis cannot be assumed to reside in CesA genes (Supplemental Table S5).
DISCUSSION
The genetic basis of the brittle stem phenotype (fs2) in two mutant lines of barley has been defined. The mutant lines were designated X054 and M245 and represented spontaneous mutants of the Japanese barley landraces Ohichi (line J755) and Shiroseto (line J156), respectively (Kokubo et al., 1989, 1991). Using a custom-designed microarray that included 1,347 gene sequences selected on the basis of their potential participation in cell wall metabolism, together with 44 standard genes, transcript analyses of RNA preparations from the stems of the mutant and parental lines revealed that the transcript abundance of relatively few genes was significantly lower in the mutant lines (Tables I and II). However, one of these “down-regulated” genes was the HvCesA4 cellulose synthase gene. Given the reduced cellulose content and reduced stem strength in these mutant lines (Fig. 1; Kokubo et al., 1989, 1991), coupled with observations that stem strength is correlated with cellulose content in maize (Appenzeller et al., 2004), it seemed likely that a lesion in the HvCesA4 gene might be responsible for the brittle stem phenotype. This possibility was supported by the observation that the barley HvCesA4 gene mapped to the same position on chromosome 1H as the fs2 gene (Fig. 5; Tsuchiya and Singh, 1973).
Therefore, the HvCesA4 genes from the parental and mutant lines were sequenced. This revealed the presence of a 964-bp insertion in the first intron of the HvCesA4 cellulose synthase gene in the mutant lines (Fig. 3). The sequence of the insertion shared approximately 76% sequence identity with the LTR of a Copia-like retrotransposon that has been designated Sasanda, and the size of the insertion is also similar to that of a solo-LTR from Sasanda (Triticeae Repeat Sequence database, GrainGenes; http://wheat.pw.usda.gov/GG2/blast.shtml). It is noteworthy that the sequences of the retroelement insertions in both the X054 and M245 lines are identical, although the spontaneous brittle stem fs2 mutants were detected in the Ohichi and Shiroseto landraces (Kokubo et al., 1989; Kimura et al., 1999). This indicates that the landraces share a common ancestry, despite the observation that they are now genetically distinct (Kimura et al., 1999). If the two landraces originated from a common ancestor in which the retroelement was inserted into the HvCesA4 gene, one might expect that single nucleotide polymorphisms (SNPs) would have accumulated in the retroelement. However, we have shown previously that the HvCesA4 genes of more than 10 mapping population parental barley lines contain no restriction enzyme polymorphisms (Burton et al., 2004). Here, sequencing the HvCesA4 genes from the genetically distant Australian malting barley variety Clipper and the North African barley landrace Sahara revealed no SNPs; the only difference is the presence of a TTTT/AAAA substitution in the promoter regions (Fig. 5). When the HvCesA4 genes from the wild-type landraces Ohichi and Shiroseto, together with the two brittle stem mutants, were sequenced in this study, no SNPs were detected in the introns or in the coding regions of the genes. Similarly, our haplotyping analyses of the HvCslF6 gene in approximately 40 barley accessions reveal just four SNPs (D.F. Marshall, J. Russell, R. Waugh, R.A. Burton, and G.B. Fincher, unpublished data). Thus, there are precedents for highly conserved CesA and Csl gene sequences in the Triticeae.
Copia-like retrotransposons and retroelements are common in the barley genome (Schulman and Kalendar, 2005), and a Sasanda retroelement has been detected in the Glu-B1 locus of wheat, where it has been implicated in the duplication of the gene encoding the Bx7 subunit of the high-Mr glutenin complex, leading to its overexpression and possibly influencing the unique rheological properties of dough made from wheat flour (Ragupathy et al., 2008; Ragupathy and Cloutier, 2008).
The location of the solo-LTR retroelement insertion in an intron of the HvCesA4 gene raises the question of why transcript levels of the gene are dramatically lower in the barley brittle stem mutants X054 and M245 (Fig. 2A). The insertion contains repetitive sequences, which are predicted to cause stable secondary structures in the corresponding mRNA (data not shown). The formation of hairpin turns in the mRNA might slow transcription of the gene in vivo and might also account for the difficulties encountered in the PCR amplification of this region of the mutant HvCesA4 gene. Furthermore, the appearance of HvCesA4 transcripts of different sizes in the mutant lines, including transcripts that contained the complete LTR retroelement (Fig. 4A), suggests that several transcripts of the region might have been present in the preparation of total RNA that was used for cDNA synthesis, and these transcripts might have originated from aberrant splicing of the intron or from errors in transcription resulting from secondary structures in the mRNA. This was confirmed through sequencing of the PCR products, which showed that missplicing had indeed occurred in the mutant lines (Fig. 4B). The consequences of the missplicing, which in many cases has left an intact intron within the mRNA, would lead to the production of a very short protein, terminating at a stop codon within the remaining first intron. The protein would consist of the 63 amino acids encoded by the first exon and eight additional amino acids from the persistent first intron. Such a truncated protein would not be expected to be active in cellulose biosynthesis. Another possibility would be the presence of a protein-binding motif within the truncated protein that would allow its aggregation with other proteins in the rosette complex. This in turn could form an unbalanced complex leading to reduced production of cellulose or the synthesis of aberrant, noncrystalline cellulose forms.
Because the LTRs of retrotransposons usually contain promoter elements, transcription terminator elements, and polyadenylation motifs (Schulman and Kalendar, 2005), it was also possible that the LTR, which is in the reverse orientation, could initiate transcription that would read across from the LTR into the HvCesA4 gene itself and hence generate antisense RNA that could silence the HvCesA4 genes in the mutant lines. However, we were unable to detect hybrid LTR-HvCesA4 transcripts resulting from transcription initiated on the retroelement and reading through into the HvCesA gene, and we conclude that the reduced levels of the HvCesA4 transcript were most likely attributable to mRNA missplicing or inhibition of transcription of the gene more generally rather than to the presence of antisense HvCesA4 mRNA.
An additional observation that might be related to the presence of the Sasanda solo-LTR retroelement in the HvCesA4 gene of the mutant lines was that one of the most strongly up-regulated features on the microarray, in both the elongation and the maturation zones, corresponded to the non-LTR gag region of a Sasanda retrotransposon (Tables II and III). This suggests that there is at least one intact, full-length Sasanda retrotransposon in the barley genome and that it is transcribed at relatively high levels in the mutant lines. It is important to note that the retrotransposon sequence was included in the cell wall microarray only because the sequence was misannotated as a rhamnogalacturonan I lyase in The Institute for Genomic Research (TIGR) databases.
Whether the presence of the solo-LTR retroelement in the HvCesA4 gene actually reflects increased transcriptional activity of the corresponding retrotransposon family that is observed in the mutant lines is a matter for speculation. It is known that retrotransposon activity and genome enlargement can occur in waves during evolutionary history and that such waves of activity in natural populations of the wild barley Hordeum spontaneum might be initiated by adverse environmental factors related to water stress, high temperatures, and irradiation stress (Faure et al., 1996; Grandbastien, 1998; Kalendar et al., 2000; Schulman and Kalendar, 2005). Increased transcriptional activity of the intact retrotransposon, as indicated by the elevated mRNA levels, might have resulted in transposition of one member of the family into the HvCesA4 gene. Subsequent recombination events would remove the intact retrotransposon from the HvCesA4 locus but would leave a solo-LTR retroelement in the gene. Activation of retrotransposons and associated genome instability can be prolonged into subsequent generations by changes in heritable DNA methylation patterns (Kaup et al., 2006).
When Q-PCR was used to confirm that transcript levels of the HvCesA4 gene were indeed much lower in the mutant lines (Fig. 2A), it was noted concurrently that the levels of HvCesA7 and HvCesA8 transcripts increased slightly (Fig. 2B). This was significant in relation to earlier observations by Burton et al. (2004) that the HvCesA4, HvCesA7, and HvCesA8 genes appeared to be coregulated in a range of barley tissues and organs. It was concluded in that earlier work that this group of three cellulose synthase genes from barley might be coordinately involved in cellulose synthesis in secondary cell walls, possibly through the formation of cellulose synthase terminal rosette complexes that contained three isoforms of cellulose synthase enzymes (Burton et al., 2004). Coordinated expression of small groups of CesA genes has been noted in other higher plant species (Tanaka et al., 2003; Taylor et al., 2003; Appenzeller et al., 2004). The relatively modest increases in levels of HvCesA7 and HvCesA8 transcripts in the barley brittle stem mutant lines that showed greatly reduced levels of HvCesA4 transcripts (Fig. 2) suggested that the mutation did not involve a transcription factor that might mediate the coordinated expression of the three barley HvCesA genes. It further suggested that any feedback mechanisms between the three genes appear to be limited in their ability to compensate for reduced CESA4 activity through up-regulating other members of the terminal rosette complex. This has clear implications in cellulose synthesis during secondary cell wall synthesis. The expression of the HvCesA4, HvCesA7, and HvCesA8 genes appears to be coordinated, but there does not seem to be an adequate mechanism for dealing with a defective or missing protein in the cellulose synthase complex, where such a mechanism would require both the up-regulation of another CesA gene and the replacement of the missing CESA enzyme in the terminal rosette complex with the product of that up-regulated CesA gene. These conclusions are consistent with earlier scanning and freeze-fracture electron microscopy data, which showed quite clearly that the barley brittle stem mutants used here have reduced numbers of terminal rosette complexes in their plasma membrane (Kimura et al., 1999). Thus, any perturbation in transcription of the HvCesA4 gene could lead to lower amounts of active HvCESA4 enzyme, and if this were to somehow prevent the formation of a functional terminal rosette complex, it would provide a logical rationale for the lower levels of crystalline cellulose in the barley brittle stem mutants. This possibility is consistent with earlier observations that the mutant rsw1 allele of Arabidopsis (Arabidopsis thaliana) is associated with a specific reduction in cellulose synthesis, disassembly of the cellulose synthase complex, and the accumulation of noncrystalline (1,4)-β-d-glucan (Arioli et al., 1998). In this connection, the barley brittle stem mutant lines might prove to be a useful system in which to investigate the possibility that single CESA enzymes, which are not associated with functional terminal rosette complexes, are nevertheless active in the plasma membrane and can catalyze the synthesis of single (1,4)-β-d-glucan chains that are deposited in the wall in a nonfibrillar, noncrystalline form.
The sampling of tissue prior to the microarray analyses was designed to provide information on the primary gene lesion in the mutant lines, through the examination of transcript abundance in the younger, elongation zones of the internodes. Indeed, transcript levels of very few genes were significantly different in the mutant and parental lines in these regions (Table II), and the lesion in HvCesA4 was identifiable. Sampling of the upper, more mature zones of the internode was expected to reveal any pleiotropic effects that might occur as a result of compensation for the lower levels of cellulose in the walls of this material and the reduced strength of the internodes in the mutant lines. Consistent with this possibility, many more genes with altered transcriptional activity were detected in the upper, maturation zones of the internode (Table III).
The up-regulated genes included genes that might be involved in lignin metabolism, such as cinnamyl alcohol dehydrogenase and peroxidases. In addition, genes encoding enzymes that could be involved in other aspects of cellulose synthesis or wall strength were up-regulated in the internodes of the mutant lines. These included a (1,4)-β-glucan endohydrolase, a xyloglucan endotransglycosylase, and Pro-rich structural proteins of the wall (Table III). The increased abundance of transcripts for xyloglucan endotransglycosylase might be related to the potential for this enzyme to catalyze the formation of covalent cross-linking of wall polysaccharides, which could reflect an attempt to reinforce the wall in the mutant lines (Hrmova et al., 2007; Fincher, 2009). The greatly increased abundance of transcripts for genes encoding putative Pro-rich wall proteins (Table III) might also serve to strengthen walls through enhancing the network of structural proteins within the wall (Bernhardt and Tierney, 2006). Up-regulation of a (1,4)-β-glucan endohydrolase gene in the mutant lines (Table III), which does not encode a Korrigan enzyme, could be related in some way to previous observations that cellulases and other polysaccharide hydrolases are known to be active during cell wall biosynthesis. The hydrolases might be involved in processes such as trimming or editing nascent polysaccharide chains or in releasing the chains from the biosynthetic enzymes (Fincher, 2009).
In summary, the custom-designed cell wall microarray has been used to demonstrate that the insertion of a retroelement in the first intron of the HvCesA4 gene causes the fs2 brittle stem phenotype in barley, for which a full genome sequence is not yet available. It is likely that the insertion of the retroelement in the HvCesA4 gene interferes with RNA splicing and that this, in part at least, is responsible for the significantly lower levels of HvCesA4 mRNA suggested by the microarray results. It also provides an explanation for the reduced levels of cellulose in walls of these brittle stem mutants. There are a large number of genes that potentially contribute to cellulose synthesis (Supplemental Table S1), and for comparative purposes we have shown that, in contrast to the fs2 mutants, the brittle stem and low cellulose phenotypes of the fs3 barley fragile stem mutant cannot be attributed to a lesion in an HvCesA gene (Supplemental Tables S4 and S5). Analyses of the crystalline cellulose contents of the fs2 mutants and their parental lines, together with full linkage analyses of cell wall polysaccharides, indicate that the reduced cellulose levels reported here and elsewhere probably result from the reduced crystallinity or molecular size of the cellulose in the wall. The total amount of (1,4)-β-glucan is not dramatically altered in leaves of the mutant lines. It has also been possible through the microarray analyses to show that the plant has a limited capacity to compensate for the down-regulation of a gene encoding a single member of the cellulose synthase complex in the fs2 mutants, insofar as the down-regulation of the HvCesA4 gene in the brittle stem barley mutants does not result in the large scale up-regulation of genes encoding partner cellulose synthases of the complex. Finally, the microarray analyses have identified a large number of genes that are probably involved in the plant's response to cell walls with reduced crystalline cellulose contents and, hence, with reduced strength.
MATERIALS AND METHODS
Plant Material
Seeds of the spontaneous fs2 barley (Hordeum vulgare) brittle mutant lines, designated X054 and M245, together with their corresponding wild-type parental lines, designated J755 (cv Ohichi) and J156 (cv Shiroseto), respectively, were obtained from the Barley Germplasm Center Research Institute for Bioresources (Okayama University; http://earth.lab.nig.ac.jp/approximatelydclust/cgi-bin/barley_germ/about.html). The fs3 mutant M382 and its parental line Kobinkatagi 4 were obtained from the same source. The seeds were sown in Horsham mix in pots and vernalized at 4°C for 6 weeks, transferred to the glasshouse, and grown under standard conditions as described by Burton et al. (2004). At Zadocks developmental stage 49 (Tottman, 1987), upper and lower zones from the fourth internodes of stems were collected for RNA isolation. Material from three individual plants was pooled as one replicate, and RNA preparations were isolated from three replicates of the stem material.
RNA Preparation and cDNA Synthesis
Total RNA was extracted from the plant material using a commercially prepared guanidine reagent, TRIzol (Invitrogen), according to the manufacturer's instructions (Burton et al., 2008). Purified RNA was treated with DNaseI using the DNA-free kit (Ambion), and RNA integrity was checked on a 1.6% (w/v) agarose gel containing ethidium bromide. Total RNA quality was assessed on an Agilent BioAnalyzer 2100 before poly(A+) RNA was isolated and used for the synthesis of cDNA for hybridization against the array.
Custom-Designed Barley Cell Wall Gene Microarray
Gene sequences potentially related to cell wall metabolism in barley were identified through examination of public EST databases and through sequences generated previously in our laboratories. Databases examined included Cell Wall Genomics (Yong et al., 2005; http://cellwall.genomics.purdue.edu/families/index.html), Cell Wall Navigator, National Center for Biotechnology Information EST (http://www.ncbi.nlm.nih.gov/dbEST/), TIGR barley gene index (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=barley), The Arabidopsis Information Resource (http://www.Arabidopsis.org/), Barleybase (http://www.barleybase.org/), and CAZy (http://afmb.cnrs-mrs.fr/CAZY/index.html). Where necessary, EST sequences were assembled into contigs (contiguous sequences) using the program ContigExpress, which is part of the Vector NTI Advance 9.1.0 (Invitrogen) suite of programs. A total of 1,347 gene sequences were identified on the basis of their potential participation in cell wall metabolism, together with 44 standard or “housekeeping” genes, and the annotations of these genes were assigned using TIGR Plant Transcript Assemblies (http://plantta.tigr.org/cgi-bin/plantta_release.pl). The microarray data are available at the National Center for Biotechnology Information under Gene Expression Omnibus accession number GSE17873.
Oligonucleotides of 60 bp were designed for each gene, and eight 1.5K chips were produced by Agilent Technologies and subjected to one-color (Cy3) hybridization. Hybridization and image extraction were performed by Agilent Technologies using standard protocols. Agilent Technologies FE 9.1 software was used for feature extraction. No gene was flagged.
For the removal of systematic errors in the data, three normalization methods were tested: fast linear Loess, which is a modified cyclic Loess normalization method (Ballman et al., 2004), 75th percentile scaling, and normalization to the 44 housekeeping genes present on the array. The measured background values were generally low, so we followed the recommendation by Zahurak et al. (2007) and did not perform background subtraction. Prior to normalization, signal intensity values of less than 1.0 were set to zero, while all other values were log2 transformed.
The statistical analysis of differential gene expression was performed using the LIMMA library implemented in R (Smyth, 2004). Differentially expressed genes between the mutant and its wild-type parent were identified at P < 0.05 with an empirical Bayes t test using false discovery rate (FDR) for multiple testing correction (Benjamini and Hochberg, 1995). An MA plot, which is a log2-based scatterplot of the average fluorescence intensity A versus the transformed ratio M, was used as a diagnostic tool to detect irregularities such as dye bias and extreme differences in hybridization intensities. For two-color microarrays, M = log2 Red-log2 Green, while A = 1/2(log2 Red + log2 Green). For single-color microarrays, this becomes M = log2 X1 – log2 X2, A = 1/2(log2 X1 +log2 X2), with X1 and X2 being the signal intensities of the first (single-color) experiment and the second experiment.
Q-PCR Validation
Q-PCR was used to validate the microarray data and was performed essentially as outlined by Burton et al. (2008). Twelve genes were selected for microarray validation. The Q-PCR mixtures were assembled with a liquid-handling CAS-1200 robot (Corbett Robotics). Three replicate PCRs for each of the cDNAs were included in every run containing 2 μL of cDNA solution, the diluted standard or water used in a reaction containing 5 μL of IQ SYBR Green PCR reagent (Bio-Rad Laboratories), 1.2 μL each of the forward and reverse primers at 4 μm, 0.3 μL of 10× SYBR Green in water, and 0.3 μL of water. The total volume of the PCRs was 10 μL. Reactions were performed in an RG 6000 Rotor-Gene Real Time Thermal Cycler (Corbett Research) for 3 min at 95°C followed by 45 cycles of 1 s at 95°C, 1 s at 55°C, 30 s at 72°C, and 15 s at the optimal acquisition temperature. Transcript levels of genes encoding glyceraldehyde 3-phosphate dehydrogenase, heat shock protein 70, cyclophilin, and α-tubulin were used as controls (Table II), and these correspond to those listed in Table IV of Burton et al. (2004). Normalization was performed using multiple control genes as described by Burton et al. (2004), and the final concentrations of mRNAs of the genes of interest are expressed as numbers of copies per μL of cDNA, normalized against the geometric means of the three control genes, which varied the least with respect to each other (Vandesompele et al., 2002).
Genetic Mapping of HvCesA4
The HvCesA4 gene was mapped by nucleotide polymorphism genotyping using the double haploid lines from the Clipper × Sahara mapping population (Karakousis et al., 2003). PCR amplification using standard methods and Platinum Taq polymerase (Invitrogen) was performed on 20 lines using primers PROMF9 and PROMR3, which produced a 411-bp fragment. Sequencing of the fragments on an ABI3700 (Australian Genome Research Facility) allowed the scoring of a polymorphism, which was assigned to chromosome 1H.
Measurement of Tissue Strength
The barley brittle stem mutants and the corresponding parental lines were grown in a glasshouse until the flag leaf was fully emerged. The second and fourth internodes from the base of the plant were analyzed for three-point flexural strength, using an Instron 5543 materials testing instrument with a span distance of 20 mm and an anvil rate of 60 mm min−1. The maximum flexural load (Newtons) required to bend the midpoint of each internode was recorded. Internodes from five replicates of each line were tested, except for line X054, where only two stems were available. For the measurement of leaf strength, the leaf below the flag leaf was removed and dried at 37°C. The leaf was held in pneumatic grips in the same Instron instrument and pulled apart at a constant rate of 30 mm min−1. The maximum load to break the leaf was used to differentiate tensile strength (Newtons).
Cellulose Content of Walls
Frozen tissue samples were treated to prepare an alcohol-insoluble residue (cell walls) by extraction of ground material with successive washes of 80% ethanol, methanol, and acetone before air drying. The amount of material recovered was quite low (as low as 4 mg for some), and as a result it was not possible to run the cellulose measurements in duplicate. Only J755 (maturation zone) and J066 (maturation zone) were analyzed in duplicate. Crystalline cellulose was estimated as described by Updegraff (1969) with some modifications. Wall material was dried in a vacuum oven at 40°C overnight and carefully weighed (10–20 mg) into 2-mL Eppendorf tubes. Acetic acid-nitric acid reagent (acetic acid:nitric acid:water, 8:1:2 [v/v/v]) was added, and the tubes were heated at 100°C for 90 min with occasional mixing. The reaction was cooled, and the tubes were centrifuged at 13,000 rpm for 5 min to pellet the cellulose. The supernatant was removed, and the cellulose pellet was washed sequentially with water, 80% (v/v) ethanol (four times), and acetone. After removal of the acetone, the pellet was air dried, dried in a vacuum oven at 40°C overnight, and weighed. The percentage of cellulose was determined from this weight as a proportion of the original starting weight.
Compositional Analysis of Cell Walls
The alcohol-insoluble cell wall preparation was suspended in 10 mm Tris-maleate buffer, pH 7.0, containing 1 mm CaCl2 and 10 mm NaCl, heated at 100°C for 5 min, and treated twice with 100 units of porcine pancreatic α-amylase at 40°C for 1 h. Ethanol was added to 80% (v/v), and polysaccharides were precipitated at 4°C overnight. The pellet was washed twice with 70% ethanol, once each with chloroform:methanol (1:1, v/v), ethanol, acetone, and methanol, and air dried. Cell walls were methylated as described by Sims and Bacic (1995) except that walls were doubly methylated before hydrolysis and reduction and acetylated according to Harris et al. (1984). The polysaccharide compositions of walls were estimated from monosaccharide composition and linkage analysis as described by Gibeaut et al. (2005) for barley walls and by Sims and Bacic (1995) for dicot walls.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number HM222644.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence of the first intron and retroelement in the HvCesA4 gene of mutant line M245.
Supplemental Table S1. Genes associated with brittle stem phenotypes and cellulose synthesis in plants.
Supplemental Table S2. Neutral monosaccharide linkage analysis of cell walls.
Supplemental Table S3. Polysaccharide composition based on neutral linkage analysis.
Supplemental Table S4. Differentially transcribed genes in the lower, elongation zones of internodes of stems from Kobinkatagi (J066) and its near isogenic fs3 mutant M382.
Supplemental Table S5. Differentially transcribed genes in the upper, maturation zones of internodes of stems from Kobinkatagi (J066) and its near isogenic fs3 mutant M382.
Supplementary Material
Acknowledgments
We thank Margie Pallotta for assistance with mapping the HvCesA4 gene and Alan Schulman for helpful suggestions. We also thank Natalie Kibble and Margaret Buchanan for their invaluable help with the manuscript.
References
- Appenzeller L, Doblin M, Barreiro R, Wang H, Niu X, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS. (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11: 287–299 [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Ballman KV, Grill DE, Oberg AL, Therneau TM. (2004) Faster cyclic Loess: normalizing RNA arrays via linear models. Bioinformatics 20: 2778–2786 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Bernhardt C, Tierney ML. (2006) Proline-rich cell-wall proteins: building blocks for an expanding cell wall? Hayashi T, , The Science and Lore of the Plant Cell Wall. Brown Walker Press, Boca Raton, FL, pp 164–170 [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN. (2007) Combining expression and comparative evolutionary analysis: the COBRA gene family. Plant Physiol 143: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB. (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol 146: 1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. (2004) The CesA gene family of barley: quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A. (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224: 1174–1184 [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. (2002) Statistical methods for identifying genes with differential expression in replicated cDNA microarray experiments. Statist Sinica 12: 111–139 [Google Scholar]
- Evert R. (2006) Esau's Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development, Ed 3 John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Faure E, Best-Belpomme M, Champion S. (1996) UVB irradiation upregulation of the Drosophila 1731 retrotransposon LTR requires the same short sequence of U3 region in a human epithelial cell line as in Drosophila cells. Photochem Photobiol 64: 807–813 [DOI] [PubMed] [Google Scholar]
- Fincher GB. (2009) Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol 149: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Pauly M, Bacic A, Fincher GB. (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221: 729–738 [DOI] [PubMed] [Google Scholar]
- Grandbastien MA. (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3: 181–187 [Google Scholar]
- Harris PJ, Henry RJ, Blakeney AB, Stone BA. (1984) An improved procedure for the methylation analysis of oligosaccharides and polysaccharides. Carbohydr Res 127: 59–73 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD. (2002) Cellulose synthase-like genes of rice. Plant Physiol 128: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Farkas V, Lahnstein J, Fincher GB. (2007) A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-beta-D-glucans. J Biol Chem 282: 12951–12962 [DOI] [PubMed] [Google Scholar]
- Islam AKMR, Shepherd KW, Sparrow DHB. (1981) Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 46: 161–174 [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97: 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis A, Gustafson JP, Chalmers KJ, Barr AR, Langridge P. (2003) A consensus map of barley integrating SSR, RFLP, and AFLP markers. J Agric Res 54: 1173–1185 [Google Scholar]
- Kaup S, Grandjean V, Mukherjee R, Kapoor A, Keyes E, Seymour CB, Mothersill CE, Schofield PN. (2006) Radiation-induced genomic instability is associated with DNA methylation changes in cultured human keratinocytes. Mutat Res 597: 87–97 [DOI] [PubMed] [Google Scholar]
- Kimura S, Sakurai N, Itoh T. (1999) Different distribution of cellulose synthesizing complexes in brittle and non-brittle strains of barley. Plant Cell Physiol 40: 335–338 [Google Scholar]
- Kokubo A, Kuraishi S, Sakurai N. (1989) Culm strength of barley: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol 91: 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo A, Sakurai N, Kuraishi S, Takeda K. (1991) Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiol 97: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al. (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Emslie D, Corcoran L, Smyth G. (2007) Normalization of boutique two-color microarrays with a high proportion of differentially expressed probes. Genome Biol 8: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragupathy R, Cloutier S. (2008) Genome organisation and retrotransposon driven molecular evolution of the endosperm Hardness (Ha) locus in Triticum aestivum cv Glenlea. Mol Genet Genomics 280: 467–481 [DOI] [PubMed] [Google Scholar]
- Ragupathy R, Naeem HA, Reimer E, Lukow OM, Sapirstein HD, Cloutier S. (2008) Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. Theor Appl Genet 116: 283–296 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman AH, Kalendar R. (2005) A movable feast: diverse retrotransposons and their contribution to barley genome dynamics. Cytogenet Genome Res 110: 598–605 [DOI] [PubMed] [Google Scholar]
- Sims IM, Bacic A. (1995) Extracellular polysaccharides from suspension-cultures of Nicotiana plumbaginifolia. Phytochemistry 38: 1397–1405 [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G. (2007) Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol 145: 1444–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Hayashi J, Hiura U. (1966) Inheritance and linkage studies in barley. III. Linkage of the gene for fragile stem-2 and orientation of the linkage map on barley chromosome 5. Ber Ohara Inst Landw Biol Okayama Univ 13: 199–212 [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottman DR. (1987) The decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol 110: 441–454 [Google Scholar]
- Tsuchiya T. (1972) Revision of linkage map of chromosome 5 in barley by means of telotrisomic analysis. J Hered 63: 373–375 [Google Scholar]
- Tsuchiya TS, Singh RJ. (1973) Further information on telotrisomic analysis in barley. Barley Genet Newsl 3: 75–77 [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DL, Buckley MJ, Helliwell CA, Wilson IW. (2003) New normalization methods for cDNA microarray data. Bioinformatics 19: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Yan C, Yan S, Zeng X, Zhang Z, Gu M. (2007) Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J Genet Genomics 34: 1019–1027 [DOI] [PubMed] [Google Scholar]
- Yong W, Link B, O'Malley R, Tewari J, Hunter CT, Lu CA, Li X, Bleecker AB, Koch KE, McCann MC, et al. (2005) Genomics of plant cell wall biogenesis. Planta 221: 747–751 [DOI] [PubMed] [Google Scholar]
- Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, Shabbeer S, Cope L. (2007) Pre-processing Agilent microarray data. BMC Bioinformatics 8: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.