Abstract
Metabolic networks reconstructed from sequenced genomes or transcriptomes can help visualize and analyze large-scale experimental data, predict metabolic phenotypes, discover enzymes, engineer metabolic pathways, and study metabolic pathway evolution. We developed a general approach for reconstructing metabolic pathway complements of plant genomes. Two new reference databases were created and added to the core of the infrastructure: a comprehensive, all-plant reference pathway database, PlantCyc, and a reference enzyme sequence database, RESD, for annotating metabolic functions of protein sequences. PlantCyc (version 3.0) includes 714 metabolic pathways and 2,619 reactions from over 300 species. RESD (version 1.0) contains 14,187 literature-supported enzyme sequences from across all kingdoms. We used RESD, PlantCyc, and MetaCyc (an all-species reference metabolic pathway database), in conjunction with the pathway prediction software Pathway Tools, to reconstruct a metabolic pathway database, PoplarCyc, from the recently sequenced genome of Populus trichocarpa. PoplarCyc (version 1.0) contains 321 pathways with 1,807 assigned enzymes. Comparing PoplarCyc (version 1.0) with AraCyc (version 6.0, Arabidopsis [Arabidopsis thaliana]) showed comparable numbers of pathways distributed across all domains of metabolism in both databases, except for a higher number of AraCyc pathways in secondary metabolism and a 1.5-fold increase in carbohydrate metabolic enzymes in PoplarCyc. Here, we introduce these new resources and demonstrate the feasibility of using them to identify candidate enzymes for specific pathways and to analyze metabolite profiling data through concrete examples. These resources can be searched by text or BLAST, browsed, and downloaded from our project Web site (http://plantcyc.org).
The ever-expanding demand for the production of food, feed, medicine, and biofuel from plants has prompted the sequencing of plant genomes and transcriptomes. To date, genome and mRNA sequences are available for a large number of plant species, and many more are under way (Duvick et al., 2008; Edwards and Batley, 2009; Liolios et al., 2009). In order to facilitate enzyme discovery and metabolic engineering, the genome sequence of each organism should be placed into its network of metabolic pathways (referred to herein as single-species metabolic pathway databases). In addition, using such systems-level annotations will enable researchers studying individual genes and mutants to contextualize their findings within the overall metabolic scheme of an organism, thereby providing a framework for assessing the broader roles of their genes of interest.
To reconstruct a single-species metabolic pathway database, a standard two-step method is to computationally infer the reactome of the organism from the enzymes present in its annotated genome and to infer the metabolic pathways of the organism from its reactome. For example, the PathoLogic component of the Pathway Tools software suite performs such inference of the reactome and the pathway complement (Karp et al., 2010). This process involves at least three components: (1) annotated enzyme sequences, (2) reference metabolic pathway databases, and (3) mappings of the annotated sequences to pathways in the reference database. Although genome-wide sequences have become available for many plant species, only a few genome-wide metabolic network reconstructions exist for plants. These include, but are not limited to, Arabidopsis (Arabidopsis thaliana) and poplar (Populusspecies) maps inferred from the KEGG reference maps (Kanehisa et al., 2008), Arabidopsis and rice (Oryza sativa) reactions and pathways inferred from the reactome human maps (Vastrik et al., 2007), and a number of databases inferred from MetaCyc (Caspi et al., 2010) such as AraCyc for Arabidopsis (Zhang et al., 2005), RiceCyc for rice (http://www.gramene.org/pathway/ricecyc.html), MedicCyc for Medicago truncatula (Urbanczyk-Wochniak and Sumner, 2007), LycoCyc for tomato (Solanum lycopersicum; http://solgenomics.net/tools/solcyc/), and ChlamyCyc for Chlamydomonas reinhardtii (May et al., 2009). The sequence annotation protocols used to create these databases are heterogeneous. Furthermore, these single-species databases were created computationally using references that contain largely nonplant pathways. For example, MetaCyc is a universal pathway database that includes a vast number of microbial pathways in addition to plant pathways. Consequently, the predicted single-species plant databases included many false-positive predictions. A varying number of these false-positive predictions remain as part of these databases, since manual validation and curation of the databases vary greatly. The lack of consistency in annotation standards and the lack of comparable quality in validation and curation hinder researchers seeking to meaningfully compare the metabolic networks of individual species housed in different metabolic databases.
We created a resource, called the Plant Metabolic Network (PMN), to address some of these shortcomings. The general approach developed at PMN includes four components: (1) PlantCyc, a reference database composed solely of plant metabolic pathways and enzymes; (2) RESD, a reference enzyme sequence database; (3) an enzyme sequence annotation pipeline using RESD; and (4) a modified pathway prediction procedure that uses both PlantCyc and MetaCyc as references in reconstructing single-species metabolic networks from the predicted enzymes. Although PlantCyc is not the first metabolic pathway database containing plant-only data (e.g. MetaCrop [Grafahrend-Belau et al., 2008]), it is by far the most comprehensive in coverage. We applied our system to the recently sequenced genome of Populus trichocarpa to build a metabolic pathway database, which we named PoplarCyc. Here, we introduce the PMN infrastructure and its components. We also compare PoplarCyc (version 1.0) with AraCyc (version 6.0). Finally, we provide some specific examples of how the PMN resources can be used to guide experimentation and discovery in studying plant metabolism.
RESULTS
Creation of PlantCyc
PlantCyc is a comprehensive plant metabolic pathway database created to house a full spectrum of plant pathways and enzymes. PlantCyc was built with all of the AraCyc pathways that have been manually validated and curated (Zhang et al., 2005), plant-specific pathways from MetaCyc (Caspi et al., 2010), curated pathways from RiceCyc (http://www.gramene.org/pathway/ricecyc.html) and MediCyc (Urbanczyk-Wochniak and Sumner, 2007), and a number of new, in-house-curated pathways from the literature. In the latest release (version 3.0), PlantCyc contains 714 pathways, with 375 in the domain of primary metabolism and 339 in secondary metabolism (Table I). Over 300 plant species are linked to one or more of these pathways. While PlantCyc provides a large set of general metabolic pathways found in many plants, it also has a large number of pathways for the biosynthesis of rare but valuable compounds. Examples of these include artemisinin and quinine (treatment of malaria), codeine and morphine (painkiller), ginsenosides (cardioprotectant), lupenol (antiinflammatory), taxol and vinblastine (anticancer); compounds for industrial materials such as resin and rubber; compounds for food flavor and perfumes such as capsaicin and piperine (chili and pepper flavor), geranyl acetate (aroma of rose), and menthol (mint). Evidence codes (Karp et al., 2004) and references are assigned to each pathway, as well as to each enzyme activity, to indicate the type and level of data support for these annotations. Three types of pathways exist in PlantCyc: (1) those described in the literature with at least some experimental evidence, (2) those described in the literature that were drawn solely from the authors’ hypothesis of the biochemical route (“paper chemistry”), and (3) those predicted computationally by mapping potential plant enzymes to microbial or animal pathways. All of the computationally predicted pathways have been manually examined, and their existence in plants was deemed probable (see “Materials and Methods”). Table I shows the types of data and the number of entries for each data type included in PlantCyc and compares it with two species-specific databases, AraCyc (Zhang et al., 2005) and PoplarCyc (this study). Notably, there are 1,974 curated enzymes with experimental evidence in PlantCyc. We also integrated 8,182 predicted orthologous enzymes of Arabidopsis, poplar, rice, tomato, and Medicago into the PlantCyc pathways. These orthologs were imported from the species-specific pathway databases. The pathways are composed of 2,619 unique reactions in which 10,156 enzymes have been assigned to 1,928 reactions. A total of 691 reactions are not yet associated with any enzyme from plants present in the PlantCyc database.
Table I. Contents of MetaCyc and the PMN databases.
| Content | MetaCyc (12.5) | PlantCyc (3.0) | PoplarCyc (1.0) | AraCyc (6.0) |
| Pathways | 1,395 | 714 | 321 | 408 |
| Primary metabolism | 1,053 | 375 | 264 | 316 |
| Secondary metabolism | 342 | 339 | 56 | 92 |
| Reactions (in pathways) | 3,973 | 2,619 | 1,212 | 1,510 |
| With enzymesa | 3,334 (84%) | 1,928 (74%) | 662 (55%) | 1,093 (72%) |
| Without enzymesa | 639 (16%) | 691 (26%) | 550 (45%) | 417 (28%) |
| Enzymesa (in pathways) | 4,501 | 10,156 | 1,807b | 2,007 |
| With experimental support | 3,991 (89%) | 1,974 (19%) | 1 | 858 (43%) |
| Without experimental support | 510 (11%) | 8,182 (81%) | 1,806 (100%) | 1,149 (57%) |
| Compounds | 7,233 | 2,679 | 1,363 | 2,630 |
| Organismsc | 1,549 | 329 | 1 | 1 |
| Citations | 17,916 | 4,803 | 903 | 2,691 |
The term “enzyme” refers to both monomers and complexes found in the databases.
The vast majority of the enzymes present in the database are from P. trichocarpa, but experimentally supported enzymes and pathways from other species or hybrids in the Populus genus can be included in PoplarCyc.
The majority of enzymes and pathways in PlantCyc are from higher plants, but a few pathways are also associated with other organisms such as cyanobacteria and algae.
Many plant secondary metabolites are species or genus specific, and PlantCyc provides a central resource to access them all under one roof. For example, glucosinolates are found almost exclusively in the order Capparales (Brassicales), which includes Arabidopsis (Daxenbichler et al., 1991), and phytoalexins called oryzalexins have only been reported in rice (Akatsuka et al., 1985). While the species-specific databases AraCyc and RiceCyc contain pathways for the biosynthesis of glucosinolates and oryzalexins, respectively, PlantCyc contains all of them. On the other hand, unlike universal pathway databases such as MetaCyc and KEGG, PlantCyc contains only plant pathways. For example, there are six variant routes for synthesizing Lys in MetaCyc, of which five are microbe specific, but only the single plant route is included in PlantCyc. Another major difference between MetaCyc and PlantCyc is that PlantCyc contains some hypothetical pathways without any associated enzymes. These pathways are proposed solely based on metabolite information. Differences between PlantCyc and MetaCyc affect their capacity to serve as reference databases for predicting pathways for a particular genome (see below).
Creation of a Reference Enzyme Sequence Database and Enzyme Annotation Pipeline
In addition to a reference plant pathway database, we created RESD to predict enzymes from a newly sequenced genome. The RESD was built by extracting sequences that are annotated to specific enzymatic activities from several curated databases, including BRENDA (Chang et al., 2009), UniProt (UniProt Consortium, 2010), MetaCyc (Caspi et al., 2010), and The Arabidopsis Information Resource (TAIR; Swarbreck et al., 2008; see “Materials and Methods”). Version 1.0 of RESD, which was used to build PoplarCyc, contained 14,187 literature-supported enzymes across all kingdoms, which are associated with four-digit Enzyme Commission (EC) numbers, Gene Ontology (GO) terms, or MetaCyc reaction identifiers.
We used the RESD to annotate the translated protein sequences from the first sequenced tree genome (version 1.1) from P. trichocarpa, a model wood-producing species (Tuskan et al., 2006). Each poplar protein sequence was queried against the RESD using BLASTP (Altschul et al., 1990). Function annotations (EC numbers, GO terms, or MetaCyc identifiers) of the best matching RESD sequence were transferred to the poplar sequence if the best hit had (1) the same four-digit EC number that was annotated by the Joint Genome Institute (JGI; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.download.ftp.html; 2,504 sequences) or (2) an E-value that was lower than the E-value threshold we defined for each enzyme class in the RESD (3,024 sequences; see “Materials and Methods”). The resulting set of annotated poplar enzymes contained 5,528 sequences. After removing enzymes that are not involved in small molecule metabolism (e.g. protein-modifying enzymes), the final annotation file contained 3,526 enzymes. The EC annotations from this study and JGI were manually reviewed by spot-checking randomly chosen sequences. For sequences that were annotated identically by JGI and PMN, the accuracy was high (10 of 10 sequences were correctly annotated). For sequences annotated differently by JGI and PMN, our annotation was more accurate than the JGI annotation (12 of 15 versus five of 15 correctly annotated, respectively).
Creation of PoplarCyc
The annotated poplar sequences were fed into PathoLogic (version 12.5) to predict metabolic pathways. PathoLogic matches the predicted functions of the newly annotated enzymes with known enzymes in a reference pathway database and assigns the predicted enzymes to corresponding reference reactions and pathways (Karp et al., 2010).
We used both PlantCyc and MetaCyc as reference databases in reconstructing the poplar metabolic network. Using PlantCyc (version 2.0; 646 pathways) as the reference database, PathoLogic assigned 2,604 enzymes to 1,018 reactions and predicted a total of 285 pathways for poplar (Table II). Of the 285 pathways, 260 were valid (see manual validation procedure in “Materials and Methods”), with a false-positive prediction rate of 7.3% (25 of 343). When MetaCyc (version 12.5; 1,395 pathways) was used as the reference database, a total of 346 pathways were predicted, in which 92 were false positives, making the false-positive prediction rate 8.4% (92 of 1,089). All but two false-positive predictions from PlantCyc were also predicted by MetaCyc (Table II). The 25 false-positive predictions from PlantCyc included pathways related to specialized/secondary metabolism or alternative metabolic routes with literature-supported evidence in some plant species but not in poplar and others, such as the biosynthesis pathway of dhurrin, a specialized cyanogenic glucoside that was found so far only in a few plant species, including sorghum (Sorghum bicolor) and sugarcane (Saccharum officinarum; Piotrowski, 2008; http://www.plantcyc.org:1555/PLANT/NEW-IMAGE?type=PATHWAY&object=PWY-861). The majority of the false-positive predictions from MetaCyc (71 of 92) were pathways not valid for plants in general, which are not present in PlantCyc. They are either unlikely to operate in plants, such as dissimilatory sulfate reduction (http://biocyc.org/META/NEW-IMAGE?type=NIL&object=DISSULFRED-PWY),or operate via a different route in plants, such as Asp degradation variant II (http://biocyc.org/META/NEW-IMAGE?type=NIL&object=MALATE-ASPARTATE-SHUTTLE-PWY). The rest of the false-positive predictions generated using MetaCyc were specialized or variant plant pathways without evidence in poplar per se (21 pathways).
Table II. Comparison of the PoplarCyc initial builds with either PlantCyc or MetaCyc as the reference database.
| Data | Reference Database Used |
|
| PlantCyc (2.0) | MetaCyc (12.5) | |
| Total no. of pathways in the reference database | 646 | 1,395 |
| Total no. of predicted pathways | 285 | 346 |
| No. of false-positive predictions (false-positive rate, FP/FP + TNa | 25 (7.3%) | 92 (8.4%) |
| Database-specific false-positive predictions | 2 | 69 |
| No. of false-negative predictions (false-negative rate, FN/TP + FNa) | 43 (14.1%) | 52 (17.0%) |
| Database-specific false-negative predictions | 6 | 13 |
FN, False negative; FP, false positive; TN, true negative; TP, true positive.
The number of false-negative predictions (valid pathways that were not predicted) is similar for both reference databases, 43 pathways from PlantCyc (false negative rate of 14.1%) and 52 pathways from MetaCyc (false negative rate of 17.0%; Table II). Using PlantCyc, PathoLogic predicted 23 valid pathways that were not predicted by using MetaCyc (Supplemental Table S1). All of these pathways are in primary metabolism. Among them, 14 did not exist in the MetaCyc version that was used. For eight pathways, PathoLogic properly predicted the plant variant of a pathway when PlantCyc was used as the reference but incorrectly predicted a nonplant variant when MetaCyc served as the reference database. The remaining one false-negative prediction appears to be caused by an incomplete annotation of enzyme synonym in MetaCyc (Supplemental Table S1). Among the 15 pathways predicted only by using MetaCyc, 12 did not exist in the PlantCyc version that was used. The remaining three false-negative predictions were due to three instances of incorrect annotations in PlantCyc. An additional 48 valid pathways were not predicted using either of the databases, because they had an insufficient number of poplar enzymes assigned to them to meet the PathoLogic prediction requirements (Supplemental Table S1).
In summary, both PlantCyc and MetaCyc databases showed similar false-positive and false-negative rates when predicting PoplarCyc, although the absolute number of false-positive predictions was much higher when using MetaCyc. While the overlap of the errors between the two databases was substantial, there were many pathways predicted only by one database, indicating the usefulness of both databases as references for predicting plant metabolic pathways. The false-positive and false-negative rates only give an estimation for pathway occurrence in poplar. The individual enzymes annotated to the pathways may still contain errors in their functional predictions.
Comparison of PoplarCyc and AraCyc
To further assess the prediction pipeline, we compared PoplarCyc (version 1.0) with AraCyc (version 6.0), which has undergone substantial curation based on experimental evidence in the literature, by using the Pathway Tools Comparative Analysis (http://www.plantcyc.org:1555/comp-genomics?tables=reaction&tables=pathway&tables=protein&orgid=ARA&orgid=POPLAR&orgids=(ARA+POPLAR+). The overall numbers of pathways, reactions, and enzymes of PoplarCyc are about 20% less than those for AraCyc (Table I). However, the portion of reactions without enzymes is much higher in PoplarCyc (45%) than in AraCyc (28%). In addition, AraCyc, after many years of manual curation from the rich corpus of Arabidopsis literature, has far more enzymes with experimental support (43% of all enzymes).
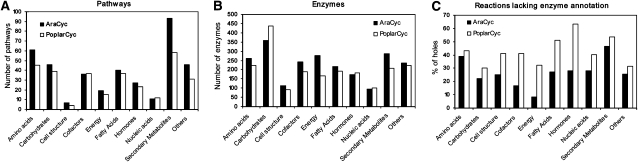
A detailed examination and comparison of pathways and enzymes across various metabolic domains are summarized in Figure 1. The distribution of pathways among different metabolic domains is similar between the two databases except for secondary metabolism. A higher portion of secondary metabolic pathways is found in AraCyc (21%) than in PoplarCyc (14%; Fig. 1A). Among the 42 secondary metabolic pathways found only in AraCyc, 16 are either specific to Arabidopsis (e.g. arabidiol biosynthesis) or specific to Brassicaceae (e.g. glucosinolate biosynthesis; Supplemental Table S2). Seventeen pathways were not predicted for poplar due to insufficient enzyme evidence in the poplar genome. Three pathways were new additions to AraCyc and not present in the version of the reference database that was used for the prediction. The remaining six were initially predicted but manually removed during the validation process (see “Materials and Methods”). On the other hand, there are six secondary metabolic pathways that are only present in PoplarCyc. They were either not predicted due to insufficient or lack of enzyme evidence in the Arabidopsis genome (TAIR9 release) or were initially predicted but removed after curator validation.
Figure 1.
Comparison of PoplarCyc and AraCyc. A, Total number of pathways in each metabolic domain. B, The number of enzymes that catalyze the reactions in pathways within each metabolic domain in the database. Because some reactions are part of more than one domain, the sum of the percentages for all of the metabolic domains will be greater than 100%. C, Percentage of reactions without any associated enzymes in each metabolic domain. “Others” includes alcohols, aldehydes, amines and polyamines, aromatic compounds, C1 compounds, carboxylates, inorganic nutrients, and unclassified.
The distribution of the enzymes across various metabolic domains (Fig. 1B) largely agrees with the distribution of the pathways, except for carbohydrate metabolism. PoplarCyc contains about 1.5-fold more carbohydrate metabolism enzymes than does AraCyc (Fig. 1B). The increased number of carbohydrate enzymes could have accommodated the growth habit of trees, such as perennial growth and large girth and height. Overrepresented poplar genes in comparison with Arabidopsis were also reported by others (Geisler-Lee et al., 2006; Tuskan et al., 2006), which include genes associated with cell wall biosynthesis, among others.
A comparison of the portion of reactions that are not associated with any enzymes in the databases (“pathway holes”) shows that AraCyc has a lower proportion of pathway holes across all metabolic domains than PoplarCyc does (Fig. 1C). The difference between the two databases is most prominent for cofactor, energy, fatty acid, and hormone metabolism. Searching for enzymes with less conserved protein sequences in poplar may be needed to fill these holes. Alternatively, these pathways may utilize different routes or enzymes in poplar from those in Arabidopsis. In general, the first version of PoplarCyc shows comparable data content to the latest version of AraCyc (6.0), indicating that there were no large errors or strong biases introduced by the prediction pipeline.
Data Availability
The PMN databases currently include PlantCyc and the single-species networks PoplarCyc and AraCyc, which can be freely accessed through the Web (http://plantcyc.org). Researchers can browse, query, and visualize the data. Users can navigate through all of the pathways, for example, from an alphabetic list or from the hierarchy ontology browser (http://www.arabidopsis.org:1555/PLANT/class-instances?object=Pathways). Information can be queried using whole words (e.g. “gibberellin”) or substrings (e.g. “gibber”) and can be queried against specific data types, such as compounds only, or all of the data types, including compounds, reactions, pathways, proteins, and genes. On pathway detail pages, graphical diagrams of the pathways can be viewed at various levels of detail through semantic zooming. On the most zoomed-in view, pathway diagrams are decorated with enzymes, genes, and compound structures. On the pathway diagrams, users can choose to either show only experimentally characterized enzymes or to include predicted enzymes, and they can elect to show all enzymes or only enzymes for a given species. All the data objects on the pathway diagrams can be clicked on to access additional information. For single-species databases, users can overlay and visualize large-scale experimental data using the Pathway Tools Omics Viewer tool (http://www.plantcyc.org:1555/ARA/expression.html). An example of using the Omics Viewer in analyzing data is demonstrated in detail below. Users can also compare summary statistics of pathways, reactions, and enzymes between the PMN databases using the Comparative Analysis tool (http://www.plantcyc.org:1555/comp-genomics).
In addition to online access, the complete PMN databases are also available for download with an “open source” license (http://www.plantcyc.org/downloads/license_agreement.faces) in the formats of flat files (http://bioinformatics.ai.sri.com/ptools/flatfile-format.html) such as Ocelot, which is readily readable by Pathway Tools, BioPAX, and SBML (Stromback and Lambrix, 2005). Installation of the PMN databases on a local computer, together with a Pathway Tools installation (http://biocyc.org/download.shtml) on the same computer, enables users to run PMN databases locally. This local database setup offers a few advantages, such as additional operations, faster speed, the opportunity to update the database with proprietary data, and the ability to perform programmatic queries.
Inferring Metabolic Functions for Specific Protein Sequences with PMN BLAST
We created two enzyme sequence databases that provide researchers with two new options for predicting metabolic function based on protein sequences. The RESD contains enzymes with experimental or other literature support and available protein sequence information (see “Materials and Methods”). Notably, this database includes enzymes from all kingdoms and some enzymes not directly involved in metabolism. In contrast, the PlantCyc Enzyme Sequence Database (PESD) includes all enzymes found in PlantCyc with known protein sequences. Both experimentally and computationally supported enzymes are included in this database. These databases can be downloaded (http://www.plantcyc.org/downloads/data_downloads.faces) or searched against user-defined query sequences using the PMN BLAST tool (http://www.plantcyc.org/tools/Blast/blast.faces). Different research questions can be addressed using these databases and search tools. For instance, the RESD has been used for the genome-wide annotation of poplar using the PMN enzyme annotation pipeline (presented in this paper).
In addition, a researcher who has experimentally identified a particular sequence of interest can quickly identify putative homologs using the PMN BLAST tool and the PESD and can view the metabolic context in which those homologs function. For instance, a recent study on chickpea (Cicer arietinum) identified a set of transcripts that are differentially expressed following exposure to Fusarium, an important fungal pathogen that typically causes a 10% to 15% loss in worldwide chickpea production each year (Ashraf et al., 2009). When one of the highly down-regulated transcripts (CaF1_WIE_16_H_10; GenBank accession no. GR915374.1) is used in a BLASTX query against the PESD, a number of enzyme sequences match with very low E-values. The top two best matching protein sequences, AT4G01850 (E-value = 8 × 10−80) and AT1G02500 (E-value = 2 × 10−79) from Arabidopsis, are annotated to participate in three biochemical pathways: ethylene biosynthesis from Met, S-adenosyl-l-methionine (SAM) biosynthesis, and the SAM cycle. AT1G02500 has experimental evidence for its biochemical function that places it in these pathways, whereas the function of AT4G01850 is only supported by computational prediction. The researcher can easily navigate to these pathway pages and learn more about the biological relevance of these putative homologs. Ethylene has known connections to plant defense (Glazebrook, 2005; Broekaert et al., 2006; Zhao and Qi, 2008), suggesting that these pathways related to the generation of ethylene and its precursor SAM merit further investigation.
Identifying Candidate Enzymes in Underannotated Species Using PlantCyc
Researchers studying plant species that are not well annotated may wish to identify enzymes that are likely to participate in a particular metabolic process. The availability of experimentally verified enzyme data from a variety of species distributed throughout the plant kingdom makes PlantCyc a useful starting point for identifying good candidate enzymes involved in a wide assortment of biochemical pathways in “orphan crops” and other understudied species. An example from cowpea (Vigna unguiculata) illustrates this use of PlantCyc.
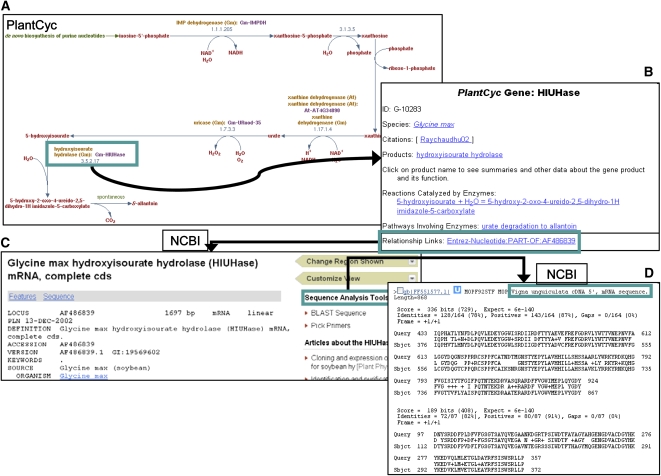
Cowpea is an important legume grown largely in western and central Africa, where its drought tolerance and ability to survive on low-quality soil make it a valuable crop for poor subsistence farmers (Timko et al., 2008). However, few resources have been dedicated to understanding and improving cowpea metabolism, despite its agronomic importance. Pathways related to nitrogen fixation and utilization may play a significant role in allowing cowpea to grow on nutrient-poor soil. Cowpea plants export the nitrogen fixed in their nodules in the form of ureides. A PlantCyc pathway describing ureide biosynthesis (http://www.plantcyc.org:1555/PLANT/new-image?type=PATHWAY&object=URSIN-PWY&detail-level=2&EXP-ONLY=T; Fig. 2A) includes several enzymes from soybean (Glycine max). Because these enzymes are already experimentally verified, and because cowpea is more closely related to soybean than to other well-studied organisms, these PlantCyc enzymes make good candidates for identifying potential cowpea homologs involved in this process. For instance, the last enzymatic step in the pathway is performed by hydroxyisourate hydrolase (HIUHase) from soybean (Fig. 2B). The detail page for the gene encoding this enzyme has a link to an Entrez gene sequence (AF486839). Clicking on the unification link (Fig. 2C) brings up the corresponding GenBank record. The soybean sequence associated with this record can be directly sent for a BLAST analysis. Following a TBLASTX search against the nonhuman, nonmouse EST data set (est_others), the best matching sequence from cowpea (FF551577) is revealed to have two regions with 78% and 82% sequence identity to the soybean HIUHase (E-value = 3 × 10−142) and is expected to encode a highly similar cowpea protein (Fig. 2D). This approach of discovering potential enzymes could help expedite the experimental characterization of metabolic pathways in understudied organisms.
Figure 2.
Using PlantCyc to find a candidate enzyme in cowpea. The ureide biosynthesis pathway shows data for several experimentally verified enzymes from soybean, shown in boldface (http://www.plantcyc.org:1555/PLANT/new-image?type=PATHWAY&object=URSIN-PWY&detail-level=2&EXP-ONLY=T). Clicking on the gene encoding the last enzyme in the pathway, HIUHase (A), opens a gene detail page that has a link to the Entrez gene database at the National Center for Biotechnology Information (B). The link provides access to the sequence deposited in GenBank. The BLAST tool can be accessed directly from this page (C). A TBLASTX analysis performed against the nonhuman, nonmouse EST data set (est_others) using the default parameters and with the species search limited to cowpea (taxid: 3917) yielded the top result shown in D. A BLAST query using the putative protein encoded by this cDNA against the RESD confirmed that HIUHase was the best hit within this data set.
Viewing Large-Scale Omics Data in a Metabolic Context
Analyzing and deriving the biological meaning of large-scale data sets from transcriptomic, proteomic, and metabolomic profiling experiments can be challenging. The Pathway Tools Omics Viewer is one tool that can help in this process by displaying experimental results on top of an overview of all the biochemical pathways present in a single species (Zhang et al., 2005). For instance, a researcher who assayed for transcript levels in a wild-type plant compared with those in a particular mutant could look at the quantitative changes in gene expression across many metabolic domains at once and gain some perspective on the overall influence of the mutated gene on metabolism. This type of analysis can help to generate hypotheses about the underlying metabolic explanations for phenotypic changes caused by various factors such as mutations and stresses. We illustrate this use case using a recent study examining the metabolic effect of a gain-of-function mutation in gibberellin (GA) signaling in poplar (Busov et al., 2006).
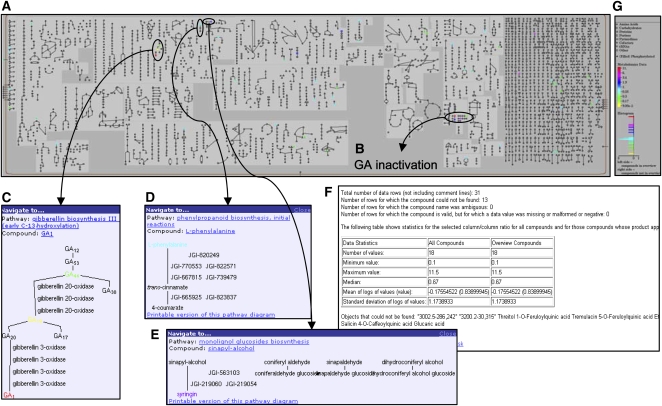
GAs play an important role in regulating plant growth and development (Sun and Gubler, 2004). Members of the DELLA family of proteins appear to act as transcriptional regulators that repress the GA signaling pathway (Sun and Gubler, 2004). Certain mutations in DELLA proteins have allowed the development of dwarf varieties of crops with improved yield (Peng et al., 1999; Ikeda et al., 2001). To better understand how this signaling pathway would affect growth in poplar, researchers generated transgenic poplar plants expressing a mutated DELLA protein from Arabidopsis, gibberellin insensitive(gai), under the control of the Arabidopsis GAI promoter (Busov et al., 2006). As in Arabidopsis, expression of the gain-of-function gai mutant in poplar appears to cause a constitutive inhibition of some GA signaling responses. Consequently, the resulting lines have stunted shoot growth and increased root growth. Relative metabolite levels were quantified from the leaves and roots of gai-expressing versus wild-type plants to learn more about the biochemical basis of these altered growth profiles. When the leaf metabolite data are displayed using the Omics Viewer, several pathways with quantitative differences in metabolite levels between the gai-expressing and wild-type plants can be readily identified (Fig. 3A). For example, the gai mutants have altered levels of specific GAs. Notably, the bioactive end product, GA1, is significantly elevated in the mutant plants, whereas two precursor GAs are present at lower levels, suggesting feedback regulation of GA metabolites as characterized in other plants (Busov et al., 2006). These changes are shown in the GA biosynthesis and GA inactivation pathways in the Omics Viewer (Fig. 3, B and C).
Figure 3.
Displaying metabolite data on the Omics Viewer. The Omics Viewer is displaying the ratio of selected metabolite levels in gai-expressing transgenic poplar leaves to wild type (WT) poplar (Populus tremula × Populus alba) leaves (for details, see Busov et al., 2006). The underlying raw data for GA levels were measured in units of ng g−1 dry weight, whereas all other compound measurements were made in units of μg Glc equivalent g−1 fresh weight. Pathways with increased and decreased relative levels of important compounds can easily be identified on the overview (A). For instance, the pathways associated with GA biosynthesis and inactivation each have several compounds with altered abundances (B and C). Clicking on any compound in the overview brings up an associated pathway popup window, as shown for the “gibberellin biosynthesis III” pathway (B) as well as for the two cell wall-related pathways that each has one compound with altered levels (D and E). These more detailed diagrams identify the enzymes and compounds depicted using shapes and lines on the Omics Viewer. Hyperlinks from the popup window to the relevant pathway and compound pages in PoplarCyc allow access to more detailed information, such as curated pathway summaries, enzyme sequence information, etc. A data table and a list of compounds from the input file that do not match any current entries in PoplarCyc are displayed below the metabolic overview (F). The color key to the right (G) indicates the magnitude of the difference in relative metabolite abundance (gai versus the wild type).
While the Omics Viewer helps to confirm the predicted effects of gai on this known target pathway, it is also useful in highlighting additional metabolic changes in the aerial tissues that might contribute to or indicate reduced shoot growth. For instance, the level of Phe, the first compound in the “phenylpropanoid biosynthesis, initial reactions” pathway, is reduced in gai leaves (Fig. 3D), which could indicate limited carbon flux toward lignin. The reduced flow through this pathway may also be reflected by the increased level of syringin, a storage form of a lignin precursor known as sinapyl alcohol (Fig. 3E). Changes in the levels of compounds in these important cell wall-related pathways may be linked to the dwarfing phenotype. Based on this initial metabolic snapshot presented within the context of the Omics Viewer, further experiments can be designed to better understand the connections and regulatory interactions between GA signaling, specific biochemical pathways, and poplar productivity in aerial tissues. While this is one very practical way to use this tool, it is just one of many options for examining different types of data sets in a metabolic context. Additional examples are provided in Table III.
Table III. Uses of Omics Viewer with various data types.
| Data Type | Situation | Solution |
| Metabolites | Compounds of known masses A, B, and C, but unknown chemical formula, have shown up in your Arabidopsis experiment | Use the Advanced Query page to look up all the chemicals with masses A, B, and C and display them using three different colors |
| Metabolites | Compounds of known, or sometimes partially known, chemical formula, but unknown structure, have appeared in your results | Use the Advanced Query page to look up all the chemicals with the known information from the chemical formula and examine them |
| Genes/proteins | The set of genes coexpressed with a gene of interest have been identified and given confidence scores | Display all of the coexpressed genes and display the confidence score using the color gradient |
| Genes/proteins | A protein interaction viewer shows all of the proteins associated with a protein of interest | View all of the interacting proteins that participate in metabolism |
| Genes/proteins | Putative targets of a transcription factor have been determined based on predicted DNA promoter elements | View all of the metabolic targets of the transcription factor |
| Genes/proteins | A set of mutants all have a particular phenotype that might be linked to metabolic defects (e.g. chlorotic leaves) | View where all of the mutants fit into metabolic pathways |
| Proteins | Spots on a two-dimensional gel with known molecular mass have been identified | Use the Advanced Query page to look up all the proteins with the desired molecular mass and display them |
| Proteins | The complete proteome of an organelle of interest has been published | View all of the metabolic proteins from the organelle |
| Transcripts | Several publicly available microarray data sets relate to a biological process of interest (e.g. drought stress) | Obtain the data sets and examine them |
DISCUSSION
We developed a general approach for facilitating the reconstruction of plant metabolic networks from sequenced genomes or transcriptomes. Four components were created for the system: (1) PlantCyc, a pan-plant reference database of metabolic pathways and enzymes; (2) RESD, a reference enzyme sequence database containing protein sequences with literature-supported enzyme activities; (3) an enzyme sequence annotation pipeline that predicts enzyme functions from predicted protein sequences based on sequence similarity to RESD sequences; and (4) a modified pathway prediction procedure that uses both PlantCyc and MetaCyc as the reference for reconstructing single-species metabolic networks from the predicted enzymes. Using such a consensus approach will make it easier to interpret the results of cross-species metabolic comparisons. The individual components of the infrastructure can also be used on their own in a number of ways.
We applied the system to the sequenced genome of poplar (Poptr 1.1 release, JGI; http://www.jgi.doe.gov/genome-projects/) and assessed its quality by comparison with the latest release of AraCyc, which has substantial support from experimental data in the literature. Overall, the predicted poplar pathway database PoplarCyc has comparable data content to AraCyc. For predicting pathways for poplar, using PlantCyc as the reference generated many fewer false-positive predictions (25 pathways) than using a universal reference such as MetaCyc (92 pathways), although the false-positive rates were similar using both databases (7.5% and 8.5%, respectively). The false-negative predictions, such as those predicted by one reference database but not the other, were also found using either PlantCyc or MetaCyc. PathoLogic considers the entire pathway population of the reference database in calculating the overall supporting evidence per pathway (Karp et al., 2010). Therefore, the calculated likelihood that a specific pathway exists can differ when PathoLogic is run using PlantCyc or MetaCyc. In some cases, the large size of the MetaCyc database can hamper “true positive” pathway prediction. For example, in PlantCyc, the last step of Gly betaine biosynthesis (http://www.plantcyc.org:1555/POPLAR/NEW-IMAGE?type=PATHWAY&object=PWY1F-353) is a unique reaction not found in other pathways in PlantCyc. Thus, the assignment of two poplar enzymes to this reaction was sufficient evidence for PathoLogic to predict the pathway when using PlantCyc as the reference. However, the reaction is not unique to this pathway in MetaCyc. It is also involved in several microbial variants of Gly betaine biosynthesis. Therefore, when MetaCyc was used as the reference database, the mapping of these two poplar enzymes to the last reaction in the pathway was not sufficient for PathoLogic to predict the existence of “Gly betaine biosynthesis.” On the other hand, using MetaCyc, PathoLogic predicted bacterial or animal pathways that may exist in poplar and other plants, such as guanosine 5′-diphosphate,3′-diphosphate biosynthesis (http://www.plantcyc.org:1555/POPLAR/NEW-IMAGE?type=NIL&object=PPGPPMET-PWY). That is, using MetaCyc, there is a greater possibility of discovering the presence of a pathway that was previously thought unlikely to occur in plants and thus was not included in PlantCyc. These results indicate that using both databases separately might be a useful way of increasing the specificity and coverage of the predicted pathways.
To maintain the high quality of the PMN databases, the initial computationally predicted database for each species must undergo manual “validation” by curators searching through primary scientific literature. This effort is required to identify which pathways are “false positives” that need to be removed and to determine which pathways are “false negatives” that were not predicted from the reference database but need to be added to the new single-species database. Manual validation requires a broad knowledge of metabolism for each new given species and is very time-consuming. Therefore, it is useful to try to reduce the number of false-positive and false-negative predictions before beginning the manual validation process. Using PlantCyc as the primary reference contributes to this effort, because while the percentage of incorrect predictions made is generally similar between MetaCyc and PlantCyc, the absolute number of false-positive pathways to review is much greater when PathoLogic uses the former. On the other hand, because MetaCyc can contribute a unique set of pathways not predicted by PathoLogic with PlantCyc, MetaCyc should not be abandoned in the prediction process. Taking advantage of our manual validation results for PoplarCyc combined with the previous work on AraCyc validation, we have built two new resources that can be used to quickly filter the results of the computational predictions. The nonplant pathways (NPPs) file includes a list of MetaCyc pathways that are likely invalid for all plants, and the universal plant pathways (UPPs) file contains a list of PlantCyc pathways that are likely present in all plants (http://www.plantcyc.org/downloads/data_downloads.faces). The two lists have been reviewed by a number of expert plant biochemists and can be used as a general guide to systematically prune false-positive predictions from and add false-negative predictions to any new plant single-species network generated by the PMN pipeline. This step should expedite the process of validation substantially. The NPPs and UPPs are also being used to update the “expected taxonomic range” field of MetaCyc and PlantCyc pathways, which should help to reduce false PathoLogic predictions in the future. Nevertheless, continual revision and further rounds of editorial review will be needed to keep the two lists up to date, accurate, and as comprehensive as possible. The NPP and UPP will continue to evolve over time, especially in response to new discoveries in the actively researched domains of plant metabolism.
While this work describes what is, to our knowledge, the first attempt at establishing a general approach for reconstructing metabolic pathway complements of plant genomes, the current version of the infrastructure has some limitations. For example, PlantCyc still has a large number of reactions without any annotated enzymes or full EC information. The current system cannot assign any putative enzymes to them. Efforts are needed to curate recent literature to fill some of these holes. Also, while the enzyme sequence annotation pipeline was aimed at improving the accuracy of the functional predictions, it could overpredict enzyme activities. Finally, the pathway prediction program, PathoLogic, currently does not take into consideration many important factors, such as the confidence level of predicted enzymes, differences in conservation levels of primary metabolism versus secondary metabolism, and evidence of the presence of metabolites in a given species. In addition, subcellular locations of pathways and enzymes are not considered when assigning putative enzymes to pathways, which can cause false-positive assignments. For example, glycolysis can operate in two different subcellular locations in plant cells, the cytosol and the plastid. Cytosolic and plastidic glycolysis share many common reactions that are catalyzed both by cytosolic and plastidic enzymes. But because PathoLogic fails to consider enzyme subcellular location information, enzymes can be wrongly assigned to both pathways instead of the correct variant. Despite these limitations, the system described here serves as a starting point to the path of “next generation” annotation scheme, upon which improvements will be built to minimize manual intervention and maximize quality.
With the vast amount of sequence data generated in recent years, namely whole genome sequences for over a dozen plant species (http://www.jgi.doe.gov/genome-projects/) and EST assemblies for over 200 species (http://www.plantgdb.org/prj/ESTCluster/), the infrastructure we described here can be readily utilized to annotate the sequences at a system level and to place the sequence of each organism into its metabolic network(s). The networks can then serve as a platform and tool to visualize and analyze large-scale omics data, predict metabolic phenotypes, study metabolic pathway evolution, and conduct comparative metabolism analyses. Several examples of research problems that can be addressed using the PMN resources have been described above to illustrate the value of both single-species databases and PlantCyc.
MATERIALS AND METHODS
PlantCyc Creation and Curation
The Pathway Tools (version 12.5) PathoLogic program was used to create PlantCyc with id = PLANT, species name = PlantCyc. The PathoLogic “Build” step was skipped so that PlantCyc contained no data at this point. The above step initialized the database schema for PlantCyc. Next, using the Pathway Tools Ontology Browser, PlantCyc was placed under the parent Multi-Organism-Groupings of the Organisms class. This converted PlantCyc from the default single-species database to a multiple-species database. All pathways and their associated reactions, compounds, enzymes, and genes of AraCyc were imported into PlantCyc using the Pathway Tools pathway export/import function. Additional plant pathways that are absent in Arabidopsis (Arabidopsis thaliana) were identified and imported from MetaCyc, which largely extended the PlantCyc coverage, especially in secondary metabolism. We also imported several pathways that were manually curated by RiceCyc and MedicCyc.
In-house manual curation is both labor-intensive and expensive. Therefore, we employ several methods of collaborative curation. We collaborate with other pathway databases such as MetaCyc, SolCyc, and RiceCyc, with which we share “to-be curated” lists to prevent redundant curation. We also curate directly into MetaCyc and then export the curated data to PlantCyc to facilitate the reuse of common building blocks such as compounds and reactions. We hold monthly meetings with MetaCyc and SolCyc curators to discuss curation and database-related issues. Another type of collaboration is with organism-specific genome databases such as SoyBase (http://soybase.org/). We have created an initial build of SoyCyc from the JGI soybean genome sequence release. We sent it to SoyBase curators, who have expertise or access to the expertise in soybean biology to review and further curate the predicted pathways. A third type of collaboration occurs with experts in specific domains of metabolism. A number of expert plant biochemists, including members of the PMN editorial board (http://www.plantcyc.org/about/editorial_board.faces), have helped the PMN by reviewing existing pathways and enzymes and identifying new literature information, especially in the fields of alkaloid, terpenoid, and glucosinolate metabolism. Finally, we encourage individual users to submit new data or make suggestions for corrections by e-mail, an online feedback form, or preformatted data submission forms. User contributions are publicly acknowledged (http://www.plantcyc.org/about/contributors.faces), and past contributors receive direct notice of new PMN releases.
Creation of a Reference Enzyme Sequence Database
Protein sequences of enzymes with experimental or other literature support were compiled from four databases: UniProt (UniProt Consortium, 2010), BRENDA (Chang et al., 2009), TAIR (Swarbreck et al., 2008), and MetaCyc (Caspi et al., 2010). From TAIR (downloaded January 29, 2009) and UniProt (gene_association.goa_uniprot.gz from December 31, 2008), we retrieved 246 and 7,615 proteins, respectively, which were annotated with GO terms that are children of the “catalytic activity” (GO:0003824) term based on experimental evidence codes IDA, IPI, IMP, IGI, and IEP. From BRENDA (brenda_dl_0702.zip), we retrieved 5,219 proteins that were assigned to the full, four-part EC numbers and also had associations to publications. From MetaCyc (version 12.5), we retrieved 1,107 UniProt identifiers of enzymes in the protein.dat file or by cross-referencing the UniProt identifiers from GenBank using the gene identifiers in the gene.dat file. All the MetaCyc proteins were also annotated with unique MetaCyc reaction identifiers.
UniProt accessions compiled from the four sources were combined to produce a nonredundant set and used to retrieve protein sequences from UniProt. If the same UniProt accession existed in multiple data sources, we took the annotation from the databases in the following order: BRENDA (5,220 sequences), GO (7,615 sequences), TAIR (246 sequences), and MetaCyc (1,006 sequences). This data set, herein named the RESD, contained 14,187 unique sequences. A total of 6,009 were associated with full EC numbers, 7,897 sequences were associated with GO terms, and 281 were associated with MetaCyc reaction identifiers. Of the 14,187 total sequences in the data set, 12,825 had only one identifier while 1,362 had more than one identifier associated with it.
Poplar Enzyme Prediction
The functional annotations of poplar (Populus trichocarpa) protein sequences were assigned based on several criteria, including sequence similarity. In the first step of the annotation process, each poplar sequence was queried against the RESD using the National Center for Biotechnology Information BLASTP program. If the top hit identified in the RESD had one or more full EC number annotation associated with it, the EC numbers were compared with the existing EC number annotations made by JGI (P. trichocarpa version 1.1) for the query protein. All EC numbers matching between the top BLAST hit and the JGI annotation were assigned to the query protein. If no EC number was shared between the top BLAST hit against the RESD and the JGI annotation, or in cases where JGI had not assigned a full EC number to the poplar sequence, additional criteria were used to determine the best annotation. Because of the varying levels of sequence diversity found for each type of enzyme in the data set, unique E-value thresholds were generated for each “functional identifier” of enzyme activity. Therefore, for each EC number, GO term, and MetaCyc reaction identifier, a specific E-value cutoff was generated by performing an all-by-all BLAST of the RESD sequences and then identifying all of the true-positive hits in which one representative sequence of a particular EC number, GO term, or MetaCyc reaction identifier matched another member with the same functional identifier. Self-hits were eliminated. The mean of the log10 of the E-values of the remaining true-positive hits was used as the cutoff for each functional identifier. Any alignment with an E-value of 0 was given a value of 10−200. From 3,805 unique functional identifiers that were represented in the RESD, 1,901 had mean E-values ranging from 10−30 to 10−200 (with a mean of 10−132 and a sd of 10−47). The remaining 1,904 functional identifiers were represented by only one sequence, and we assigned a “global” threshold of 10−132, equivalent to the mean of the established thresholds, to these functional identifiers. For query proteins that failed to receive a functional identifier in the first step of the annotation process, these E-value cutoffs were used to prune the list of candidates generated through the BLASTP analysis. We removed candidate sequences that matched the query with an E-value greater than the functional identifier's mean E-value. The remaining matching sequences were ranked by an ascending order of E-values, and the functional identifier(s) associated with the top matching sequence were used to annotate the poplar sequence.
The resulting set of annotated poplar sequences contained 5,528 sequences representing 1,073 functional identifiers. Each poplar protein was assigned a common name based on its EC number, GO term, or MetaCyc reaction identifier annotations. If the reference sequence had multiple annotations, all annotations were used to assign the poplar sequence. To remove enzymes not involved in small molecule metabolism (e.g. protein-modifying enzymes), protein names were compared with a blacklist of nonenzymatic names in a case-insensitive string/word match search using a Perl script. This filtering step resulted in the removal of 2,002 sequences and produced a final set of 3,526 enzymes representing 943 functional identifiers (420 EC numbers, 489 GO identifiers, and 34 MetaCyc reaction identifiers) that were used as input for the PathoLogic pathway prediction program.
PoplarCyc Creation
The PathoLogic input file was prepared according to the instructions in the Pathway Tools (version 12.5) user's guide. An initial build of PoplarCyc using the default reference database, MetaCyc, was created following the user's guide. Currently, MetaCyc is the built-in reference database used by PathoLogic. One can choose to use either MetaCyc as the sole reference database or to supplement MetaCyc with another reference database such as PlantCyc. In either case, all MetaCyc pathways are included as the reference pathways. To use PlantCyc as the sole reference, before running PathoLogic, all MetaCyc pathways that did not exist in PlantCyc were deleted and PlantCyc was chosen as the supplemental reference. In this way, only PlantCyc pathways served as the reference pathways. Following this step, the regular PathoLogic procedure was followed to create a PoplarCyc build with PlantCyc as the sole reference.
PoplarCyc Validation
The pathways in the initial build were validated by curators based on a preliminary literature search. Specifically, curators looked up literature for (1) any evidence about the pathway or its enzymes from poplar, (2) any indication that the pathway is likely to be common to all plants, and (3) any evidence for the presence, in poplar, of unique compounds in the pathway. If none of the above evidence was found, we checked for the existence of any P. trichocarpa enzymes annotated to unique reactions in the pathway and the quality of the enzyme annotation. A pathway was considered invalid and was consequently removed from the database if none of the above criteria was met. We also deleted pathways that are known to occur in bacteria but unlikely to exist in plants, and we used these to generate a PMN NPP list. During this refinement process, we also added a number of pathways from PlantCyc or MetaCyc that were not predicted by PathoLogic in the initial build. These included (1) adding pathways that are expected to be present in all plants and (2) adding the probable pathways for which the presence of a key enzyme or a compound is reported in a poplar species. The former class of pathways was also used to create a PMN UPP list. These lists can be employed to automatically accept or reject pathways for inclusion in future single-species database builds.
Viewing Large-Scale Omics Data in a Metabolic Context
All data values were obtained from Table III and Table V of Busov et al. (2006) and used to generate a data file in Excel. Calculated ratios provided by the authors were entered in column B (referred to as column 1 in the Omics Viewer input page). These values represent the ratio of absolute metabolite levels present in gai-overexpressing leaves relative to wild-type leaves from two different experiments that were performed using different treatment and analytic methods (for details, see Busov et al., 2006). Four GA3 compounds from Table III and 10 compounds from Table V were removed from the analysis because they were not significantly different from the wild type, with P < 0.05. A tab-delimited text file containing the remaining 31 compounds plus additional data from Busov et al. (2006) and explanatory text is available as Supplemental File S1. The data file was displayed on the Omics Viewer (http://www.plantcyc.org:1555/POPLAR/expression.html) using the following settings: dataset: Populus trichocarpa; relative data values; a single data column; one-centered scale; compound names and/or identifiers; data column = 1; color scheme = full color spectrum, computed from data provided (default); display type = paint data on cellular overview chart (default).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. False-negative predictions in the initial builds of PoplarCyc.
Supplemental Table S2. Comparison of secondary metabolism pathways in PoplarCyc and AraCyc.
Supplemental File S1. Omics Viewer input file.
Supplementary Material
Acknowledgments
We are grateful to Tom Meyer, Larry Ploetz, and Shanker Singh for their help in software, hardware, and database maintenance; to Tomer Altman, Joe Dale, Fred Gilham, Pallavi Kaipa, Markus Krummenacker, Liviu Popescu, and Suzanne Paley for excellent Pathway Tools support; to summer interns Ricardo Leitao and Michael Ahn for their assistance in curating compounds and reactions; to Dr. Kun He for advising the interns; and to Drs. Clint Chapple, Markus Piotrowski, Nick Smirnoff, Ruth Welti, Brenda Winkel, Oliver Yu, and Rita Zrenner for valuable feedback on the UPPs and NPPs.
References
- Akatsuka T, Kodama O, Sekido H, Kono Y, Takeuchi S. (1985) Novel phytoalexins (oryzalexins A, B and C) isolated from rice blast leaves infected with Pyricularia oryzae. Part 1: Isolation, characterization and biological activities of oryzalexins. Agric Biol Chem 49: 1689–1694 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ashraf N, Ghai D, Barman P, Basu S, Gangisetty N, Mandal MK, Chakraborty N, Datta A, Chakraborty S. (2009) Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics 10: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delaure SL, De Bolle MF, Cammue BP. (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Busov V, Meilan R, Pearce DW, Rood SB, Ma C, Tschaplinski TJ, Strauss SH. (2006) Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 224: 288–299 [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. (2010) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 38: D473–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. (2009) BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res 37: D588–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxenbichler ME, Spencer GF, Carlson DG, Rose GB, Brinker AM, Powell RG. (1991) Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry 30: 2623–2638 [Google Scholar]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. (2008) PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res 36: D959–D965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Batley J. (2009) Plant genome sequencing: applications for crop improvement. Plant Biotechnol J 8: 2–9 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunnerås S, et al. (2006) Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Grafahrend-Belau E, Weise S, Koschutzki D, Scholz U, Junker BH, Schreiber F. (2008) MetaCrop: a detailed database of crop plant metabolism. Nucleic Acids Res 36: D954–D958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley S, Krieger CJ, Zhang P. (2004) An evidence ontology for use in pathway/genome databases. Pac Symp Biocomput 9: 190–201 [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, et al. (2010) Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform 11: 40–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. (2009) The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 38: D346–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Christian JO, Kempa S, Walther D. (2009) ChlamyCyc: an integrative systems biology database and Web-portal for Chlamydomonas reinhardtii. BMC Genomics 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Piotrowski M. (2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69: 2655–2667 [DOI] [PubMed] [Google Scholar]
- Stromback L, Lambrix P. (2005) Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics 21: 4401–4407 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timko MP, Rushton PJ, Laudeman TW, Bokowiec MT, Chipumuro E, Cheung F, Town CD, Chen X. (2008) Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38: D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Sumner LW. (2007) MedicCyc: a biochemical pathway database for Medicago truncatula. Bioinformatics 23: 1418–1423 [DOI] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. (2007) Reactome: a knowledge base of biologic pathways and processes. Genome Biol 8: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY. (2005) MetaCyc and AraCyc: metabolic pathway databases for plant research. Plant Physiol 138: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Qi X. (2008) Signaling in plant disease resistance and symbiosis. J Integr Plant Biol 50: 799–807 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PMN databases currently include PlantCyc and the single-species networks PoplarCyc and AraCyc, which can be freely accessed through the Web (http://plantcyc.org). Researchers can browse, query, and visualize the data. Users can navigate through all of the pathways, for example, from an alphabetic list or from the hierarchy ontology browser (http://www.arabidopsis.org:1555/PLANT/class-instances?object=Pathways). Information can be queried using whole words (e.g. “gibberellin”) or substrings (e.g. “gibber”) and can be queried against specific data types, such as compounds only, or all of the data types, including compounds, reactions, pathways, proteins, and genes. On pathway detail pages, graphical diagrams of the pathways can be viewed at various levels of detail through semantic zooming. On the most zoomed-in view, pathway diagrams are decorated with enzymes, genes, and compound structures. On the pathway diagrams, users can choose to either show only experimentally characterized enzymes or to include predicted enzymes, and they can elect to show all enzymes or only enzymes for a given species. All the data objects on the pathway diagrams can be clicked on to access additional information. For single-species databases, users can overlay and visualize large-scale experimental data using the Pathway Tools Omics Viewer tool (http://www.plantcyc.org:1555/ARA/expression.html). An example of using the Omics Viewer in analyzing data is demonstrated in detail below. Users can also compare summary statistics of pathways, reactions, and enzymes between the PMN databases using the Comparative Analysis tool (http://www.plantcyc.org:1555/comp-genomics).
In addition to online access, the complete PMN databases are also available for download with an “open source” license (http://www.plantcyc.org/downloads/license_agreement.faces) in the formats of flat files (http://bioinformatics.ai.sri.com/ptools/flatfile-format.html) such as Ocelot, which is readily readable by Pathway Tools, BioPAX, and SBML (Stromback and Lambrix, 2005). Installation of the PMN databases on a local computer, together with a Pathway Tools installation (http://biocyc.org/download.shtml) on the same computer, enables users to run PMN databases locally. This local database setup offers a few advantages, such as additional operations, faster speed, the opportunity to update the database with proprietary data, and the ability to perform programmatic queries.