Abstract
Volatile methyl esters are common constituents of plant volatiles with important functions in plant defense. To study the biosynthesis of these compounds, especially methyl anthranilate and methyl salicylate, we identified a group of methyltransferases that are members of the SABATH enzyme family in maize (Zea mays). In vitro biochemical characterization after bacterial expression revealed three S-adenosyl-l-methionine-dependent methyltransferases with high specificity for anthranilic acid as a substrate. Of these three proteins, Anthranilic Acid Methyltransferase1 (AAMT1) appears to be responsible for most of the S-adenosyl-l-methionine-dependent methyltransferase activity and methyl anthranilate formation observed in maize after herbivore damage. The enzymes may also be involved in the formation of low amounts of methyl salicylate, which are emitted from herbivore-damaged maize. Homology-based structural modeling combined with site-directed mutagenesis identified two amino acid residues, designated tyrosine-246 and glutamine-167 in AAMT1, which are responsible for the high specificity of AAMTs toward anthranilic acid. These residues are conserved in each of the three main clades of the SABATH family, indicating that the carboxyl methyltransferases are functionally separated by these clades. In maize, this gene family has diversified especially toward benzenoid carboxyl methyltransferases that accept anthranilic acid and benzoic acid.
Volatile compounds have important roles in the reproduction and defense of plants. Volatiles can attract pollinators and seed dispersers (Dobson and Bergström, 2000; Knudsen et al., 2006) or function as indirect defense compounds that attract natural enemies of herbivores (Dicke, 1994; Degenhardt et al., 2003; Howe and Jander, 2008). A well-studied example for the role of volatiles in plant defense is the tritrophic interaction between maize (Zea mays) plants, their lepidopteran herbivores, and parasitoid wasps of the herbivores. After damage by larvae of Spodoptera species, maize releases a complex volatile blend containing different classes of natural products (Turlings et al., 1990; Turlings and Benrey, 1998a). This volatile blend can be used as a cue by parasitic wasps to find hosts for oviposition (Turlings et al., 1990, 2005). After parasitization, lepidopteran larvae feed less and die upon emergence of the adult wasp, resulting in a considerable reduction in damage to the plant (Hoballah et al., 2002, 2004). The composition of the maize volatile blend is complex, consisting of terpenoids and products of the lipoxygenase pathway, along with three aromatic compounds: indole, methyl anthranilate, and methyl salicylate (Turlings et al., 1990; Degen et al., 2004; Köllner et al., 2004a). In the last decade, several studies have addressed the biosynthesis of terpenoids (Shen et al., 2000; Schnee et al., 2002, 2006; Köllner et al., 2004b, 2008a, 2008b) and indole (Frey et al., 2000, 2004) in maize. The formation of methyl anthranilate and methyl salicylate, however, has not been elucidated.
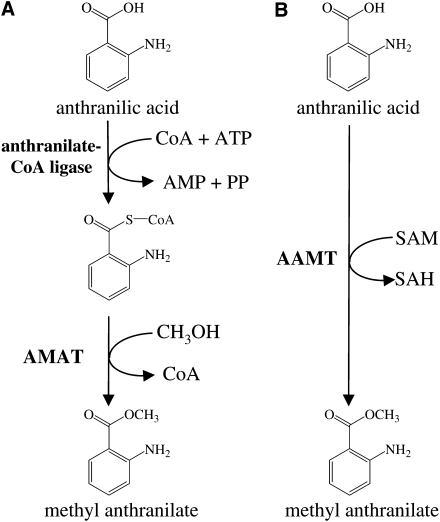
Methyl anthranilate and methyl salicylate are carboxyl methyl esters of anthranilic acid, an intermediate of Trp biosynthesis, and the plant hormone salicylic acid, respectively. Our understanding of methyl anthranilate biosynthesis in plants is very limited. The only enzyme that has been described to be involved in methyl anthranilate synthesis is the anthraniloyl-CoA:methanol acyltransferase in Washington Concord grape (Vitis vinifera; Wang and De Luca, 2005). In contrast, the biosynthesis of methyl salicylate has been well studied in several plant species, such as Clarkia brewerii (Ross et al., 1999), Arabidopsis (Arabidopsis thaliana; Chen et al., 2003), and rice (Oryza sativa; Xu et al., 2006; Koo et al., 2007; Zhao et al., 2010). In all these species, methyl salicylate is synthesized by the action of S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase (SAMT). The apparent homology of SAMTs from different plant species suggests that methyl salicylate formation in maize, a species closely related to rice, is also catalyzed by an SAMT. SAMT enzymes are considered part of a larger family of methyltransferases called SABATH methyltransferases (D'Auria et al., 2003). The SABATH family also includes methyltransferases producing other methyl esters such as methyl benzoate, methyl jasmonate, and methyl indole-3-acetate (Seo et al., 2001; Effmert et al., 2005; Qin et al., 2005; Song et al., 2005; Zhao et al., 2007). An activity forming methyl anthranilate has not been described in the SABATH family, despite the striking structural similarity between methyl anthranilate and methyl salicylate or methyl benzoate. Two different classes of enzymes, methanol acyl transferases and methyltransferases, therefore, might be responsible for methyl anthranilate biosynthesis in maize (Fig. 1). Some of the SABATH methyltransferases have been shown previously to have methyltransferase activity in vitro using anthranilic acid as substrate (Chen et al., 2003; Zhao et al., 2010), but the biological relevance of such activity is unknown.
Figure 1.
The biosynthesis of methyl anthranilate from anthranilic acid can proceed over two pathways. Pathway A has been documented in grape, while pathway B is demonstrated here. AMAT, Anthraniloyl-CoA:methanol acyltransferase; SAH, S-adenosyl-l-homocysteine.
In our ongoing attempt to investigate the biosynthesis and function of maize volatiles, we have studied the biosynthesis of the aromatic methyl esters, methyl salicylate and methyl anthranilate, and their regulation by herbivory. Biochemical characterization of maize benzenoid carboxyl methyltransferases of the SABATH family led to the discovery of a group of anthranilic acid methyltransferases (AAMTs). Homology-based structural modeling combined with site-directed mutagenesis identified the residues critical for the binding of the anthranilic acid substrate. Such functionally important residues are responsible for the diversification and evolution of benzenoid carboxyl methyltransferases in plants.
RESULTS
Identification of Methyltransferase Activities Involved in Maize Volatile Biosynthesis
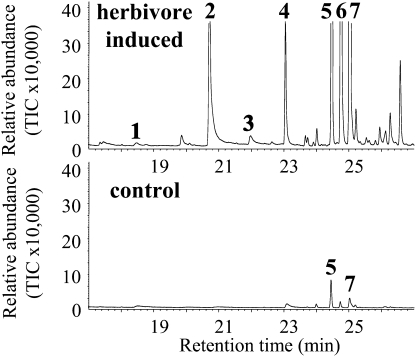
To study the production of benzenoid methyl esters in response to herbivory, we analyzed the volatile emissions of maize cv Delprim damaged by larvae of the herbivore Spodoptera littoralis. Besides indole, methyl anthranilate and methyl salicylate were the only other aromatic components emitted within a complex blend of monoterpenes and sesquiterpenes in response to leaf damage (Fig. 2). To elucidate the biosynthetic pathway of these methyl esters, we tested for the presence of S-adenosyl-l-methionine (SAM)-dependent methyltransferase activity for anthranilic acid, salicylic acid, and other substrates in maize leaves. In undamaged control plants, only trace amounts of SAM-dependent methyltransferase activity were found (Fig. 3). Herbivore damage led to a strongly induced activity with both anthranilic acid and benzoic acid (Fig. 3), suggesting that an herbivore-induced SAM-dependent methyltransferase is involved in the formation of methyl anthranilate in maize.
Figure 2.
Volatiles released by maize after herbivore attack. Volatiles from 2-week-old plants of maize cv Delprim were collected after no treatment (control) or after 16 h of feeding by Egyptian cotton leaf worm (herbivore induced). After separation by GC, the peaks were identified by MS as methyl salicylate (peak 1), indole (peak 2), methyl anthranilate (peak 3), geranyl acetate (peak 4), (E)-β-caryophyllene (peak 5), (E)-α-bergamotene (peak 6), and (E)-β-farnesene (peak 7). TIC, Total ion current.
Figure 3.
SAM-dependent methyltransferase activity in undamaged (ctr) and herbivore-damaged (ind) maize seedlings. Plant crude protein extracts were incubated with the methyl group donor [14C]SAM and different acid substrates. The formed methyl esters were extracted from the assays and quantified using a scintillation counter. AA, Anthranilic acid; BA, benzoic acid; SA, salicylic acid; IAA, indole-3-acetic acid; IPA, indole-3-propionic acid; IBA, indole-3-butyric acid; CinA, cinnamic acid; CouA, o-coumaric acid; JA, jasmonic acid; 3ABA, 3-aminobenzoic acid; 4ABA, 4-aminobenzoic acid; boil. ctr, control with heat-denatured enzyme.
Identification of the Benzenoid Carboxyl Methyltransferase Genes from Maize
To identify putative AAMT and SAMT genes, we screened The Institute for Genomic Research Maize Genome Database (http://maize.tigr.org/) as well as the Maize Genome Resource (www.maizesequence.org) for sequences with similarity to SAMTs that have been identified in other plants. Eight sequences representing complete open reading frames (ORFs) were identified and designated as omt1, omt2, omt3, omt4 (GRMZM2G143871), omt5 (GRMZM2G133996), omt6 (GRMZM2G050307), omt7 (GRMZM2G050321), and omt8. Since omt1, omt2, and omt3 are characterized as AAMTs below, we preferred to designate these genes more specifically as aamt1, aamt2, and aamt3, respectively. In addition, we identified two pseudogenes with similarity to samt and bamt genes. One of these pseudogenes, designated omt9 (GRMZM2G303419), contained a stop codon within the last exon. The ORF of the second pseudogene, omt10 (GRMZM2G405947), was interrupted by two frame-shift mutations in the second and fourth exons, respectively. Four of the putative omt genes, aamt1, aamt2, aamt3, and omt8, could be amplified as complete ORFs from cDNA made from herbivore-induced leaves of the maize cv Delprim. All other genes, omt4, omt5, omt6, and omt7, could not be amplified from cDNA in repeated experiments, suggesting that these genes are not expressed in leaves of the maize cv Delprim after herbivore feeding. Sequence analysis of several independently amplified clones of aamt1 revealed two sequences that differed in three nucleotide positions that lead to two altered amino acids. Because the maize cv Delprim is a hybrid line, both cDNAs are likely alleles of one locus and were named aamt1-Del1 and aamt1-Del2. However, sequence analysis of several aamt2, aamt3, and omt8 clones from two independent PCRs provided only one sequence for each gene.
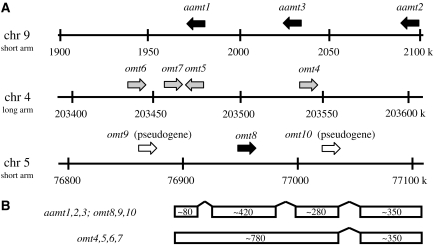
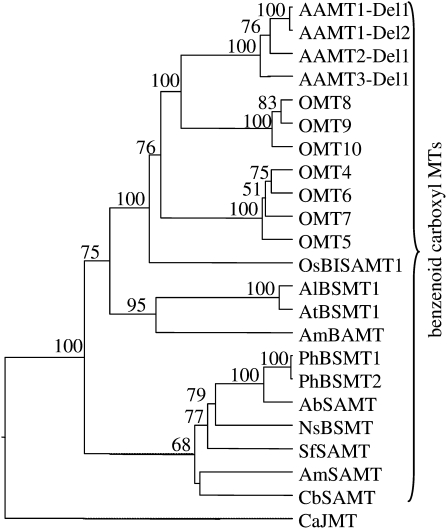
The gene family of benzenoid carboxyl methyltransferases in maize forms three groups that are located on chromosome 4 (omt4, omt5, omt6, and omt7), on chromosome 9 (aamt1, aamt2, and aamt3), and on chromosome 5 (omt8, omt9, and omt10; Fig. 4A). The genomic sequences of genes located on chromosome 4 only contain one intron in the C-terminal half of the gene, while all other putative benzenoid carboxyl methyltransferase genes of maize contain two additional introns in the N-terminal part (Fig. 4B). A dendrogram analysis of the ORFs showed well-defined clades for each of the chromosomal groups, indicating that these groups have been formed by successive gene duplications and diversification (Fig. 5). The first clade comprises the three aamt genes that display a high sequence similarity (90%–92%) with each other. The second clade is formed by omt8 through omt10. The third clade was formed by the genes with only one intron, omt4 through omt7. Interestingly, most of the genes of the second and third clades are not expressed (omt4, omt5, omt6, and omt7) or are pseudogenes (omt9 and omt10), which indicates that they have no function in response to herbivory or within the maize plant. It is possible, though, that functional alleles of these genes are present in other germplasm.
Figure 4.
Chromosomal locations of the benzenoid carboxyl methyltransferase gene family in the maize genome. A, Sections of the maize chromosomes with approximate locations of the SABATH-like genes and pseudogenes. Black arrows represent genes expressed in maize cv Delprim after herbivory; gray arrows represent genes without detectable expression; white arrows represent pseudogenes. B, Exon-intron structure of maize SABATH-like genes.
Figure 5.
Dendrogram analysis of maize AAMT1-Del1, AAMT1-Del2, AAMT2-Del1, AAMT3-Del1, OMT4, OMT5, OMT6, OMT7, OMT8, OMT9, and OMT10 with different methyltransferases (MTs) specific for carboxyl groups of benzenoids and jasmonic acid. The analysis was conducted using a neighbor-joining algorithm. Bootstrap values are shown in percentage and were generated with a sample of 1,000. Accession numbers are as follows: PhBSMT1, AAO45012; PhBSMT2, AAO45013; AbSAMT, BAB39396; NsBSMT, CAF31508; SfSAMT, CAC33768; AmSAMT, AAN40745; CbSAMT, AAF00108; AlBSMT1, AAP57211; AtBSMT1, NP_187755; AmBAMT, AAF98284; CaJMT, ABB02661; OsBISAMT1, AAS18419. Sequences of OMT4, OMT5, OMT6, OMT7, OMT9, and OMT10 were obtained from the Maize Genome Database (www.maizesequence.org) with the following accession numbers: OMT4, GRMZM2G143871; OMT5, GRMZM2G133996; OMT6, GRMZM2G050307; OMT7, GRMZM2G050321; OMT9, GRMZM2G303419; OMT10, GRMZM2G405947. The ORFs of OMT9 and OMT10 were reconstructed in silico by the deletion of a stop codon (OMT9) or the deletion of two frame-shift mutations (OMT10).
Sequence comparison with SAM-dependent methyltransferases from other plants revealed that the identified maize methyltransferases are similar to benzenoid carboxyl methyltransferases of the SABATH family, which are specific for carboxyl groups of small molecules (Fig. 5). The highest amino acid sequence similarity was observed with AbSAMT from Atropa belladonna (41%; Fukami et al., 2002) and the OsBISAMT1 enzyme from rice (46%; Xu et al., 2006; Koo et al., 2007).
Biochemical Characterization of the Methyltransferases AAMT1, AAMT2, AAMT3, and OMT8
Since the aamt genes constitute a subclass of the SABATH genes that had not been characterized previously, we analyzed the substrate specificities and biochemical properties of AAMT1-Del1, AAMT1-Del2, AAMT2, and AAMT3. The genes were cloned and expressed with an N-terminal His tag in Escherichia coli, and the purified enzymes were incubated with the 14C-labeled methyl group donor 5′-SAM and different potential acid substrates. Labeled products were extracted and analyzed using a scintillation counter. Product formation was observed for AAMT1-Del1, AAMT2, and AAMT3, with the highest enzyme activity for anthranilic acid (Table I). AAMT3 also accepted benzoic acid as a substrate, but with decreased reaction velocity in comparison with anthranilic acid. However, AAMT1-Del2 showed no enzymatic activity with all tested substrates (data not shown).
Table I. Relative activity of recombinant and purified maize AAMT enzymes and OMT8 with various substrates.
Values are averages of four independent measurements. All substrates were tested at a 1.5 mm concentration, and methyltransferase activity was determined by measuring the radioactivity of the transferred 14C-methyl group from SAM. The relative activity of the three AAMTs with anthranilic acid was set arbitrarily at 100%. nd, Not determined.
| Substrate | AAMT1 Relative Activity | AAMT2 Relative Activity | AAMT3 Relative Activity | OMT8 Relative Activity |
| % | ||||
| Anthranilic acid | 100 | 100 | 100 | 0 |
| Benzoic acid | 3 | 3 | 26 | 100 |
| Salicylic acid | 0 | 0 | 2 | 0 |
| Indole-3-acetic acid | 0 | 0 | 2 | nd |
| Jasmonic acid | 0 | 0 | 1 | nd |
| Cinnamic acid | 0 | 0 | 0 | 0 |
| Coumaric acid | 0 | 0 | 0 | 0 |
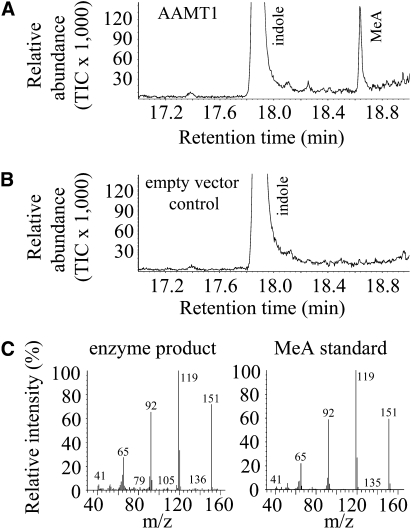
Because both functional groups of anthranilic acid, the carboxyl group and the amino group, could act as potential target sites for methyltransferases, SAM-dependent methylation could result in the formation of O-methyl anthranilate as well as N-methyl anthranilate. Therefore, we analyzed the enzyme product formed from anthranilic acid using gas chromatography-mass spectrometry (GC-MS). A comparison of retention time and mass spectrum of the enzyme product with those from an authentic O-methyl anthranilate standard revealed that the enzymes produced exclusively the O-methyl ester (Fig. 6; results shown only for AAMT1).
Figure 6.
GC-MS analysis of methyl anthranilate (MeA) produced by AAMT1. A, The enzyme was expressed in E. coli, extracted, partially purified, and incubated with the substrates SAM and anthranilic acid. The resulting methyl ester was collected with a solid-phase microextraction (SPME) fiber and analyzed by GC-MS. B, GC trace resulting from an empty-vector control experiment. A crude protein extract from E. coli carrying the empty expression vector was incubated with the substrates SAM and AA and analyzed as described above. TIC, Total ion current. C, Comparison of the mass spectrum from the enzyme-produced methyl anthranilate and the mass spectrum derived from an authentic methyl anthranilate standard.
The kinetic properties of AAMT1, AAMT2, and AAMT3 determined using the purified His-tagged enzymes and 14C-labeled SAM under linear reaction velocity and substrate saturation are summarized in Table II. The Km values for the cosubstrate SAM were in the range reported for other plant methyltransferases, whereas the Km values for anthranilic acid were two to 10 times higher than Km values of salicylic acid for plant SAMT enzymes (Effmert et al., 2005). However, the calculated kcat/Km values for AAMT1 and AAMT2 were in the range reported for other SAM-dependent methyltransferases (Table II; Effmert et al., 2005). The removal of the N-terminal His tag via thrombin digestion did not influence the kinetic properties of the recombinant enzymes (data not shown).
Table II. Kinetic properties of AAMT1, AAMT2, and AAMT3.
AA, Anthranilic acid; BA, benzoic acid; nd, not determined.
| Enzyme | Km for AA | kcat for AA | kcat/Km for AA | Km for BA | kcat for BA | kcat/Km for BA | Km for SAM |
| μm | s−1 | mm−1 s−1 | μm | s−1 | mm−1 s−1 | μm | |
| AAMT1 | 641 | 0.45 | 0.7 | 2,025 | 0.04 | 0.02 | 79 |
| AAMT2 | 311 | 0.37 | 1.2 | nd | nd | – | 94 |
| AAMT3 | >2,000 | nd | – | nd | nd | – | 76 |
OMT8 was the only member of the second group of benzenoid carboxyl methyltransferases that was expressed in the maize tissues we studied. The enzyme accepted only benzoic acid to produce methyl benzoate (Table I). Because the genes of group 3 (omt4–omt7) are apparently not expressed in maize cv Delprim, we tried to amplify them from the maize inbred line B73. While amplification of omt4, omt5, and omt7 failed, the complete ORF of omt6 could be amplified and cloned. However, heterologous expression of the OMT6 allele from B73 did not result in any activity with the tested substrates (data not shown).
Herbivore Feeding Induces aamt1 and aamt3 Expression via Jasmonic Acid Signaling
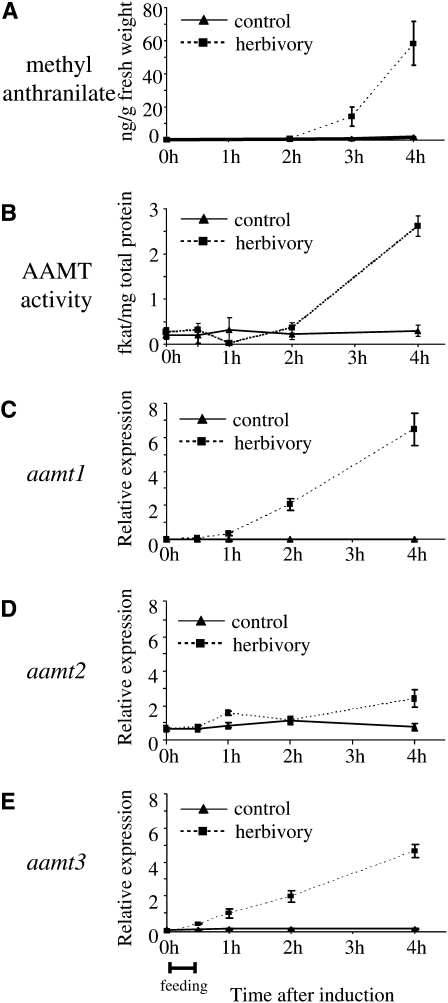
To test whether AAMT1, AAMT2, and AAMT3 are involved in herbivore-induced biosynthesis of methyl anthranilate, we measured the transcript accumulation of these genes at certain time points in an interval of 4 h after caterpillar feeding in maize plants of cv Delprim. In parallel, material from the same plants was used to determine AAMT activity in crude protein extracts as well as the accumulation of the compound methyl anthranilate in the leaf tissue. The hexane extraction of leaf material showed that methyl anthranilate accumulation in leaves was induced by 30 min of S. littoralis feeding. Methyl anthranilate release started after 2 h and increased continuously during the next 2 h (Fig. 7A). No accumulation occurred in uninduced plants. A corresponding time course was found for SAM-dependent methyltransferase activity in leaves, which was first detectable after 2 h of caterpillar feeding (Fig. 7B). Quantitative PCR analysis showed that aamt1 and aamt3 were induced by herbivory (Fig. 7, C and E). Transcript accumulation of aamt1 and aamt3 started after 1 h and after 30 min, respectively, of herbivore feeding. In control plants, no transcripts of aamt1 and only small amounts of aamt3 transcripts could be detected. In contrast to aamt1 and aamt3, the transcripts of aamt2 increased only slightly at 4 h of herbivore feeding (Fig. 7D).
Figure 7.
Expression analysis of aamt genes in comparison with methyl anthranilate accumulation and AAMT activity after herbivore damage. Plants were treated with S. littoralis larvae caged for 30 min on a single leaf. Leaf material was harvested at different time points after feeding and was split into equal parts for the different analyses. Means and se are shown (n = 4). A, Methyl anthranilate accumulation after herbivore damage in maize leaves. Leaf material was extracted with hexane, and the extracts were analyzed using GC-MS. B, AAMT activity in herbivore-damaged maize leaves. Plant crude protein extracts were incubated with the methyl group donor [14C]SAM and anthranilic acid. The formed methyl ester was extracted from the assays and quantified using a scintillation counter. C to E, Transcript accumulation of aamt1 (C), aamt2 (D), and aamt3 (E) after herbivore damage. Expression was analyzed using quantitative PCR. The relative expression levels were calculated as the expression levels of the respective genes divided by the geometric mean of the expression levels of the two reference genes (for details, see “Materials and Methods”).
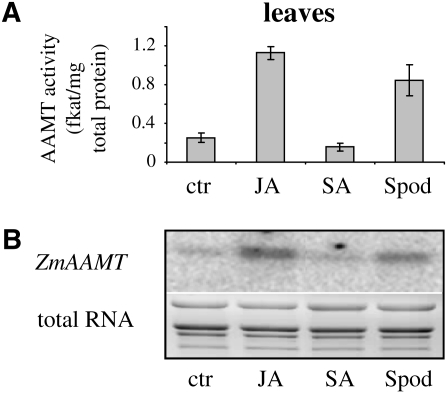
The emission of terpene volatiles from maize can be induced by treatment with jasmonic acid (Hopke et al., 1994). To test whether methyl anthranilate biosynthesis is also influenced by jasmonic acid, 10-d-old seedlings were treated with jasmonic acid, and methyltransferase activity in crude extracts as well as aamt transcript accumulation were measured. The results showed that both methyltransferase activity (Fig. 8A) and transcript accumulation of aamt genes (Fig. 8B) were strongly increased after jasmonic acid treatment comparable to those in caterpillar-damaged seedlings. In contrast to this, salicylic acid treatment did not influence the biosynthesis of methyl anthranilate (Fig. 8).
Figure 8.
AAMT activity correlates with transcript accumulation of aamt genes after different plant treatments. Plants were treated with jasmonic acid (JA), salicylic acid (SA), or S. littoralis larvae (Spod). Leaf material was harvested and split into two parts for different analyses. A, AAMT activity. Plant crude protein extracts were incubated with the methyl group donor [14C]SAM and anthranilic acid. The formed methyl ester was extracted from the assays and quantified using a scintillation counter. Means and se are shown (n = 4). B, Transcript accumulation of aamt genes. RNA isolated from leaf material was hybridized with a probe specific for maize aamt genes. The bottom panel shows the total RNA of the ethidium bromide-stained RNA gel. ctr, Control plants without treatment.
The Concentration of the Anthranilic Acid Substrate Accumulates upon Herbivory
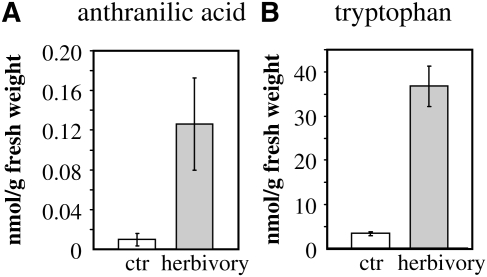
Regulation of methyl anthranilate production might not only be dependent on enzyme concentration but on the availability of the substrate. Anthranilic acid marks a branch point between primary and secondary metabolism in plants. Anthranilic acid is produced from chorismic acid and is further converted to the aromatic amino acid Trp. After herbivore feeding, anthranilic acid is also converted to methyl anthranilate, a secondary metabolite. To test the availability of the substrate anthranilic acid for both pathways in herbivore-induced maize, we measured the levels of anthranilic acid and Trp in herbivore-damaged plants and control maize plants of cv Delprim. LC-MS and HPLC measurements showed that the amount of both compounds was about 8-fold increased in caterpillar-damaged leaves in comparison with control leaves (Fig. 9). This indicates a much greater availability of anthranilic acid to the AAMTs in herbivore-induced plants.
Figure 9.
Levels of anthranilic acid (A) and Trp (B) in herbivore-damaged maize plants (herbivory) in comparison with undamaged control plants (ctr). Anthranilic acid was extracted with methanol from leaf material and analyzed using LC-MS. Trp present in a crude leaf extract was derivatized with mercaptoethanol and O-phthaldialdehyde and analyzed by HPLC coupled to a fluorescence detector. Means and se are shown (n = 5).
Tyr-246 and Gln-167 Are Involved in Substrate Specificity of AAMT1
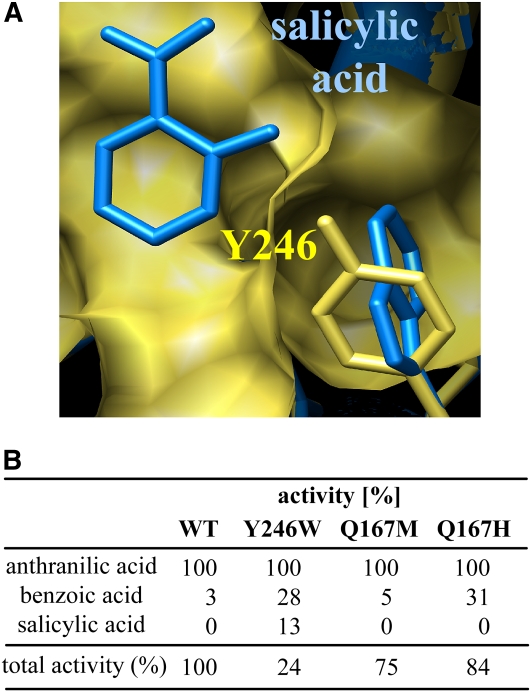
A sequence comparison between the maize AAMT1 protein with SAM-dependent methyltransferases from other plants showed a high similarity of AAMT1 to salicylic acid methyltransferases and benzoic acid methyltransferases (Fig. 5). However, AAMT1 possessed no enzymatic activity with salicylic acid and nearly no activity with benzoic acid (Table I), suggesting structural changes in the active site responsible for the different substrate specificity in comparison with SAMT and BAMT enzymes. Based on the crystal structure of SAMT from C. brewerii (CbSAMT), several amino acids involved in substrate binding were recently identified (Zubieta et al., 2003; Supplemental Fig. S1). To find amino acids responsible for substrate specificity of AAMT1, we looked for active site amino acids that are highly conserved in SAMT enzymes but are different at corresponding positions in maize AAMTs. One of these residues, Tyr-246 of AAMT1, was further investigated by structure modeling. An overlay of the three-dimensional structure of CbSAMT including the substrate salicylic acid with the modeled structure of AAMT1 showed that the position of the side chain of Tyr-246 from AAMT1 could prevent binding and methylation of salicylic acid (Fig. 10A). Mutagenesis of Tyr-246 to Trp, which is highly conserved in SAMT enzymes at this position, resulted in an enzyme that could accept salicylic acid as a substrate (Fig. 10B). However, the activity in the presence of salicylic acid was only 13% relative to anthranilic acid (100%), and the total activity of the mutant measured in the presence of anthranilic acid was decreased to approximately 24% relative to the wild-type enzyme.
Figure 10.
A, Model of part of the AAMT1 active site cavity based on the structure of C. brewerii SAMT (Zubieta et al., 2003) with the proposed position of salicylic acid. The surface of the active site cavity and the residue Tyr-246 of AAMT1 are shown in yellow. The structure of CbSAMT was overlaid, but only the bound substrate salicylic acid and the residue Trp-226 were visualized (blue). B, Relative activity of wild-type AAMT1 (WT) in comparison with several AAMT1 mutant enzymes with various substrates. Values are averages of four independent measurements. All substrates were tested at a 1.5 mm concentration, and methyltransferase activity was determined by measuring the radioactivity of the transferred 14C-methyl group from SAM. The relative activity of the three AAMTs with anthranilic acid was set arbitrarily at 100%.
A recent publication demonstrated the importance of an active site Met for substrate specificity of SAMT enzymes (Barkman et al., 2007). Using in vitro mutagenesis, Barkman and coworkers (2007) demonstrated that a change from Met to His at position 156 of SAMT from Datura wrightii (DwSAMT) resulted in an increased ability to use benzoic acid as a substrate relative to salicylic acid. This finding coincides with the observation that most SAMT-type enzymes contain a Met at this position, whereas in BAMT-type enzymes, there is a His at the corresponding position (Effmert et al., 2005). Interestingly, AAMT1 and AAMT3 also differ at this position. Whereas AAMT3 contains a His (His-164) and was able to convert benzoic acid to methyl benzoate, the corresponding position in AAMT1 is occupied by a Gln residue and the enzyme showed a strongly reduced activity with benzoic acid relative to AAMT3 (Table I; Supplemental Fig. S1). To analyze whether the high substrate specificity of AAMT1 is due to the presence of Gln at position 167, we altered this amino acid residue to His and Met by site-directed mutagenesis. As expected, the mutant AAMT1 Q167H showed an increased activity with benzoic acid comparable to the wild-type AAMT3 enzyme (Fig. 10B). The change from Gln to Met did not influence substrate specificity. Both mutants were not able to accept the substrate salicylic acid to produce methyl salicylate (Fig. 10B).
DISCUSSION
Methyl Anthranilate Is Formed by aamt Genes, a Separate Group of Benzenoid Carboxyl Methyltransferases
The formation of volatile methyl esters in plants has been shown to be mediated either by methyltransferases or acyl transferases. For methyl anthranilate formation, only an acyl transferase activity has been described to date condensing anthraniloyl-CoA and methanol (Wang and DeLuca, 2005). However, some SAMT- and BAMT-type methyltransferases were also shown to form low levels of methyl anthranilate in vitro by reaction of the free acid and SAM (Chen et al., 2003; Zhao et al., 2010). Here, we characterized a family of SAM-dependent methyltransferases from maize that exhibit a high specificity for anthranilic acid. These aamt genes form a clearly defined subgroup within the benzenoid carboxyl methyltransferases of maize. Both aamt1 and aamt3 display transcript levels that correlate well with the enzyme activity and product accumulation in maize after damage by an herbivore. Since AAMT3 has a Km of greater than 2 mm and the anthranilate concentration in planta is in the micromolar range, the enzyme AAMT1 appears to be responsible for most of the methyl anthranilate production in the plant. In addition, the low transcript levels of aamt2 might encode an enzyme that carries out methyl anthranilate formation when the anthranilic acid concentration increases strongly after herbivory. The low affinity of AAMT3 for salicylic acid might be sufficient to produce the small amounts of methyl salicylate released after herbivory. The up-regulation of both the enzyme concentration and the substrate concentration coincides with the onset of methyl anthranilate release after herbivore damage in maize. Likewise, the concentration of Trp is elevated, indicating that the production of many phenolic compounds is strongly induced after herbivory. An herbivore-induced increase in the expression of Trp biosynthesis genes has also been observed in poplar (Populus species) and rice, indicating that these plants also form higher amounts of phenolic products during defense against herbivores (Ralph et al., 2006; Yuan et al., 2008).
Anthranilic acid marks a branching point between methyl anthranilate formation and Trp biosynthesis. The Km values observed in the maize AAMTs are rather high (greater than 300 μm) and may prevent the enzymes from efficient conversion of low concentrations of anthranilic acid. This would allow a constant flux of anthranilic acid into Trp biosynthesis in vivo, thereby ensuring that this essential amino acid is available even when the methyl anthranilate is produced in response to herbivore damage.
The herbivore-induced methyl salicylate may be produced by AAMT3, which shows affinity for the corresponding substrate. If so, AAMT3 has a dual activity, making both methyl anthranilate and methyl salicylate. Dual activity has been detected for a number of SABATH genes, for instance the Arabidopsis BSMT, which produces both methyl salicylate and methyl benzoate (Chen et al., 2003). However, the activity of AAMT3 with salicylic acid is rather low (Table I), raising the possibility that there might be additional enzymes responsible for methyl salicylate formation. Sequence analysis identified four maize genes (omt4, omt5, omt6, and omt7) with similarity to rice SAMT and an intron-exon pattern identical to OsBISAMT (Fig. 5). Although these genes could be candidates for a methyl salicylate-producing activity, none of them showed expression in herbivore-damaged maize tissues. There are no additional candidates for methyl salicylate formation, since no additional members of the maize SABATH family have been discovered by genomic sequencing.
Methyl Anthranilate Might Be Involved in Indirect Plant Defense
The emission of the structurally similar benzenoid methyl esters from plants is often induced by biotic factors like herbivory (Chen et al., 2003; van den Boom et al., 2004; Leitner et al., 2005; Yuan et al., 2008) or pathogen infestation (Huang et al., 2003; Cardoza and Tumlinson, 2006). The biosynthesis of methyl salicylate involves SAMT enzymes in several plants (Fukami et al., 2002; Chen et al., 2003; Koo et al., 2007; Kwon et al., 2009). For example, Chen and coworkers (2003) demonstrated that the transcript accumulation of the Arabidopsis AtSAMT gene was increased upon herbivory and jasmonic acid treatment. In contrast to the Arabidopsis gene, the OsBISAMT1 gene of rice was induced by pathogen infestation and salicylic acid treatment (Xu et al., 2006) and jasmonic acid (Zhao et al., 2010). Both the salicylate-dependent and jasmonate-dependent induction were also observed with a SAMT from A. belladonna, which was suggested to play a dual role in defense against herbivores and pathogens (Kwon et al., 2009). We observed that the transcript levels of maize AAMT1 and AAMT3 were only increased by herbivory and jasmonate application, but not by salicylate treatment. Most likely, the jasmonate-dependent induction mechanism is similar to that of other plant volatile compounds, the terpenes. It was observed that the sesquiterpenes (E)-β-farnesene and (E)-β-caryophyllene are induced with the same kinetics within 2 to 3 h after herbivore damage (Turlings et al., 1998b). Both (E)-β-farnesene and (E)-β-caryophyllene were shown to play important roles in the indirect defense of the plant. In combination with (E)-α-bergamotene, (E)-β-farnesene is emitted by leaves after attack by larvae of Egyptian cotton leaf worm (S. littoralis). The volatiles can attract parasitic wasps that use the lepidopteran larvae as host (Schnee et al., 2006; Köllner et al., 2008a). Essential to the attractive effect is an initial learning experience that associates the volatile with the presence of the host larvae. While methyl anthranilate might provide parasitic wasps with a very reliable indicator for herbivore damage, its functional role in such interactions has not been proven. Experimental evidence is only available for the behavior of individuals of Cotesia marginiventris, which had no prior experience with plant volatiles. These “naive” parasitoids did not alter their behavior toward maize plants with strongly reduced emission of indole and methyl anthranilate (D'Alessandro et al., 2006). A second parasitoid species, Microplitis rufiventris, was even slightly repelled by volatiles containing both indole and methyl anthranilate. However, the attraction of beneficial insects by methyl anthranilate was implicated in other plant-insect interactions. The chloropid fly Thaumatomyia glabra, a predator of several root-feeding aphids, and the thrips parasitoid Ceranisus menes were both attracted to pure methyl anthranilate (Landolt et al., 2000; Murai et al., 2000; James, 2005). The role of methyl anthranilate in parasitoid attraction to maize needs to be elucidated in further studies.
In addition, herbivore-induced plant volatiles can enhance the induction of defense reactions upon future insect attacks, both in the plant itself and in neighboring plants (Heil and Kost, 2006; Heil and Silva Bueno, 2007; Ton et al., 2007; Frost et al., 2008). In maize, Ton and coworkers (2007) demonstrated that volatiles boost direct and indirect resistance of nearby plants in a process termed “priming.” Much of the priming effect can be attributed to the so-called group of “green leafy volatiles” that comprise (Z)-3-hexenal, (Z)-3-hexen-1-ol, and (Z)-3-hexen-1-yl acetate (Ruther and Fürstenau, 2005). Because methyl anthranilate is only emitted in small amounts from herbivore-damaged maize, its role as a priming signal in plant communication has not been elucidated.
A Single Amino Acid Affects Substrate Specificity of Benzenoid Carboxyl Methyltransferases within the SABATH Family
The AAMT enzymes presented here are distinguished from most of the BAMTs and SAMTs by a Tyr residue that determines the substrate specificity of this subgroup. The elucidation of the C. brewerii SAMT structure indicated that a Trp residue in position 226 forms part of the salicylic acid-binding site (Zubieta et al., 2003). This amino acid residue is located next to a small pocket that provides space for the hydroxyl group of salicylic acid. While this Trp residue is conserved among most of the SAMT and BAMT enzymes, the maize AAMTs have a Tyr in the corresponding position. Our model of AAMT1 structure suggests that the Tyr hydroxyl group reduces the size of the neighboring pocket, forcing the anthranilic acid substrate to change its orientation in the reaction center (Fig. 10). A similar change of substrate orientation was observed in another benzenoid carboxyl methyltransferase, indolic acid methyltransferase AtIAMT1 of Arabidopsis, which binds the indole-3-acetic acid substrate in a conformation different from that of salicylic acid in CbSAMT. The binding pocket of the AtIAMT1 is also strongly reduced compared with that of CbSAMT, which affects the substrate specificity of the enzyme (Zhao et al., 2008). These comparisons of methyltransferase binding sites suggest that the substrate specificity of the enzymes is mostly determined by single amino acid residues in the active center. The resulting substrate specificity is very high and might enable the plants to closely control the methylation of structurally similar compounds.
Evolution and Diversification of Benzenoid Carboxyl Methyltransferases within the Maize SABATH Gene Family
The plant species that have been fully sequenced contain a large gene family encoding SABATH methyltransferases (Zhao et al., 2008). In maize, the benzenoid carboxyl methyltransferases as part of the SABATH gene family are organized in three well-defined clades (Fig. 5). One clade contains the AAMT enzymes that have not been described in other plants, to our knowledge, until this point. The three maize AAMTs are highly homologous to each other, suggesting that they are the consequence of recent gene duplications. New functionalization of duplicated genes may result from either changes in expression or changes in function of coded proteins (Zhang and Kishino, 2004). The expression of AAMT2 is different from those of AAMT1 and AAMT3 (Fig. 7), suggesting that the evolution of biological functions of AAMTs is partly driven by the changes in expression patterns of individual genes. In addition, AAMT3 showed more broad-spectrum activity compared with AAMT1 and AAMT2, implying that the evolution of maize AAMTs also has occurred at the biochemical activity level. The second clade comprises omt8 and the closely related pseudogenes omt9 and omt10. The enzyme OMT8 accepted benzoic acid as a substrate and produced methyl benzoate (Table I). However, methyl benzoate was not detected in the volatile bouquet of herbivore-induced maize, suggesting low expression levels of omt8 or low substrate concentrations. The third clade contains the genes omt4, omt5, omt6, and omt7. These genes are distinguished by differences in the exon-intron pattern, which suggests that they arose from gene duplications after separation from the other clades. Since the exon-intron pattern of the rice gene OsBISAMT is similar to that of the two other maize clades, it can be speculated that the genes in this clade lost the first two introns. Interestingly, the genes of this clade have a Met residue in the position that corresponds to 167 in AAMT1, indicating that they are related to SAMTs of other plants that accept salicylic acid as substrate. Surprisingly, maize omt4, omt5, omt6, and omt7 genes were not expressed under the conditions tested, although herbivore-damaged maize released low levels of methyl salicylate. Functional characterization of these omt genes in other maize varieties or under other growth conditions might help understand the biochemical and biological roles of these genes.
A striking general feature of the benzenoid carboxyl methyltransferase gene family in maize is the high number of inactive genes. This has also been observed for the family of terpene synthases in maize and might be a common property of gene families of secondary metabolism that may be caused both by the evolution and the breeding of this highly polymorphic species (Köllner et al., 2004b; Degenhardt et al., 2009).
MATERIALS AND METHODS
Plant and Insect Material
Seeds of maize (Zea mays ‘Delprim’) were obtained from Delley Samen und Pflanzen. Plants were grown in commercially available potting soil (Tonsubstrat; Klasmann) in a climate-controlled chamber with a 16-h photoperiod, 1 mmol m−2 s−1 photosynthetically active radiation, a temperature cycle of 22°C/18°C (day/night), and 65% relative humidity. After 12 to 14 d, the plants were carefully unearthed to inflict minimal damage to the roots and transferred into glass beakers containing 50 mL of water. For induction treatments, either jasmonic acid (Sigma-Aldrich) or salicylic acid (Sigma-Aldrich) was added to the water to a final concentration of 250 μm. Plants were treated for 16 h.
Eggs of Spodoptera littoralis (Lepidoptera: Noctuidae) were obtained from Aventis and were reared on an artificial wheat germ diet (Heliothis mix; Stonefly Industries) for about 10 to 15 d at 22°C under illumination of 750 μmol m−2 s−1. For herbivory treatments, three third instar larvae were enclosed on the middle portion of each plant in a cage made out of two halves of a petri dish (9 cm diameter) with a circle cut out of each side and covered with gauze to allow for ventilation (Degenhardt and Gershenzon, 2000).
Plant Volatile Extracts
For volatile extraction, 600 to 800 mg of leaf material was extracted with 2 mL of hexane containing 10 ng μL−1 nonyl acetate as an internal standard for 10 min at room temperature. The hexane phase was removed, and a 2-μL aliquot was injected into a GC-MS apparatus.
Plant Protein Extracts
Plants were harvested and immediately frozen in liquid nitrogen. Leaf material was ground in a mortar to a fine powder. Tissue powder (0.2 g) was extracted in 1 mL of extraction buffer (50 mm Tris-HCl [pH 7.5], 5 mm β-mercaptoethanol, 1 mm EDTA, 10% [v/v] glycerol, 0.5 mm phenylmethylsulfonyl fluoride, 25 mm NaHSO3, 5 mm sodium ascorbate, 10% [w/v] polyvinylpyrrolidone 40,000, and 25% [w/v] amberlite XAD) for 1 h at 4°C with moderate shaking. After centrifugation, the supernatant was transferred to a new tube and stored at −20°C. The protein concentration was determined by the method of Bradford using the Bio-Rad reagent with bovine serum albumin as standard.
cDNA Construction
Ten-day-old maize plants of cv Delprim were subjected to herbivory by S. littoralis for 4 h. One gram of leaf material was ground in a mortar to a fine powder in liquid nitrogen and added to 10 mL of Trizol Reagent (GIBCO BRL). The mixture was treated with a Polytron (Kinematika) for 1 min and incubated for 3 min on ice. Total RNA was isolated according to the manufacturer's instructions. From about 80 μg of total RNA, the mRNA was isolated utilizing poly(T)-coated ferromagnetic beads (Dynal). The mRNA was transcribed into cDNA while constructing a Marathon RACE library according to the manufacturer's instructions (BD Biosciences).
Identification and Isolation of Methyltransferase cDNAs
To identify putative SABATH genes in maize, we did BLAST searches of The Institute for Genomic Research maize database (http://maize.tigr.org/) as well as the Maize Genome Resource (www.maizesequence.org) using CbSAMT from Clarkia brewerii (Ross et al., 1999), SfSAMT from Stephanotis floribunda (Pott et al., 2004), and AmSAMT from Antirrhinum majus (Negre et al., 2002). Eight maize sequences representing complete ORFs with high similarity to plant salicylic acid methyltransferases were identified. Four of these sequences could be amplified from a cDNA library made from herbivore-induced leaves of the maize cv Delprim using the primer pairs SF1/SF2 (aamt1, aamt3), SK1/SK2 (aamt2), and OMT8-1/OMT8-2 (omt8). Primer sequences are shown in Supplemental Table S1. The PCR products were introduced into the sequencing vector pCR4-TOPO (Invitrogen) and fully sequenced. Each clone was isolated at least twice in independent experiments to avoid errors introduced by the polymerase and the sequencing reaction. Sequence alignments were performed with the DNASTAR suite of programs.
Site-Directed Mutagenesis
For site-directed mutagenesis, the QuickChange site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions. The PCR-based mutagenesis of aamt1-Del1 was performed with the ORF cloned into the expression vector pASK-IBA7 (IBA Biologics) as an BsaI fragment with an N-terminal Strep tag and primers containing the desired mutations (Supplemental Table S1). The mutagenized constructs were fully sequenced.
Heterologous Expression and Purification of AAMT1, AAMT2, AAMT3, and OMT8
For expression with an N-terminal 8× His tag in Escherichia coli, the ORFs of aamt1-Del1, aamt1-Del2, aamt2-Del1, and aamt3-Del1 were amplified with the primer pairs SF7/SF8 (aamt1, aamt3) and SK12/SK13 (aamt2-Del1). PCR products were cloned as EcoRI/HindIII fragments into the expression vector pHIS8-3. The constructs were introduced into the E. coli strain BL21 (DE3) (Invitrogen). For expression of omt8 with an N-terminal Strep tag, the ORF was amplified with the primer pair OMT8-1/OMT8-2 and cloned as a BsmBI fragment into the vector pASK-IBA7 (IBA Biologics). Liquid cultures of the bacteria harboring the expression constructs were grown at 37°C to an optical density at 600 nm of 0.6. Expression of the recombinant proteins from pHIS8-3 constructs in BL21 (DE3) was induced by the addition of isopropyl-β-thiogalactopyranoside to a final concentration of 1 mm, and the expression of pASK-IBA7 constructs in TOP10 cells was induced with 200 μg L−1 anhydrotetracycline (IBA Biologics). To obtain crude extracts, after 20 h of incubation at 18°C the cells were collected by centrifugation and disrupted by a 4× 30-s treatment with a sonicator (Bandelin UW2070) in chilled extraction buffer (50 mm Tris-HCl [pH 7.5] with 5 mm sodium ascorbate, 0.5 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, and 10% [v/v] glycerol). The cell fragments were removed by centrifugation at 14,000g, and the supernatant was desalted into reaction buffer (50 mm Tris-HCl [pH 7.5], 100 mm KCl, 5 mm β-mercaptoethanol, and 10% [v/v] glycerol) by passage through a Econopac 10DG column (Bio-Rad).
To purify the recombinant proteins, the E. coli cells were harvested by centrifugation, resuspended in chilled lysis buffer (50 mm Tris-HCl [pH 7.5], 500 mm NaCl, 20 mm imidazole [pH 8.0], 10 mm β-mercaptoethanol, 1% [v/v] Tween 20, and 10% [v/v] glycerol) and stirred for 2 h at 4°C with lysozyme (0.5 mg mL−1). After sonication and centrifugation, the supernatant was passed over a nickel-nitrilotriacetic acid agarose column (Qiagen) and washed with 10 bed volumes of chilled lysis buffer and 10 bed volumes of chilled wash buffer (50 mm Tris-HCl [pH 7.5], 500 mm NaCl, 20 mm imidazole [pH 8.0], 10 mm β-mercaptoethanol, and 10% [v/v] glycerol). The recombinant His-tagged proteins were eluted with 3 bed volumes of chilled elution buffer (50 mm Tris-HCl [pH 7.5], 500 mm NaCl, 250 mm imidazole [pH 8.0], 10 mm β-mercaptoethanol, and 10% [v/v] glycerol) and transferred into reaction buffer (50 mm Tris-HCl [pH 7.5], 100 mm KCl, 5 mm β-mercaptoethanol, and 10% [v/v] glycerol) using a NAP 5 column (Amersham Biosciences). The protein concentration was determined by the method of Bradford using the Bio-Rad reagent with bovine serum albumin as standard.
Enzyme Assays and Km Value Determination
To determine the SAM-dependent methyltransferase activity in plant extracts, assays containing 188 μL of crude extract, 6 μL of acid substrate (50 mm, dissolved in dimethyl sulfoxide [DMSO]), and 6 μL of [methyl-14C]SAM (51.4 mCi mmol−1, in a 9:1 [v/v] mixture of sulfuric acid [pH 2.0] and ethanol; Hartmann) were performed. Assays were overlaid with 1 mL of pentane to trap volatile products and incubated for 1 h at 25°C. The reaction was stopped by mixing, and 0.5 mL of the pentane layer was taken for measurement of radioactivity by liquid scintillation counting in 2 mL of Lipoluma cocktail (Packard Bioscience) using a Packard Tricarb 2300TR liquid scintillation counter (3H efficiency = 61%). For investigation of the substrate specificity of OMT enzymes, 1 μL of purified protein, 3 μL of acid substrate (50 mm, dissolved in DMSO), 3 μL of [14C]SAM, and 93 μL of reaction buffer were mixed, overlaid with 1 mL of pentane, incubated for 1 h at 25°C, and analyzed as described above. For GC-MS analysis, 92 μL of crude E. coli extract, 3 μL of acid substrate (50 mm), and 5 μL of unlabeled SAM (10 mm; Sigma-Aldrich) were mixed in a Teflon-sealed, screw-capped 1-mL GC glass vial. A solid-phase microextraction fiber consisting of 100-μm polydimethylsiloxane (Supelco) was placed into the head space of the vial for 30 min of incubation at 30°C. For analysis of the adsorbed reaction products, the solid-phase microextraction fiber was directly inserted into the injector of the gas chromatograph.
In all kinetic studies, appropriate enzyme concentrations and incubation times were chosen so that the reaction velocity was linear during the incubation time period. Assays contained 2 μL of [14C]SAM (390 μm), 10 μL of SAM (10 mm; unlabeled), 2 μL of acid substrate dissolved in DMSO in appropriate concentrations, 5 μL of purified enzyme, and 81 μL of reaction puffer. The Km values were determined using seven substrate concentrations with four repetitions each.
GC
A Hewlett-Packard model 6890 gas chromatograph was employed with the carrier gas helium at 1 mL min−1, splitless injection (injector temperature, 220°C; injection volume, 2 μL), a Chrompack CP-SIL-5 CB-MS column [(5%-phenyl)-methylpolysiloxane, 25 m × 0.25 mm i.d. × 0.25 μm film thickness; Varian), and a temperature program from 40°C (3-min hold) at 5°C min−1 to 240°C (3-min hold). The coupled mass spectrometer was a Hewlett-Packard model 5973 with a quadrupole mass selective detector, transfer line temperature of 230°C, source temperature of 230°C, quadrupole temperature of 150°C, ionization potential of 70 eV, and a scan range of 40 to 350 atomic mass units. Methyl anthranilate was identified by comparison of retention time and mass spectrum with those of an authentic reference compound (Sigma-Aldrich). Approximate quantification of methyl anthranilate was performed using a calibration curve made from four different concentrations of an authentic standard.
Determination of Gene Transcript Levels
Total RNA was extracted from maize leaves using the RNeasy kit from Qiagen according to the manufacturer's specifications. RNA (2.75 μg) was DNase treated in a 10-μL reaction using 1 μL of DNase from Promega. DNase-treated RNA (3 μL) corresponding to about 1 μg of RNA was reverse transcribed in a 20-μL reaction with SuperScriptIII from Invitrogen according to the manufacturer's specifications using a mix of anchored oligo(dT)12–18 and random primers (Invitrogen). To minimize pipetting errors, 5 μL of the generated cDNA was used in a 1:5 dilution as a template for quantitative PCR.
All quantitative PCRs were run in triplicate in 20-μL reactions using the Brilliant SYBR Green QPCR Master Mix from Stratagene. Final primer concentration was 0.25 μm. The specificity of all primers was verified by sequencing the respective PCR products. Supplemental Table S2 summarizes the information on primer sequences, PCR product size, and position. Diluted cDNA (5 μL) was used as a template. The following PCR protocol was used for all genes: initial incubation at 95°C for 10 min, followed by 40 cycles of amplification (95°C for 30 s, 56°C for 30 s, and 72°C for 1 min, plate read), and a melting curve from 56°C to 95°C.
Gene expression levels were quantified using the standard curve method. The standard curve was generated using pooled cDNA in equal amounts from all samples. The standard curve was constructed from PCRs using 3 μL, 1 μL, 1/3 μL, 1/9 μL, and 1/27 μL in 5 μL as a template. All sample expression levels were calculated as multiples of the cDNA pool. Two reference genes, putative apt1A (for adenine phosphoribosyltransferase 1; TA172777_4577) and rpb1 (for maize RNA polymerase II largest subunit; AF519538), were used to quantify the cDNA in each sample. Both reference genes showed equal expression levels in control plants and herbivore-damaged plants, with cycle threshold value differences in the range of 1 to 2. The relative expression level in each sample was calculated as the expression level of the respective gene divided by the geometric mean of the expression levels of the two reference genes.
Northern-Blot Hybridization
Plant RNA was prepared with the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. A 1,150-bp fragment was used as a probe, generated by linear PCR with the primer SF4 (Supplemental Table S1) and the complete ORF of AAMT1-Del1 as a template. The probe was labeled with [32P]ATP using the Strip-EZ PCR procedure (Ambion). Blotting on a Nytran-Plus nylon membrane (Schleicher & Schuell), hybridization, and washing were carried out following standard procedures. The blots were visualized using a phosphorimager.
Modeling
A model of the three-dimensional structure of AAMT1 was generated using the Swiss-model server (http://www.expasy.org; Schwede et al., 2003). For modeling, the AAMT1-Del1 amino acid sequence was fitted to the template structure of C. brewerii SAMT (Zubieta et al., 2003). The resulting model was visualized with the program UCSF-Chimera (Pettersen et al., 2004).
Anthranilic Acid and Trp Analysis
Trp present in a crude plant extract was derivatized with mercaptoethanol and O-phthaldialdehyde, yielding a fluorescent isoindole (Sarwar and Botting, 1993), and analyzed by HPLC coupled to a fluorescence detector. Trp content was determined in 100 mg of plant material according to the protocol described previously by de Kraker et al. (2007).
Anthranilic acid was extracted with methanol and analyzed by LC-MS. Leaves of caterpillar-damaged plants and control plants were harvested, frozen in liquid nitrogen, and pulverized. Powder (0.3 g) was extracted with 1 mL of methanol (50% [v/v] in water) for 1 h at 4°C. After centrifugation at 16,000g at 4°C, the supernatant was transferred to a fresh tube and an 80-μL sample was injected into an HPLC system (Agilent HP1200 series) coupled to an Esquire-6000 system electrospray ionization-ion trap mass detector (Bruker Daltonics) operated in positive mode in the mass-to-charge ratio range 70 to 210, with skimmer voltage of −60 V, capillary exit voltage of 101.3 V, capillary voltage of −3,000 V, nebulizer pressure of 35 p.s.i., drying gas of 11 L min−1, and gas temperature of 330°C. Elution was accomplished using an EC 250/4.6 Nucleodur Sphinx RP column (25 cm × 4.6 mm, 5 μm; Macherey-Nagel) with a gradient of 0.2% (v/v) formic acid (solvent A) and acetonitrile (solvent B) at a flow rate of 0.8 mL min−1 at 25°C as follows: 10% to 58% (v/v) B (12 min), 58% to 100% (v/v) B (10 s), 100% (v/v) B (3 min), 100% to 10% (v/v) B (10 s), and 10% (v/v) B (4 min, 50 s). Flow coming from the column was diverted in the ratio 4:1 before reaching the electrospray ionization unit.
Sequences were deposited in GenBank (http://www.ncbi.nlm.nih.gov) with the accession numbers HM242244 (aamt1-Del1), HM242245 (aamt1-Del2), HM242246 (aamt2-Del1), HM242247 (aamt3-Del1), and HM242248 (omt8-Del1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the deduced amino acid sequences of maize AAMT genes with related genes from other plant species.
Supplemental Table S1. Primer sequences for cloning, site-directed mutagenesis, and northern blot.
Supplemental Table S2. Primer sequences for quantitative PCR.
Supplementary Material
Acknowledgments
We thank Jens Wurlitzer for excellent technical assistance. We are indebted to Michael Reichelt and John D'Auria for advice on LC-MS analysis and biochemical assays.
References
- Barkman TJ, Martins TR, Sutton E, Stout JT. (2007) Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol Biol Evol 24: 1320–1329 [DOI] [PubMed] [Google Scholar]
- Cardoza YJ, Tumlinson JH. (2006) Compatible and incompatible Xanthomonas infections differentially affect herbivore-induced volatile emission by pepper plants. J Chem Ecol 32: 1755–1768 [DOI] [PubMed] [Google Scholar]
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36: 577–588 [DOI] [PubMed] [Google Scholar]
- D'Alessandro M, Held M, Triponez Y, Turlings TCJ. (2006) The role of indole and other shikimic acid derived maize volatiles in the attraction of two parasitic wasps. J Chem Ecol 32: 2733–2748 [DOI] [PubMed] [Google Scholar]
- D'Auria JC, Chen F, Pichersky E. (2003) The SABATH family of MTS in Arabidopsis thaliana and other plant species. Romeo JT, , Integrative Phytochemistry: From Ethnobotany to Molecular Ecology. Recent Advances in Phytochemistry, Vol 37 Elsevier, Oxford, pp 253–283 [Google Scholar]
- Degen T, Dillmann C, Marion-Poll F, Turlings TCJ. (2004) High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant Physiol 135: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Gershenzon J. (2000) Demonstration and characterization of (E)-nerolidol synthase from maize: a herbivore-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta 210: 815–822 [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Gershenzon J, Baldwin IT, Kessler A. (2003) Attracting friends to feast on foes: engineering terpene emission to make crop plants more attractive to herbivore enemies. Curr Opin Biotechnol 14: 169–176 [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Kollner TG, Gershenzon J. (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J. (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143: 970–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicke M. (1994) Local and systemic production of volatile herbivore-induced terpenoids: their role in plant-carnivore mutualism. J Plant Physiol 143: 465–472 [Google Scholar]
- Dobson HEM, Bergström G. (2000) The ecology and evolution of pollen odors. Plant Syst Evol 222: 63–87 [Google Scholar]
- Effmert U, Saschenbrecker S, Ross J, Negre F, Fraser CM, Noel JP, Dudareva N, Piechulla B. (2005) Floral benzenoid carboxyl methyltransferases: from in vitro to in planta function. Phytochemistry 66: 1211–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M, Spiteller D, Boland W, Gierl A. (2004) Transcriptional activation of Igl, the gene for indole formation in Zea mays: a structure-activity study with elicitor-active N-acyl glutamines from insects. Phytochemistry 65: 1047–1055 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Pare PW, Schmelz EA, Tumlinson JH, Gierl A. (2000) An herbivore elicitor activates the gene for indole emission in maize. Proc Natl Acad Sci USA 97: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CJ, Mescher MC, Dervinis C, Davis JM, Carlson JE, De Moraes CM. (2008) Priming defense genes and metabolites in hybrid poplar by the green leaf volatile cis-3-hexenyl acetate. New Phytol 180: 722–733 [DOI] [PubMed] [Google Scholar]
- Fukami H, Asakura T, Hirano H, Abe K, Shimomura K, Yamakawa T. (2002) Salicylic acid carboxyl methyltransferase induced in hairy root cultures of Atropa belladonna after treatment with exogeneously added salicylic acid. Plant Cell Physiol 43: 1054–1058 [DOI] [PubMed] [Google Scholar]
- Heil M, Kost C. (2006) Priming of indirect defences. Ecol Lett 9: 813–817 [DOI] [PubMed] [Google Scholar]
- Heil M, Silva Bueno JC. (2007) Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc Natl Acad Sci USA 104: 5467–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah ME, Kollner TG, Degenhardt J, Turlings TCJ. (2004) Costs of induced volatile production in maize. Oikos 105: 168–180 [Google Scholar]
- Hoballah MEF, Tamo C, Turlings TCJ. (2002) Differential attractiveness of induced odors emitted by eight maize varieties for the parasitoid Cotesia marginiventris: is quality or quantity important? J Chem Ecol 28: 951–968 [DOI] [PubMed] [Google Scholar]
- Hopke J, Donath J, Blechert S, Boland W. (1994) Herbivore-induced volatiles: the emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a β-glucosidase and jasmonic acid. FEBS Lett 352: 146–150 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Huang J, Cardoza YJ, Schmelz EA, Raina R, Engelberth J, Tumlinson JH. (2003) Differential volatile emissions and salicylic acid levels from tobacco plants in response to different strains of Pseudomonas syringae. Planta 217: 767–775 [DOI] [PubMed] [Google Scholar]
- James DG. (2005) Further field evaluation of synthetic herbivore-induced plant volatiles as attractants for beneficial insects. J Chem Ecol 31: 481–495 [DOI] [PubMed] [Google Scholar]
- Knudsen JT, Eriksson R, Gershenzon J, Stahl B. (2006) Diversity and distribution of floral scent. Bot Rev 72: 1–120 [Google Scholar]
- Köllner TG, Held M, Lenk C, Hiltpold I, Turlings TCJ, Gershenzon J, Degenhardt J. (2008a) A maize (E)-β-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J. (2004a) The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distribution. Phytochemistry 65: 1895–1902 [DOI] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J. (2004b) The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Li S, Svatos A, Schneider B, Gershenzon J, Degenhardt J. (2008b) Protonation of a neutral (S)-beta-bisabolene intermediate is involved in (S)-beta-macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J Biol Chem 283: 20779–20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo YJ, Kim MA, Kim EH, Song JT, Jung C, Moon JK, Kim JH, Seo HS, Song SI, Kim JK, et al. (2007) Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol Biol 64: 1–15 [DOI] [PubMed] [Google Scholar]
- Kwon S, Hamada K, Matsuyama A, Yasuda M, Nakashita H, Yamakawa T. (2009) Biotic and abiotic stresses induce AbSAMT1, encoding S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, in Atropa belladonna. Plant Biotechnol 26: 207–215 [Google Scholar]
- Landolt PJ, Wixson T, Remke LJ, Lewis RR, Zack RS. (2000) Methyl anthranilate attracts males of Thaumatomyia glabra (Meigen) (Diptera: Chloropidae). J Kans Entomol Soc 73: 189–194 [Google Scholar]
- Leitner M, Boland W, Mithofer A. (2005) Direct and indirect defences induced by piercing-sucking and chewing herbivores in Medicago truncatula. New Phytol 167: 597–606 [DOI] [PubMed] [Google Scholar]
- Murai T, Imai T, Maekawa M. (2000) Methyl anthranilate as an attractant for two thrips species and the thrips parasitoid Ceranisus menes. J Chem Ecol 26: 2557–2565 [Google Scholar]
- Negre F, Kolosova N, Knoll J, Kish CM, Dudareva N. (2002) Novel S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme responsible for biosynthesis of methyl salicylate and methyl benzoate, is not involved in floral scent production in snapdragon flowers. Arch Biochem Biophys 406: 261–270 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF-Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pott MB, Hippauf F, Saschenbrecker S, Chen F, Ross J, Kiefer I, Slusarenko A, Noel JP, Pichersky E, Effmert U, et al. (2004) Biochemical and structural characterization of benzenoid carboxyl methyltransferases involved in floral scent production in Stephanotis floribunda and Nicotiana suaveolens. Plant Physiol 135: 1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin GJ, Gu HY, Zhao YD, Ma ZQ, Shi GL, Yang Y, Pichersky E, Chen HD, Liu MH, Chen ZL, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al. (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Ross JR, Nam KH, D'Auria JC, Pichersky E. (1999) S-Adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys 367: 9–16 [DOI] [PubMed] [Google Scholar]
- Ruther J, Fürstenau B. (2005) Emission of herbivore-induced volatiles in absence of a herbivore—response of Zea mays to green leaf volatiles and terpenoids. Z Naturforsch C 60: 743–756 [DOI] [PubMed] [Google Scholar]
- Sarwar G, Botting HG. (1993) Evaluation of liquid chromatographic analysis of nutritionally important amino acids in food and physiological samples. J Chromatogr 615: 1–22 [DOI] [PubMed] [Google Scholar]
- Schnee C, Kollner TG, Gershenzon J, Degenhardt J. (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-beta-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol 130: 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Kollner TG, Held M, Turlings TCJ, Gershenzon J, Degenhardt J. (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA 103: 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen BZ, Zheng ZW, Dooner HK. (2000) A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: characterization of wild-type and mutant alleles. Proc Natl Acad Sci USA 97: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Kim DG, Lee SH. (2005) Isolation and characterization of a jasmonic acid carboxyl methyltransferase gene from hot pepper (Capsicum annuum L.). J Plant Biol 48: 292–297 [Google Scholar]
- Ton J, D'Alessandro M, Jourdie V, Jakab G, Karlen D, Held M, Mauch-Mani B, Turlings TCJ. (2007) Priming by airborne signals boosts direct and indirect resistance in maize. Plant J 49: 16–26 [DOI] [PubMed] [Google Scholar]
- Turlings TCJ, Benrey B. (1998a) Effects of plant metabolites on the behavior and development of parasitic wasps. Ecoscience 5: 321–333 [Google Scholar]
- Turlings TCJ, Jeanbourquin PM, Held M, Degen T. (2005) Evaluating the induced-odour emission of a Bt maize and its attractiveness to parasitic wasps. Transgenic Res 14: 807–816 [DOI] [PubMed] [Google Scholar]
- Turlings TCJ, Lengwiler UB, Bernasconi ML, Wechsler D. (1998b) Timing of induced volatile emissions in maize seedlings. Planta 207: 146–152 [Google Scholar]
- Turlings TCJ, Tumlinson JH, Lewis WJ. (1990) Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 30: 1251–1253 [DOI] [PubMed] [Google Scholar]
- van den Boom CEM, Van Beek TA, Posthumus MA, De Groot A, Dicke M. (2004) Qualitative and quantitative variation among volatile profiles induced by Tetranychus urticae feeding on plants from various families. J Chem Ecol 30: 69–89 [DOI] [PubMed] [Google Scholar]
- Wang J, De Luca V. (2005) The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J 44: 606–619 [DOI] [PubMed] [Google Scholar]
- Xu RR, Song FM, Zheng Z. (2006) OsBISAMT1, a gene encoding S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, is differentially expressed in rice defense responses. Mol Biol Rep 33: 223–231 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Kollner TG, Wiggins G, Grant J, Degenhardt J, Chen F. (2008) Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J 55: 491–503 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kishino H. (2004) Genomic background predicts the fate of duplicated genes: evidence from the yeast genome. Genetics 166: 1995–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Ferrer JL, Ross J, Guan J, Yang Y, Pichersky E, Noel JP, Chen F. (2008) Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol 146: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Guan J, Ferrer J-L, Engle N, Chern M, Ronald P, Tschaplinski TJ, Chen F. (2010) Emission, biosynthesis and regulation of insect-induced methyl salicylate and methyl benzoate from rice. Plant Physiol Biochem 48: 279–287 [DOI] [PubMed] [Google Scholar]
- Zhao N, Guan J, Lin H, Chen F. (2007) Molecular cloning and biochemical characterization of indole-3-acetic acid methyltransferase from poplar. Phytochemistry 68: 1537–1544 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15: 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.